Published online Oct 7, 2021. doi: 10.3748/wjg.v27.i37.6262
Peer-review started: April 8, 2021
First decision: June 26, 2021
Revised: July 6, 2021
Accepted: September 1, 2021
Article in press: September 1, 2021
Published online: October 7, 2021
Processing time: 174 Days and 0.1 Hours
Genome-wide association studies from Asia indicate that HLA-DP and HLA-DQ loci are important in persistent hepatitis B virus (HBV) infections. One of the key elements for HBV-related carcinogenesis is persistent viral replication and inflammation.
To examine genetic and nongenetic factors with persistent HBV infection and viral load in families with hepatocellular carcinoma (HCC).
The HCC families included 301 hepatitis B surface antigen (HBsAg) carriers and 424 noncarriers born before the nationwide vaccination program was initiated in 1984. Five HBV-related single nucleotide polymorphisms (SNPs) — rs477515, rs9272105, rs9276370, rs7756516, and rs9277535 — were genotyped. Factors associated with persistent HBV infection and viral load were analyzed by a generalized estimating equation.
In the first-stage persistent HBV study, all SNPs except rs9272105 were associated with persistent infection. A significantly higher area under the reciprocal operating characteristic curve for nongenetic factors vs genetic factors (P < 0.001) suggests that the former play a major role in persistent HBV infection. In the second-stage viral load study, we added 8 HBsAg carriers born after 1984. The 309 HBsAg carriers were divided into low (n = 162) and high viral load (n = 147) groups with an HBV DNA cutoff of 105 cps/mL. Sex, relationship to the index case, rs477515, rs9272105, and rs7756516 were associated with viral load. Based on the receiver operating characteristic curve analysis, genetic and nongenetic factors affected viral load equally in the HCC family cohort (P = 0.3117).
In these east Asian adults, the mechanism of persistent HBV infection-related SNPs was a prolonged viral replication phase.
Core Tip: Hepatitis B virus (HBV)-related single nucleotide polymorphisms (SNPs) have been identified in East Asians. We evaluated five SNPs and nongenetic factors associated with HBV infection in a hepatocellular carcinoma family cohort. The factors were correlated with hepatitis B surface antigen (HBsAg) in the first-stage and with HBV viral load in the second-stage. The SNPs, sex, generation, and index case HBsAg contributed to persistent HBV infection. Neonatal tolerance and SNPs in the HLA loci were both independently associated with persistent HBV infection. A prolonged HBV replication phase in parents could be the main mechanism of persistent HBV infection in children in East Asia.
- Citation: Hsieh AR, Fann CSJ, Lin HC, Tai J, Hsieh SY, Tai DI. Hepatitis B virus persistent infection-related single nucleotide polymorphisms in HLA regions are associated with viral load in hepatoma families. World J Gastroenterol 2021; 27(37): 6262-6276
- URL: https://www.wjgnet.com/1007-9327/full/v27/i37/6262.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i37.6262
Chronic hepatitis B is a global disease, with the highest prevalence in Africa and Asia[1,2]. Hepatitis B virus (HBV) is highly infectious[3,4], and those who are infected early in life are likely to develop a persistent infection[5-7]. Intra-familial spread of infection is common, resulting in the clustering of chronic hepatitis B surface antigen (HBsAg) carriers and hepatocellular carcinoma (HCC) in families[8-10]. Recent genome-wide association studies (GWASs) in Japan, Korea, Saudi Arabia, China, and Taiwan have consistently shown that single nucleotide polymorphisms (SNPs) at the HLA-DP and HLA-DQ loci play important roles in persistent HBV infection[11-19]. However, risk alleles of HBV-related SNPs are not present in the majority of Africans[20,21], so the high prevalence of HBsAg carriers in Africa cannot be completely explained by the SNPs.
It is well known that clearance of the hepatitis B e antigen (HBeAg) occurs earlier in African than in Asian HBsAg carriers[22-25]. In east Asia, the annual HBeAg seroconversion rate is < 2% in children younger than 3 years of age and around 5% in children older than 3 years of age[22,23]. On the contrary, an HBeAg annual clearance rate of 14%-16% has been found in Euro-Mediterranean and African children[24,25]. HBeAg clearance is associated with a decreased viral load and results in a decrease of perinatal infections and the development of chronic persistent HBV infection[7,23]. We propose that persistent HBV infection-related SNPs may be one of the reasons for the prolonged HBV replication phase in east Asians. To evaluate this hypothesis, we analyzed the HBV-related SNP and demographic data obtained from HCC families. HCC families are known to have higher perinatal transmission and a longer HBV replication phase than the general population[9,10]. We expect that the genetic and nongenetic factors characteristic of HCC families may help us to understand the nature of persistent HBV infection.
Our study was approved by the institutional review board of Chang Gung Memorial Hospital, Taiwan (IRB 104-2596). Written informed consent was obtained from all participants. All experiments and data comparisons were carried out in compliance with relevant laws and guidelines, and complied with the ethical standards of the Declaration of Helsinki.
Patients with HCC who were diagnosed at Chang Gung Memorial Hospital, Lin-Kou Medical Center were included as index cases. From 2003 to 2007, relatives of the patients were prospectively invited to complete a liver disease survey. The details of the survey can be seen in our previous report[10]. Briefly, after confirmation of their relation to the index HCC patient, the relatives received a structured questionnaire and underwent assessments of their liver biochemistry, alpha-fetoprotein, viral markers, and HBV genotyping. Peripheral blood samples were collected for host genome analysis.
We calculated sample sizes and statistical power to detect genetic effects in the study. The calculation considered the impact the minor allele frequency (MAF, from 0.1 to 0.4), odds ratio (OR, from 1.05 to 3), statistical power (from 0.5 to 0.9) and mea
Four genetic variants (rs477515, rs9276370, rs7756516, rs9277535) associated with persistent HBV infection that were previously identified[17] were included in the analysis. One additional HCC-related SNP (rs9272105) previously identified in China was also included[26]. Genomic DNA was extracted from peripheral blood cells using MagNA Pure LC DNA isolation kits with automated DNA isolation instruments (MagNA Pure LC II; Roche Diagnostics, Mannheim, Germany). Triple-SNP (rs477515, rs9272105, rs9277535) genotyping was performed with TaqMan Genotyping assays (Applied Biosystems, Foster City, CA, United States). Two SNPs (rs7756516, rs9276370) were genotyped with a Sequenom MassARRAY System (Sequenom, San Diego, CA, United States). The TaqMan assays were carried out by Vita Genomics (New Taipei City, Taiwan), and the Sequenom MassARRAY assays were performed by the Academia Sinica National Genotyping Center (Taipei, Taiwan). The overall genotype call rate was > 95%.
The statistical analyses were performed with SAS version 8.2 for UNIX (SAS Institute, Cary, NC, United States), PLINK (http://zzz.bwh.harvard.edu/plink/) (http://zzz.bwh.harvard.edu/plink/summary.shtml), R 2.15.1 (http://www.r-project.org/), and the Family-Based Association Test software (http://www.biostat.harvard.edu/~fbat/fbat.htm)[27]. A two-tailed P value < 0.05 was considered statistically sig
Individual locus analysis: We assessed the association of SNPs with persistent HBV infection or viral load in an additive genetic model using univariate and multivariate logistic regression of the data from unrelated male participants. In the family analysis, relatives included individuals living in the same household. First- and second-stage analyses were conducted with a generalized estimating equation (GEE) that included data correlated with a binary response (e.g., to HBsAg status and HBV DNA level) using an exchangeable working correlation structure[28,29]. Univariate and mul
Weighted genetic risk score calculation: The weighted genetic risk score (WGRS) was calculated for the SNPs that were significantly associated with persistent infection or viral load. We assumed that each SNP was independently associated with risk according to an additive genetic model. The WGRS was calculated by multiplying the number of risk alleles at each polymorphic locus (0, 1, or 2) by each person for the corresponding relative logarithm of the OR (wi) from the multivariate individual locus analysis and rescaling it with the factor m/∑iwi, as follows: WGRS = (m/∑iwi)·∑wini, where m is the number of statistically significant SNPs and ni is the number of risk alleles for SNPi[30]. We divided the continuous WGRS into quartiles (Q1-4) and compared the risks among them.
Evaluation of genetic and nongenetic factors: We analyzed factors associated with persistent HBV infection or viral load using the logistic regression model unrelated participants and the GEE method for family data. Three prediction models were used: (1) The genetic model included only SNPs and WGRS; (2) The nongenetic model included only demographic data; and (3) The mixed model included both genetic and nongenetic variables. The contribution of the WGRS was evaluated using the area under the receiver operating characteristic curve (AUC), net reclassification improvement (NRI) method[31], and integrated discrimination improvement (IDI)[32] with the prediction model with and without the WGRS. To assess the demographic impact of including the WGRS in the model, an AUC of 0.5 indicated no discrimination and an AUC of 1 indicated perfect discrimination. The NRI indicated the proportion of subjects reclassified correctly (NRI > 0) or incorrectly (NRI < 0) into the various risk categories. An IDI > 0 indicated a statistically significant prediction of improvement as a result of adding variables to the model.
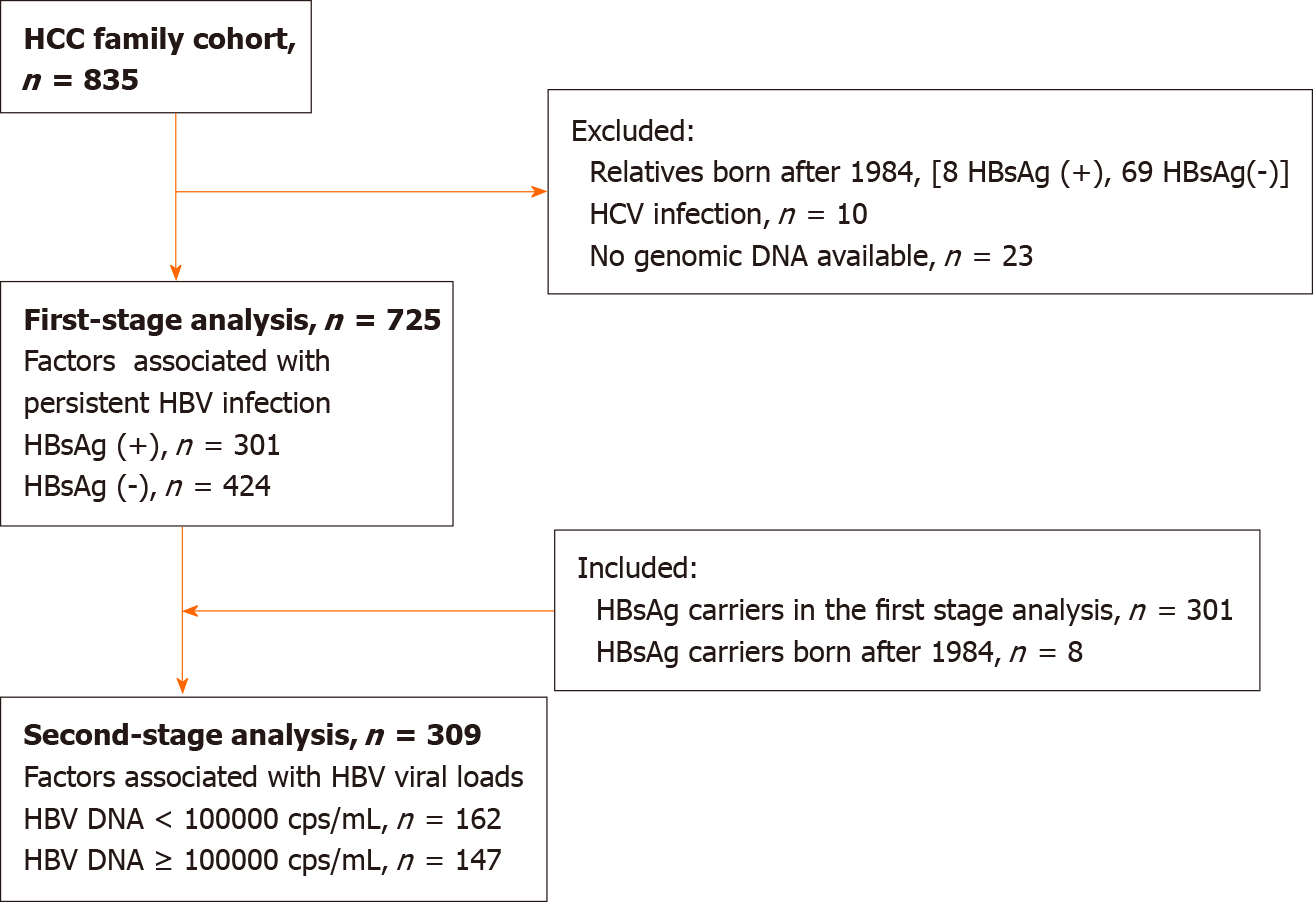
The HCC family cohort included 835 participants (Figure 1), of whom 301 HBsAg-positive and 424 of HBsAg-negative family members were selected for the first-stage HBV infection-persistence analysis. We excluded those born after the nationwide vaccination program was initiated in 1984. In the second-stage viral load study, we added 8 HBsAg carriers born after 1984 (Figure 1). A cohort of 309 HBsAg carriers was divided into high (n = 147) and low (n = 162) viral load groups using an HBV DNA cutoff of 105 cps/mL.
Risk factors associated with being an HBsAg carrier were identified in the first-stage analysis. Demographic factors, which included age, sex, index case sex, relation to the index case, index HBsAg, and maternal HBsAg, are shown in Table 1. Age (OR = 1.018, P = 0.0013), sex (OR = 1.641, P = 0.0001), relation to the index case (OR = 3.203, P < 0.0001; index generation compared with children and grandchildren), index HBsAg (OR = 4.913, P < 0.0001), maternal HBsAg (OR = 3.31, P < 0.0001), and serum glutamic pyruvic transaminase (SGPT) (OR = 1.017, P < 0.0001) were significantly associated with persistent HBV infection. The associations remained significant after controlling for sex and age.
| HBsAg | ||||||
| Category | Positive | Negative | OR (95%CI) | Adjusted OR (95%CI)1 | P value | Adjusted P value2 |
| Total family members, n | 301 | 424 | ||||
| Age in yr, mean ± SD | 44.23 ± 13.84 | 41.25 ± 14.97 | 1.018 (1.007-1.03) | 1.017 (1.006-1.028) | 0.0013 | 0.0030 |
| Sex, n (%) | ||||||
| Male | 182 (60.47) | 203 (47.88) | 1.641 (1.279-2.107) | 1.57 (1.225-2.011) | 0.0001 | 0.0004 |
| Female | 119 (39.53) | 221 (52.12) | 1 | 1 | ||
| Index sex, n (%) | ||||||
| Male | 226 (75.08) | 309 (72.88) | 1.207 (0.726-2.006) | 1.147 (0.681-1.93) | 0.4685 | 0.6061 |
| Female | 75 (24.92) | 115 (27.12) | 1 | 1 | ||
| Relation to index, n (%) | ||||||
| Children and grandchildren | 146 (48.50) | 319 (75.24) | 1 | 1 | ||
| Parent generation | 7 (2.33) | 15 (3.54) | 0.7 (0.302-1.622) | 1.472 (0.533-4.065) | 0.4059 | 0.4559 |
| Index generation | 148 (49.17) | 90 (21.23) | 3.203 (2.282-4.498) | 4.861 (2.923-8.083) | < 0.0001 | < 0.0001 |
| Index status, n (%) | ||||||
| HBsAg- | 70 (23.26) | 257 (60.61) | 1 | 1 | ||
| HBsAg+ | 231 (76.74) | 167 (39.39) | 4.913 (3.209-7.522) | 5.928 (3.747-9.377) | < 0.0001 | < 0.0001 |
| Mother’s status, n (%) | ||||||
| HBsAg- | 85 (28.24) | 239 (56.37) | 1 | 1 | ||
| HBsAg+ | 91 (30.23) | 45 (10.61) | 3.31 (1.894-5.783) | 3.296 (1.891-5.746) | < 0.0001 | < 0.0001 |
| Unknown | 125 (41.53) | 140 (33.02) | 2.305 (1.568-3.39) | 1.87 (1.202-2.91) | <.0001 | 0.0055 |
| SGPT, mean ± SD | 49.98 ± 65.39 | 25.83 ± 24.92 | 1.017 (1.011-1.022) | 1.015 (1.009-1.020) | < 0.0001 | < 0.0001 |
| rs477515 (MAF = 0.1552) Chr6: 326019141 | ||||||
| TT (reference) | 5 (1.66) | 20 (4.72) | ||||
| TC | 59 (19.60) | 116 (27.36) | ||||
| CC | 237 (78.74) | 288 (67.92) | 1.377 (1.036-1.831) | 1.38 (1.034-1.842) | 0.0274 | 0.0285 |
| rs9272105 (MAF = 0.4282) Chr6: 326322221 | ||||||
| GG (reference) | 54 (17.94) | 84 (19.86) | ||||
| GA | 137 (45.51) | 207 (48.94) | ||||
| AA | 110 (36.54) | 132 (31.21) | 1.054 (0.859-1.293) | 1.031 (0.844-1.261) | 0.6126 | 0.7639 |
| rs9276370 (MAF = 0.1159) Chr6: 327395181 | ||||||
| GG (reference) | 3 (1.00) | 13 (3.07) | ||||
| GT | 39 (12.96) | 97 (22.88) | ||||
| TT | 259 (86.05) | 314 (74.06) | 1.790 (1.258-2.547) | 1.759 (1.228-2.519) | 0.0012 | 0.0021 |
| rs7756516 (MAF = 0.1166) Chr6: 327561401 | ||||||
| CC (reference) | 3 (1.00) | 13 (3.07) | ||||
| CT | 42 (13.95) | 95 (22.41) | ||||
| TT | 256 (85.05) | 316 (74.53) | 1.654 (1.166-2.346) | 1.612 (1.123-2.313) | 0.0048 | 0.0096 |
| rs9277535 (MAF = 0.3234) Chr6: 330870841 | ||||||
| AA (reference) | 21 (6.98) | 61 (14.39) | ||||
| AG | 114 (37.87) | 191 (45.05) | ||||
| GG | 166 (55.15) | 172 (40.57) | 1.519 (1.204-1.916) | 1.493 (1.182-1.886) | 0.0004 | 0.0008 |
The SNPs rs477515 (OR = 1.377, P = 0.0274), rs9276370 (OR = 1.790, P = 0.0012), rs7756516 (OR = 1.654, P = 0.0048), and rs9277535 (OR = 1.519, P = 0.0004) were significantly associated with chronic HBV infection (Table 1). The ORs remained statistically significant after controlling for sex and age. HCC families carrying more risk alleles had an increased OR (Table 2, upper panel). Compared with participants with a WGRS in Q1, those with scores in Q2 and Q3–4 had higher risks of HBsAg positivity (Q2 OR = 1.878, P = 0.0014; Q3-4 OR = 2.538, P < 0.0001).
| Study | WGRS quartile | OR (95%CI) | P value |
| Family first stage: Persistent HBV infection | Q1 (WGRS ≤ 6.166) | 1 | |
| Q2 (WGRS = 6.166-7.083) | 1.878 (1.277-2.762) | 0.0014 | |
| Q3,4 (WGRS > 7.083)1 | 2.538 (1.742-3.698) | < 0.0001 | |
| Cochran-Armitage trend test | < 0.0001 | ||
| Family second stage: Viral load | Q1 (WGRS ≤ 4.583) | 1 | |
| Q2 (WGRS = 4.583-5.291) | 2.204 (1.253-3.878) | 0.0061 | |
| Q3,4 (WGRS > 5.291)2 | 3.156 (1.780-5.595) | < 0.0001 | |
| Cochran-Armitage trend test | < 0.0001 | ||
Results of the multivariate GEE analysis of the risk factors associated with persistent HBV infection are shown in Table 3. In the nongenetic model, sex, index generation, and index and maternal index HBsAg were associated with persistent HBV infection. In the genetic model, rs9277535 and WGRS were associated with persistent HBV infection. In the mixed model, all the risk factors were significant (male sex P = 0.0205; index generation P = 0.0001; index HBsAg P < 0.0001; maternal HBsAg P = 0.0072; rs9277535 P = 0.0029; WGRS P = 0.0012; Table 3).
| Nongenetic model | Mixed model | |||
| Variable | OR (95%CI) | P value | OR (95%CI) | P value |
| Sex, male | 1.458 (1.048-2.027) | 0.0250 | 1.514 (1.075-2.133)1/1.487 (1.063-2.081)2 | 0.01771/0.02052 |
| Index sex, male | 0.915 (0.532-1.573) | 0.7475 | 0.853 (0.496-1.466)1/0.838 (0.488-1.442)2 | 0.56481/0.52382 |
| Age in yr | 1.001 (0.984-1.018) | 0.9392 | 1.000 (0.982-1.017)1/0.998 (0.981-1.016)2 | 0.96081/0.85822 |
| Relation to index | ||||
| Parent generation | 0.523 (0.176-1.555) | 0.2434 | 0.560 (0.182-1.719)1/0.603 (0.198-1.833)2 | 0.31081/0.37262 |
| Index generation | 3.385 (1.836-6.239) | < 0.0001 | 3.344 (1.766-6.331)1/3.493 (1.860-6.559)2 | 0.00021/0.00012 |
| Index’s HBsAg+ | 5.077 (3.103-8.308) | < 0.0001 | 4.919 (2.980-8.119)1/4.756 (2.912-7.766)2 | < 0.00011/< 0.00012 |
| Mother’s status | ||||
| HBsAg+ | 2.597 (1.332-5.064) | 0.0051 | 2.459 (1.270-4.760)1/2.517 (1.284-4.933)2 | 0.00761/0.00722 |
| Unknown | 1.395 (0.788-2.469) | 0.2538 | 1.412 (0.790-2.522)1/1.413 (0.795-2.511)2 | 0.24441/0.23872 |
| AUC (95%CI) | 0.786 (0.752-0.820) | < 0.0001 | ||
| Genetic model | ||||
| rs477515 | 1.303 (0.969-1.753) | 0.0802 | 1.121 (0.770-1.631) | 0.5507 |
| rs9276370 | 2.741 (0.766-9.812) | 0.1211 | 3.040 (0.623-14.839) | 0.1693 |
| rs7756516 | 0.592 (0.171-2.042) | 0.4064 | 0.516 (0.104-2.554) | 0.4177 |
| rs9277535 | 1.575 (1.244-1.995) | 0.0002 | 1.535 (1.157-2.035) | 0.0029 |
| AUC (95%CI) | 0.632 (0.593-0.671) | < 0.0001 | 0.798 (0.765-0.831) | < 0.0001 |
| WGRS | 1.322 (1.162-1.505) | < 0.0001 | 1.269 (1.099-1.465) | 0.0012 |
| AUC (95%CI) | 0.620 (0.580-0.660) | < 0.0001 | 0.795 (0.762-0.829) | < 0.0001 |
| IDI (95%CI) | 0.017 (0.009-0.026) | < 0.0001 | ||
| NRI (95%CI) | 0.330 (0.192-0.467) | < 0.0001 | ||
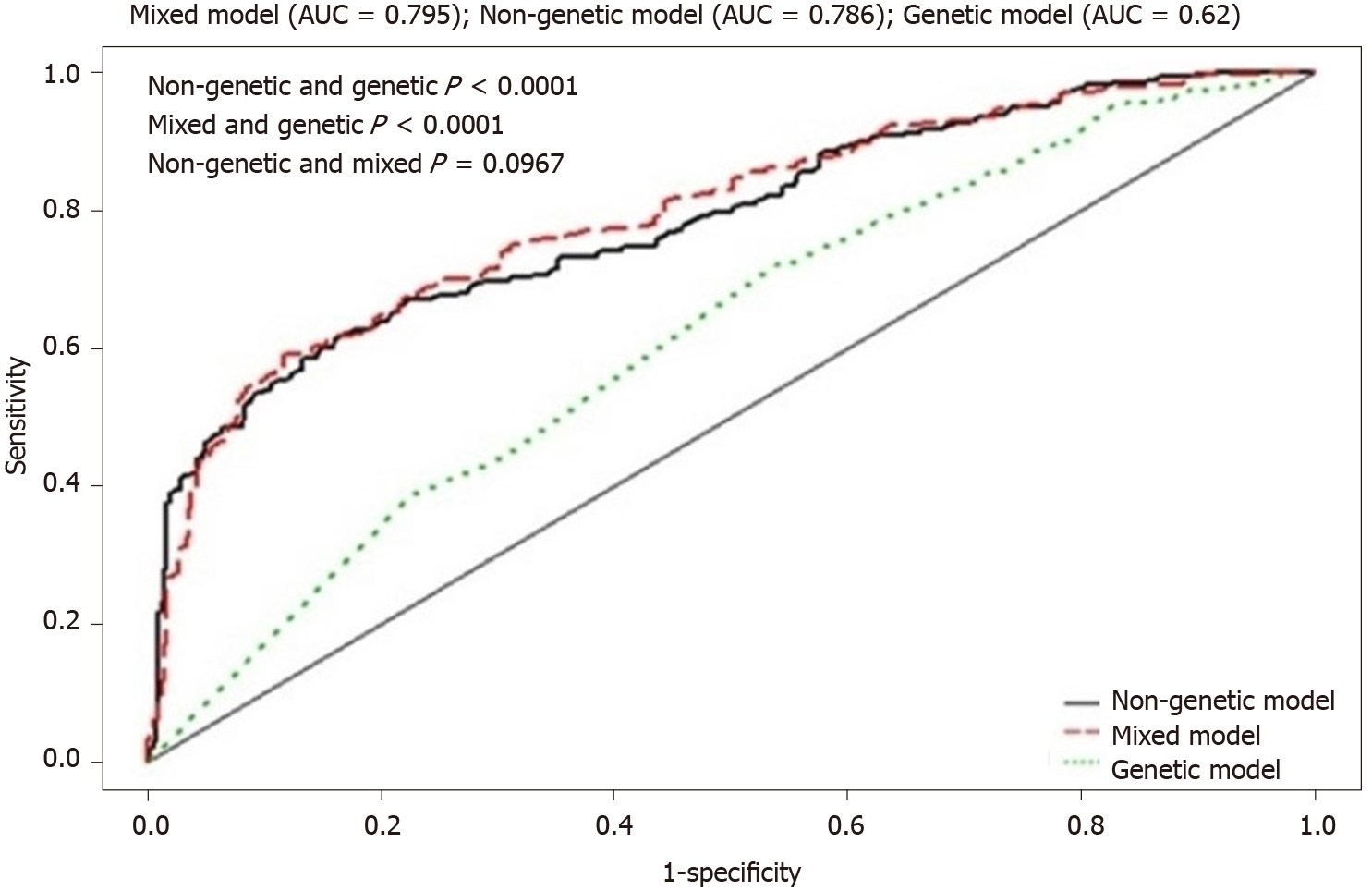
The AUC for persistent HBV infection (Table 3) was 0.786 (P < 0.0001) in the nongenetic model and 0.620 (P < 0.0001) in the genetic model. Although the SNPs were identified by GWAS in unrelated subjects, the AUC data suggest that nongenetic factors were more important than genetic factors for the development of persistent HBV infection (P < 0.0001; Figure 2). The combination of genetic and nongenetic factors resulted in an AUC of 0.795 (P < 0.0001; Figure 2 and Table 3). The IDI was 0.017 (95%CI: 0.009-0.026, P < 0.0001) and the NRI was 0.330 (95%CI: 0.192-0.467, P < 0.0001). The IDI and NRI values indicated statistically significant predicted improvement in the mixed, relative to the nongenetic model (Table 3).
Factors associated with the HBV viral load were evaluated in HBsAg-positive families (Table 4). In that group, male sex (OR = 1.922, P = 0.0078), relation to the index case (OR = 2.033, P = 0.0029), index HBsAg (OR = 2.508, P = 0.0036), and SGPT (OR = 1.010, P = 0.0105) were significantly associated with the HBV viral load. The associations remained statistically significant after controlling for sex. HBV genotypes were also evaluated in HCC families, and of the participants with known HBV genotypes, the prevalence of genotype C was higher in those with high viral loads (41/143, 28.7%) than in those with low viral loads (15/90, 16.7%, P = 0.0431). The difference was marginally significant in multivariate analysis (P = 0.0515; Table 4).
| HBV DNA | |||||||
| Category | ≥ 105 cps/mL | < 105 cps/mL | OR (95%CI) | Adjusted OR (95% CI)2 | P value | Adjusted P value2 | |
| Total members | 147 | 162 | |||||
| Age in yr, mean ± SD | 45.03 ± 14.18 | 41.82 ± 14.21 | 1.017 (1-1.035) | 1.017 (0.999-1.035) | 0.0538 | 0.0668 | |
| Sex, n (%) | |||||||
| Male | 100 (68.03) | 88 (54.32) | 1.922 (1.188-3.111) | 1.914 (1.187-3.087) | 0.0078 | 0.0078 | |
| Female | 47 (31.97) | 74 (45.68) | 1 | 1 | |||
| Relation to index, n (%) | |||||||
| Children and grandchildren generation | 58 (39.46) | 95 (58.64) | 1 | 1 | |||
| Parent generation | 5 (3.4) | 2 (1.23) | 3.683 (0.866-15.656) | 5.056 (1.259-20.3) | 0.0775 | 0.0223 | |
| Index generation | 84 (57.14) | 65 (40.12) | 2.033 (1.274-3.246) | 1.845 (1.144-2.977) | 0.0029 | 0.0121 | |
| Index’s status, n (%) | |||||||
| HBsAg- | 21 (14.29) | 49 (30.25) | 1 | 1 | |||
| HBsAg+ | 126 (85.71) | 113 (69.75) | 2.508 (1.351-4.657) | 2.492 (1.324-4.692) | 0.0036 | 0.0047 | |
| Mother’s status, n (%) | |||||||
| HBsAg- | 43 (29.25) | 43 (26.54) | 1 | 1 | |||
| HBsAg+ | 46 (31.29) | 51 (31.48) | 0.874 (0.467-1.634) | 0.91 (0.485-1.707) | 0.6724 | 0.7693 | |
| Unknown | 58 (39.46) | 68 (41.98) | 0.855 (0.491-1.49) | 0.857 (0.488-1.503) | 0.5804 | 0.5898 | |
| HBV genotype (BGT230), n (%) | |||||||
| Unknown3 | 3 (2.05) | 72 (44.44) | 0.03 (0.009-0.104) | 0.029 (0.008-0.1) | < 0.0001 | < 0.0001 | |
| B | 102 (69.86) | 75 (46.3) | 1 | 1 | |||
| C | 41 (28.08) | 15 (9.26) | 2.042 (1.023-4.079) | 2.066 (0.995-4.288) | 0.0431 | 0.0515 | |
| SGPT, mean ± SD | 63.92 ± 79.63 | 37.02 ± 45.22 | 1.010 (1.002-1.018) | 1.009 (1.001-1.017) | 0.0105 | 0.0260 | |
| rs477515 (MAF = 0.1149) Chr6: 326019141 | |||||||
| TT (reference) | 1 (0.68) | 4 (2.47) | |||||
| TC | 15 (10.2) | 46 (28.4) | |||||
| CC | 131 (89.12) | 112 (69.14) | 3.107 (1.708-5.653) | 3.195 (1.746-5.847) | 0.0002 | 0.0002 | |
| rs9272105 (MAF = 0.4078) Chr6: 326322221 | |||||||
| GG (reference) | 20 (13.61) | 36 (22.22) | |||||
| GA | 59 (40.14) | 81 (50) | |||||
| AA | 68 (46.26) | 45 (27.78) | 1.747 (1.256-2.428) | 1.75 (1.247-2.456) | 0.0009 | 0.0012 | |
| rs9276370 (MAF = 0.07605) Chr6: 327395181 | |||||||
| GG (reference) | 1 (0.68) | 3 (1.85) | |||||
| GT | 14 (9.52) | 25 (15.43) | |||||
| TT | 132 (89.8) | 134 (82.72) | 1.747 (0.933-3.272) | 1.679 (0.901-3.131) | 0.0811 | 0.1029 | |
| rs7756516 (MAF = 0.08091) Chr6: 327561401 | |||||||
| CC (reference) | 0 (0) | 4 (2.47) | |||||
| CT | 16 (10.88) | 26 (16.05) | |||||
| TT | 131 (89.12) | 132 (81.48) | 1.951 (1.078-3.53) | 1.875 (1.029-3.417) | 0.0272 | 0.0400 | |
| rs9277535 (MAF = 0.2589) Chr6: 330870841 | |||||||
| AA (reference) | 13 (8.84) | 8 (4.94) | |||||
| AG | 45 (30.61) | 73 (45.06) | |||||
| GG | 89 (60.54) | 81 (50) | 1.235 (0.849-1.797) | 1.303 (0.888-1.911) | 0.2703 | 0.1767 | |
Of the five SNPs included in the analysis, rs477515 (OR = 3.107, P = 0.0002), rs9272105 (OR = 1.747, P = 0.0009), and rs7756516 (OR = 1.951, P = 0.0272) were significantly associated with HBV viral load. The associations remained significant after controlling for sex (Table 4). Participants carrying more risk alleles had higher ORs for HBV viral load (Table 2, lower panel) and compared with patients having a WGRS in Q1, those in Q2 (OR = 2.204, P = 0.0061) and Q3-4 (OR = 3.156, P < 0.0001) had higher odds of having an HBV viral load.
The results of multivariate GEE analysis of factors associated with the HBV viral load in the genetic, nongenetic, and mixed models are shown in Table 5. In the nongenetic model, the risk of HBV viral load was higher in males (OR = 1.955, P = 0.0162) and in those with index HBsAg positivity (OR = 2.219, P = 0.0187). In the genetic model, the risk allele rs477515 (OR = 2.246, P = 0.0159) and the WGRS (OR = 1.644, P < 0.0001) were significantly different between the groups with high and low viral loads. In the mixed model, sex, rs477515, and WGRS were significantly different in the groups with high and low viral loads (Table 5).
| Nongenetic model | Mixed model | |||
| Variable | OR (95%CI) | P value | OR (95%CI) | P value |
| SNP | ||||
| Sex, male | 1.955 (1.132-3.376) | 0.0162 | 1.918 (1.101-3.341)1/1.911 (1.097-3.328)2 | 0.02141/0.02232 |
| Index sex, male | 0.903 (0.481-1.699) | 0.7527 | 0.914 (0.488-1.713)1/0.912 (0.482-1.725)2 | 0.77861/0.77712 |
| Age in yr | 1.012 (0.987-1.037) | 0.3363 | 1.007 (0.983-1.032)1/1.008 (0.983-1.033)2 | 0.56581/0.54262 |
| Relation to index | ||||
| Parent generation | 4.182 (0.68-25.731) | 0.1228 | 4.343 (0.81-23.285)1/4.091 (0.75-22.316)2 | 0.08661/0.10362 |
| Index generation | 1.7 (0.844-3.423) | 0.1372 | 1.851 (0.912-3.756)1/1.797 (0.894-3.611)2 | 0.08811/0.09992 |
| Index HBsAg+ | 2.219 (1.142-4.31) | 0.0187 | 1.734 (0.853-3.526)1/1.816 (0.907-3.636)2 | 0.12831/0.09182 |
| Mother’s status | ||||
| HBsAg+ | 0.828 (0.407-1.684) | 0.6021 | 0.766 (0.361-1.623)1/0.763 (0.360-1.619)2 | 0.48621/0.48142 |
| Unknown | 0.537 (0.275-1.045) | 0.0673 | 0.549 (0.272-1.107)1/0.559 (0.278-1.125)2 | 0.0941/0.10302 |
| AUC (95%CI) | 0.674 (0.614-0.734) | < 0.0001 | ||
| Genetic model | ||||
| rs477515 | 2.246 (1.164-4.333) | 0.0159 | 2.242 (1.113-4.515) | 0.0238 |
| rs9272105 | 1.386 (0.965-1.991) | 0.0775 | 1.266 (0.866-1.849) | 0.2232 |
| rs7756516 | 1.385 (0.765-2.509) | 0.2826 | 1.379 (0.753-2.524) | 0.2977 |
| AUC (95%CI) | 0.638 (0.579-0.698) | < 0.0001 | 0.705 (0.648-0.763) | < 0.0001 |
| WGRS | 1.644 (1.317-2.052) | < 0.0001 | 1.567 (1.250-1.965) | < 0.0001 |
| AUC (95%CI) | 0.632 (0.573-0.692) | < 0.0001 | 0.704 (0.646-0.761) | < 0.0001 |
| IDI (95%CI) | 0.042 (0.019-0.065) | 0.0003 | ||
| NRI (95%CI) | 0.440 (0.236-0.644) | < 0.0001 | ||
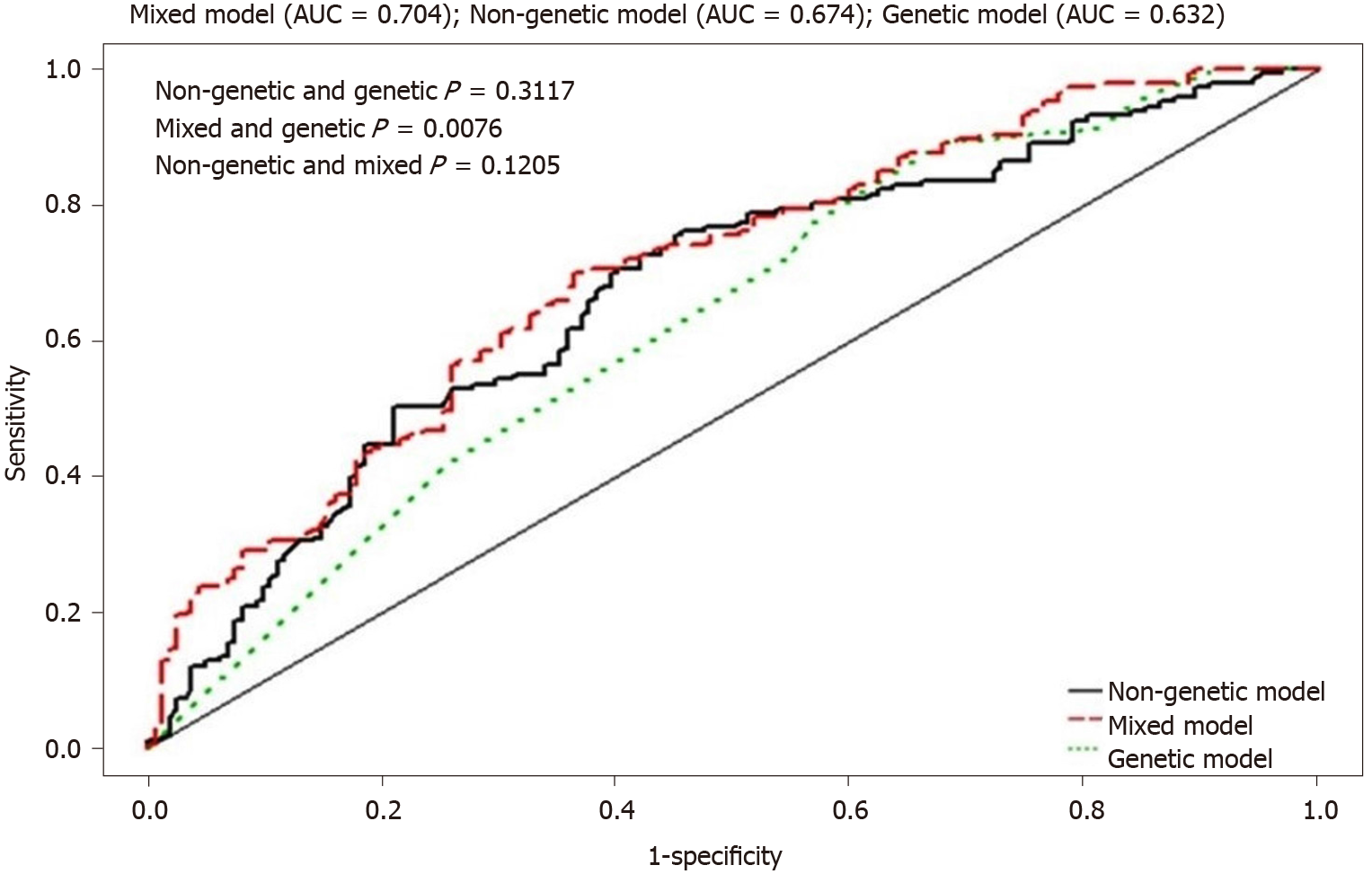
The AUC of the HBV viral load was 0.674 (P < 0.0001) for the nongenetic model, 0.632 (P < 0.0001) for the genetic model, and 0.704 (P < 0.0001) for the mixed model (Figure 3 and Table 5). The results suggest that both genetic and nongenetic factors had an effect on HBV viral load. Both the IDI (0.042, 95%CI: 0.019-0.065, P = 0.0003) and the NRI (0.440, 95%CI: 0.236–0.644, P < 0.0001) indicated that the mixed model represented a significant improvement (Table 5).
In this HCC family cohort, we found that both genetic and nongenetic factors were significantly associated with persistent HBV infection. In addition, HBV-related SNPs in the HLA-DP and HLA-DQ regions were associated with HBV viral load. GWASs conducted in diverse Asian populations have revealed that the HLA-DP and -DP loci play roles in persistent HBV infection[10-19]. We evaluated persistent HBV infection in the first-stage HCC family study. Expression of four of the five HBV-related SNPs differed significantly between the HBsAg carriers and the noncarriers. When only the risk alleles of the four SNPs were included in the univariate analysis, the OR for persistence was significant if the WGRS was > 7 (Table 2, upper panel). In the genetic model, multivariate GEE analysis found that expression of one SNP (rs9277535) and the WGRS were significantly different between HBsAg carriers and noncarriers, and the differences remained significant in the presence of nongenetic factors (Table 3). Regression analysis showed that HBV-related SNPs were associated with persistent HBV infection in these HCC families. This is the first study to confirm that SNPs identified by GWAS were associated with persistent HBV infection in a family cohort.
Nongenetic factors also affected persistent HBV infection. Age, sex, generation, index HBsAg status, and maternal HBsAg status all differed significantly between HBsAg carriers and noncarriers (Table 1). The AUC was 0.786 in the nongenetic model, 0.620 in the genetic model, and 0.795 in the mixed model (Table 3). The ROC analysis thus implied that nongenetic factors contributed more to persistent HBV infection than genetic factors did (genetic vs nongenetic factors P < 0.0001 and mixed vs nongenetic factors P < 0.0001; Figure 2). The results are consistent with exposure to HBV early in life and an important influence on the persistence of HBV infection[5-7]. Our overall findings indicate that the HCC family members may have been exposed to HBV early in life because of the high HBsAg prevalence in the index cases and/or their mothers. Accounting for both genetic and nongenetic cofactors, the prevalence of HBsAg was 41.5% (301/725) in this HCC family cohort.
In the presence of SNPs identified in a GWAS, nongenetic factors remain important in persistent HBV infection. The persistence of infection induced by the SNPs might depend on a delay in clearance of the HBeAg. It is known that in HBsAg carriers, HBeAg clearance occurs earlier in African than in Asian populations[22-25]. That means East Asians of reproductive age are likely to have higher HBV viral loads and a higher rate of perinatal HBV infection of their babies[7,10,22,23]. Perinatal infection usually persists as a chronic infection[7,23]. As African women usually clear HBeAg before reproductive age[24,25], the viral load during pregnancy is likely to be lower than that in East Asians, which would decrease the chance of perinatal HBV infection[24]. We suspect that a prolonged HBV replication phase in parents could be the mechanism of persistent HBV infection associated with SNPs.
Univariate analysis of the factors associated with HBV viral load in the HCC family cohort revealed that three of the five SNPs (rs477515, rs9272105, rs7756516) differed significantly between the high and low viral load groups (Table 4). The cumulative effect of the WGRS was also greater in the high viral load group (Table 3, lower panel). Multivariate GEE analysis found that the rs477515 SNP (OR = 2.242, P = 0.0238) and WGRS (OR = 1.567, P < 0.0001) were independently associated with a high viral load in the mixed model (Table 5). Our data thus support the prevailing view that the SNPs associated with persistent HBV infection promote persistent HBV replication. The mean ages of our study groups ranged from 41.25-45.03 years (Tables 1 and 4). Persistent high viral loads in these age groups were likely to have resulted in perinatal transmission of chronic HBV infection during the reproductive age.
Our previous study demonstrated that nongenetic factors influenced the HBV viral load in HCC families[10]. In this study, we observed that sex, generation, and index HBsAg cases were associated with a high viral load in the nongenetic model (Table 4). We also compared the relative contributions of genetic and nongenetic factors associated with viral load in the HCC family cohort. The AUCs of the viral load were 0.674 in the nongenetic model and 0.632 in the genetic model. The AUC in the mixed model was up to 0.704 (Table 5). Therefore, both genetic and nongenetic factors were associated with HBV viral load in the HCC family cohort. It should be noted that we included only SNPs in the HLA region. The association of other loci, such as polymorphisms of interferon gamma, complement factor B, CD40, and INST10, which have also been reported to be associated with HBV viral load, was not investigated[33-35].
One of the five SNPs we evaluated, rs9277535, was reported by Tao et al[36] to be associated with more aggressive liver disease, but it was reported by Li et al[37] not to be associated with disease progression. Our previous GWAS revealed that rs9276370 was associated with HBV therapeutic response[17]. Univariate analysis found that the two SNPs were not significantly associated with viral load in this HCC family cohort. Two previous studies found that rs477515 was associated with HBV vaccine response[38,39], and that SNP was found to be associated with viral load in this cohort. Li et al[26] reported that rs9272105 was associated with HCC in a GWAS, and univariate analysis found that it was associated with viral load in this HCC family cohort. All these previous reports suggest that a single SNP provides a small contribution to HBV viral loads. Persistent HBV replication seems to be determined by multiple genetic and nongenetic risk factors.
This study provides information that may help to establish more accurate models of disease through the incorporation of genetic and nongenetic factors, but it was limited by the relatively small number of HCC families. Another limitation was that HBV genotype studies were not available in patients with low viral loads. HBV genotype C has been associated with a lower HBeAg clearance rate than genotype B[40]. We found a high adjusted OR (2.066, P = 0.0515) for the association of genotype C with a high viral load relative to a low viral load in this HCC family cohort (Table 4).
We conclude that SNPs associated with persistent HBV infection prolong the replication phase in the parent generation and increase the burden of persistent infection in the offspring generation.
Genome-wide association studies (GWASs) in Asian populations indicate that the HLA-DP and HLA-DQ loci are involved in the persistence of hepatitis B virus (HBV) infections. Persistent viral replication and inflammation are key influencers in HBV-related carcinogenesis.
HBV-related single nucleotide polymorphisms (SNPs) have been identified in east Asian populations but are uncommon in African populations. Different mechanisms may drive persistent infection in those regions.
We examined genetic and nongenetic factors associated with persistent HBV infection and viral load in families with hepatocellular carcinoma (HCC).
HCC families were enrolled. Five HBV-related SNPs (rs477515, rs9272105, rs9276370, rs7756516, and rs9277535) were genotyped. Factors associated with persistent HBV infection and viral load were identified with the use of generalized estimating equations.
In the first-stage persistent HBV study, all SNPs except rs9272105 were associated with persistent infection. A significantly higher contribution of nongenetic than genetic factors (P < 0.001) to persistent HBV infection was found. In the second-stage viral load study, sex, relationship with index case, rs477515, rs9272105, and rs7756516 were associated with viral load. Receiver operating characteristic curve, and genetic and nongenetic factors had equal effects on viral load in the HCC family cohort (P = 0.3117).
GWAS identified SNPs that have roles in persistent HBV infection and HBV viral loads in an HCC family cohort. Nongenetic factors were more important than genetic factors in persistent HBV infection but had equal contributions to HBV viral load. HBV-related SNPs resulting in high viral loads in parents may drive persistent infection in East Asian populations. The mechanism of persistent HBV infection-related SNPs involves a prolonged viral replication phase in East Asian adults.
Termination of the HBV replication phase before pregnancy will be a therapeutic goal in East Asian countries.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: American Association for the Study of Liver Diseases, No. 139352; The Gastroenterological Society of the Taiwan, No. 398; The Ultrasound Society of the R.O.C., No. G355.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Taiwan
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Wang L S-Editor: Gao CC L-Editor: A P-Editor: Liu JH
| 1. | Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546-1555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1806] [Cited by in RCA: 1995] [Article Influence: 199.5] [Reference Citation Analysis (4)] |
| 2. | Hou J, Liu Z, Gu F. Epidemiology and Prevention of Hepatitis B Virus Infection. Int J Med Sci. 2005;2:50-57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 245] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 3. | Kingsley LA, Rinaldo CR Jr, Lyter DW, Valdiserri RO, Belle SH, Ho M. Sexual transmission efficiency of hepatitis B virus and human immunodeficiency virus among homosexual men. JAMA. 1990;264:230-234. [PubMed] |
| 4. | Kane A, Lloyd J, Zaffran M, Simonsen L, Kane M. Transmission of hepatitis B, hepatitis C and human immunodeficiency viruses through unsafe injections in the developing world: model-based regional estimates. Bull World Health Organ. 1999;77:801-807. [PubMed] |
| 5. | Beasley RP. Rocks along the road to the control of HBV and HCC. Ann Epidemiol. 2009;19:231-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 6. | Edmunds WJ, Medley GF, Nokes DJ, Hall AJ, Whittle HC. The influence of age on the development of the hepatitis B carrier state. Proc Biol Sci. 1993;253:197-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 283] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 7. | Burk RD, Hwang LY, Ho GY, Shafritz DA, Beasley RP. Outcome of perinatal hepatitis B virus exposure is dependent on maternal virus load. J Infect Dis. 1994;170:1418-1423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 144] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Sung JL, Chen DS. Geographical distribution of the subtype of hepatitis B surface antigen in Chinese. Gastroenterol Jpn. 1977;12:58-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 9. | Liu X, Baecker A, Wu M, Zhou JY, Yang J, Han RQ, Wang PH, Jin ZY, Liu AM, Gu X, Zhang XF, Wang XS, Su M, Hu X, Sun Z, Li G, Fu A, Jung SY, Mu L, He N, Li L, Zhao JK, Zhang ZF. Family history of liver cancer may modify the association between HBV infection and liver cancer in a Chinese population. Liver Int. 2019;39:1490-1503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 10. | Hsieh AR, Fann CS, Yeh CT, Lin HC, Wan SY, Chen YC, Hsu CL, Tai J, Lin SM, Tai DI. Effects of sex and generation on hepatitis B viral load in families with hepatocellular carcinoma. World J Gastroenterol. 2017;23:876-884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 11. | Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H, Puseenam A, Sura T, Daigo Y, Chayama K, Chantratita W, Nakamura Y, Matsuda K. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 430] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 12. | Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, Takahashi A, Kamatani Y, Miki D, Abe H, Tsunoda T, Kamatani N, Chayama K, Nakamura Y, Matsuda K. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet. 2011;20:3884-3892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 189] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 13. | Hu Z, Liu Y, Zhai X, Dai J, Jin G, Wang L, Zhu L, Yang Y, Liu J, Chu M, Wen J, Xie K, Du G, Wang Q, Zhou Y, Cao M, Liu L, He Y, Wang Y, Zhou G, Jia W, Lu J, Li S, Yang H, Shi Y, Zhou W, Shen H. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet. 2013;45:1499-1503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (1)] |
| 14. | Nishida N, Sawai H, Matsuura K, Sugiyama M, Ahn SH, Park JY, Hige S, Kang JH, Suzuki K, Kurosaki M, Asahina Y, Mochida S, Watanabe M, Tanaka E, Honda M, Kaneko S, Orito E, Itoh Y, Mita E, Tamori A, Murawaki Y, Hiasa Y, Sakaida I, Korenaga M, Hino K, Ide T, Kawashima M, Mawatari Y, Sageshima M, Ogasawara Y, Koike A, Izumi N, Han KH, Tanaka Y, Tokunaga K, Mizokami M. Genome-wide association study confirming association of HLA-DP with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PLoS One. 2012;7:e39175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 15. | Kim YJ, Kim HY, Lee JH, Yu SJ, Yoon JH, Lee HS, Kim CY, Cheong JY, Cho SW, Park NH, Park BL, Namgoong S, Kim LH, Cheong HS, Shin HD. A genome-wide association study identified new variants associated with the risk of chronic hepatitis B. Hum Mol Genet. 2013;22:4233-4238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 16. | Al-Qahtani AA, Al-Anazi MR, Abdo AA, Sanai FM, Al-Hamoudi W, Alswat KA, Al-Ashgar HI, Khalaf NZ, Eldali AM, Viswan NA, Al-Ahdal MN. Association between HLA variations and chronic hepatitis B virus infection in Saudi Arabian patients. PLoS One. 2014;9:e80445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 17. | Chang SW, Fann CS, Su WH, Wang YC, Weng CC, Yu CJ, Hsu CL, Hsieh AR, Chien RN, Chu CM, Tai DI. A genome-wide association study on chronic HBV infection and its clinical progression in male Han-Taiwanese. PLoS One. 2014;9:e99724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 18. | Huang YH, Liao SF, Khor SS, Lin YJ, Chen HY, Chang YH, Huang YH, Lu SN, Lee HW, Ko WY, Huang C, Liu PC, Chen YJ, Wu PF, Chu HW, Wu PE, Tokunaga K, Shen CY, Lee MH. Large-scale genome-wide association study identifies HLA class II variants associated with chronic HBV infection: a study from Taiwan Biobank. Aliment Pharmacol Ther. 2020;52:682-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 19. | Zeng Z, Liu H, Xu H, Lu H, Yu Y, Xu X, Yu M, Zhang T, Tian X, Xi H, Guan L, Zhang J, O'Brien SJ; HBVstudy consortium. Genome-wide association study identifies new loci associated with risk of HBV infection and disease progression. BMC Med Genomics. 2021;14:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Tai DI, Jeng WJ, Lin CY. A global perspective on hepatitis B-related single nucleotide polymorphisms and evolution during human migration. Hepatol Commun. 2017;1:1005-1013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 21. | Tai DI, Tai J. The role of genetic factors in HBV-related HCC: perspectives from local genetic backgrounds and clinical epidemiology. Hepatoma Res. 2020;6:74. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Chang MH, Hsu HY, Hsu HC, Ni YH, Chen JS, Chen DS. The significance of spontaneous hepatitis B e antigen seroconversion in childhood: with special emphasis on the clearance of hepatitis B e antigen before 3 years of age. Hepatology. 1995;22:1387-1392. [PubMed] |
| 23. | Chang MH, Sung JL, Lee CY, Chen CJ, Chen JS, Hsu HY, Lee PI, Chen DS. Factors affecting clearance of hepatitis B e antigen in hepatitis B surface antigen carrier children. J Pediatr. 1989;115:385-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 86] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 24. | Hadziyannis SJ. Natural history of chronic hepatitis B in Euro-Mediterranean and African countries. J Hepatol. 2011;55:183-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (1)] |
| 25. | Iorio R, Giannattasio A, Cirillo F, D' Alessandro L, Vegnente A. Long-term outcome in children with chronic hepatitis B: a 24-year observation period. Clin Infect Dis. 2007;45:943-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 26. | Li S, Qian J, Yang Y, Zhao W, Dai J, Bei JX, Foo JN, McLaren PJ, Li Z, Yang J, Shen F, Liu L, Li S, Pan S, Wang Y, Li W, Zhai X, Zhou B, Shi L, Chen X, Chu M, Yan Y, Wang J, Cheng S, Shen J, Jia W, Liu J, Wen Z, Li A, Zhang Y, Zhang G, Luo X, Qin H, Chen M, Wang H, Jin L, Lin D, Shen H, He L, de Bakker PI, Zeng YX, Wu M, Hu Z, Shi Y, Zhou W. GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 2012;8:e1002791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 167] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 27. | Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9:301-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 594] [Cited by in RCA: 611] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 28. | Miyake K, Yang W, Hara K, Yasuda K, Horikawa Y, Osawa H, Furuta H, Ng MC, Hirota Y, Mori H, Ido K, Yamagata K, Hinokio Y, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Wang HY, Tanahashi T, Nakamura N, Takeda J, Maeda E, Yamamoto K, Tokunaga K, Ma RC, So WY, Chan JC, Kamatani N, Makino H, Nanjo K, Kadowaki T, Kasuga M. Construction of a prediction model for type 2 diabetes mellitus in the Japanese population based on 11 genes with strong evidence of the association. J Hum Genet. 2009;54:236-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 29. | Song YM, Sung J, Yang S, Choe YH, Chang YS, Park WS. Factors associated with immunoprophylaxis failure against vertical transmission of hepatitis B virus. Eur J Pediatr. 2007;166:813-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 92] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 30. | Ding K, Bailey KR, Kullo IJ. Genotype-informed estimation of risk of coronary heart disease based on genome-wide association data linked to the electronic medical record. BMC Cardiovasc Disord. 2011;11:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Pencina MJ, D'Agostino RB Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2030] [Cited by in RCA: 2017] [Article Influence: 144.1] [Reference Citation Analysis (0)] |
| 32. | Pencina MJ, D'Agostino RB Sr, D'Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157-172; discussion 207-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4325] [Cited by in RCA: 5065] [Article Influence: 297.9] [Reference Citation Analysis (0)] |
| 33. | Ben Selma W, Laribi AB, Alibi S, Boukadida J. Association of an IFN-γ variant with susceptibility to chronic hepatitis B by the enhancement of HBV DNA replication. Cytokine. 2021;143:155525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 34. | Jiang DK, Ma XP, Yu H, Cao G, Ding DL, Chen H, Huang HX, Gao YZ, Wu XP, Long XD, Zhang H, Zhang Y, Gao Y, Chen TY, Ren WH, Zhang P, Shi Z, Jiang W, Wan B, Saiyin H, Yin J, Zhou YF, Zhai Y, Lu PX, Gu X, Tan A, Wang JB, Zuo XB, Sun LD, Liu JO, Yi Q, Mo Z, Zhou G, Liu Y, Sun J, Shugart YY, Zheng SL, Zhang XJ, Xu J, Yu L. Genetic variants in five novel loci including CFB and CD40 predispose to chronic hepatitis B. Hepatology. 2015;62:118-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 35. | Li Y, Si L, Zhai Y, Hu Y, Hu Z, Bei JX, Xie B, Ren Q, Cao P, Yang F, Song Q, Bao Z, Zhang H, Han Y, Wang Z, Chen X, Xia X, Yan H, Wang R, Zhang Y, Gao C, Meng J, Tu X, Liang X, Cui Y, Liu Y, Wu X, Li Z, Wang H, Hu B, He M, Gao Z, Xu X, Ji H, Yu C, Sun Y, Xing B, Yang X, Tan A, Wu C, Jia W, Li S, Zeng YX, Shen H, He F, Mo Z, Zhou G. Genome-wide association study identifies 8p21.3 associated with persistent hepatitis B virus infection among Chinese. Nat Commun. 2016;7:11664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 36. | Tao J, Su K, Yu C, Liu X, Wu W, Xu W, Jiang B, Luo R, Yao J, Zhou J, Zhan Y, Ye C, Yuan W, Jiang X, Cui W, Li MD, Li L. Fine mapping analysis of HLA-DP/DQ gene clusters on chromosome 6 reveals multiple susceptibility loci for HBV infection. Amino Acids. 2015;47:2623-2634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 37. | Li J, Yang D, He Y, Wang M, Wen Z, Liu L, Yao J, Matsuda K, Nakamura Y, Yu J, Jiang X, Sun S, Liu Q, Song Q, Chen M, Yang H, Tang F, Hu X, Wang J, Chang Y, He X, Chen Y, Lin J. Associations of HLA-DP variants with hepatitis B virus infection in southern and northern Han Chinese populations: a multicenter case-control study. PLoS One. 2011;6:e24221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 38. | Pan L, Zhang L, Zhang W, Wu X, Li Y, Yan B, Zhu X, Liu X, Yang C, Xu J, Zhou G, Xu A, Li H, Liu Y. A genome-wide association study identifies polymorphisms in the HLA-DR region associated with non-response to hepatitis B vaccination in Chinese Han populations. Hum Mol Genet. 2014;23:2210-2219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 39. | Deng Y, Li P, Liu W, Pu R, Yang F, Song J, Yin J, Han X, Li C, Zhao J, Wang H, Cao G. The genetic polymorphism down-regulating HLA-DRB1 enhancer activity facilitates HBV persistence, evolution and hepatocarcinogenesis in the Chinese Han population. J Viral Hepat. 2020;27:1150-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 40. | Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J Med Virol. 2004;72:363-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 139] [Article Influence: 6.6] [Reference Citation Analysis (0)] |