Published online Feb 28, 2019. doi: 10.3748/wjg.v25.i8.1012
Peer-review started: December 6, 2018
First decision: January 6, 2019
Revised: January 18, 2019
Accepted: January 26, 2019
Article in press: January 26, 2019
Published online: February 28, 2019
Processing time: 85 Days and 13.6 Hours
The lethal-7 (let-7) family members and their targets are involved in the development and progression of tumors. Let-7-related polymorphisms have been reported to be associated with tumorigenesis and prognosis. In gastric cancer, however, the related studies are limited.
To investigate the role of let-7-related microRNA polymorphisms in the tumorigenesis and prognosis of gastric cancer in a Chinese population.
A total of 898 gastric cancer patients and 992 tumor-free controls were recruited into this study from 2008 to 2013. Gastric cancer patients were followed periodically. Ten single nucleotide polymorphisms (SNPs) in the let-7 gene region or their target mRNAs were genotyped using the MassARRAY system and their associations with the risk for or overall survival of gastric cancer were analyzed.
All the ten SNPs were in Hardy-Weinberg equilibrium. The C allele of the rs3811463 polymorphism in the 3’-untranslated region (UTR) of LIN28A was associated with a lower risk of gastric cancer [odds ratio (OR) = 0.74, 95% confidence interval (CI): 0.61-0.88, P = 0.001] after adjustment for age and Helicobacter pylori status. Seven hundred and thirty-five gastric cancer patients who had undergone radical tumorectomy were included in the survival analysis and their 5-year survival rate was 53.9% (95%CI: 50.1%-57.6%). The rs10889677 in the 3’-UTR of IL23R was corresponded to the prognosis of gastric cancer in a dose-response manner, in which the death risk increased by 25% [hazard ratio (HR) = 1.25, 95%CI: 1.04-1.45, P = 0.011] with each increase in the number of C alleles after controlling for other potential clinicopathological parameters.
The let-7-related polymorphism rs3811463 in LIN28A is associated with the susceptibility to gastric cancer and the let-7-related polymorphism rs10889677 in IL23R is associated with the prognosis of gastric cancer.
Core tip: This study included a relatively large number of gastric cancer patients and tumor-free controls and explored the relationship of ten lethal-7 (let-7)-related single nucleotide polymorphisms with gastric carcinogenesis and prognosis. The results showed that the LIN28A rs3811463 polymorphism of the let-7 target was associated with the development of gastric cancer and that the IL23R rs10889677 polymorphism was related to the overall survival of gastric cancer patients in a dose-dependent manner. This study adds evidence that polymorphisms represent a genetic factor that modifies the susceptibility to and prognosis of gastric cancer.
- Citation: Jia ZF, Cao DH, Wu YH, Jin MS, Pan YC, Cao XY, Jiang J. Lethal-7-related polymorphisms are associated with susceptibility to and prognosis of gastric cancer. World J Gastroenterol 2019; 25(8): 1012-1023
- URL: https://www.wjgnet.com/1007-9327/full/v25/i8/1012.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i8.1012
Gastric cancer (GC) is the fifth most common malignancy and the third leading cause of cancer-related death worldwide[1]. It is estimated that there were 1 million new cases in 2018, and nearly half of the new cases were in China[1]. Despite advances in the diagnosis and treatment of GC in recent decades, the prognosis for GC patients is still poor, especially in China, with the five-year survival rate of only approximately 30%[2]. GC is a heterogeneous disease with distinct clinical, epidemiological, and molecular features and is thought to be a multifactorial disease influenced by environmental factors, microbial infection, and the host’s genetic background[3]. Recently, increasing numbers of studies have identified that genetic variations, of which single nucleotide polymorphisms (SNPs) are the most common type, play an important role in the development and progression of tumors including GC[4,5].
MicroRNAs (miRNAs) are endogenous small noncoding RNAs that bind the mRNAs of the target genes to inhibit their translation and/or induce their decay[6]. It is estimated that more than 30% of human genes, involved in nearly all human physical and pathophysiological processes, are regulated by miRNAs[7]. The miRNA lethal-7 (let-7) is the first miRNA identified in humans, and more than ten members of the let-7 family have been identified, including let-7a-1, let-7a-2, let-7a-3, let-7b, let-7c, let-7d, let-7e, let-7f-1, let-7f-2, let-7g, and let-7i[8]. Let-7 often exhibits tumor-suppressor functions in tumorigenesis such as inhibiting proliferation, inducing apoptosis, and suppressing the invasion and metastasis of cancer cells[8]. The let-7 family members are often downregulated in several cancers, thus derepressing the oncogenic targets such as K-ras, LIN28A, c-Myc, and HMGA2[9].
Previous studies have reported that miRNA-related SNPs could modulate the risk of tumors by altering the production of mature miRNAs or by affecting the binding affinity of miRNAs to their targets[10,11]. Xie et al[12] found that hepatocellular carcinoma patients carrying the C allele of rs10877887 in the promotor region of let-7i had a significantly increased death risk compared to patients with the TT genotype. The rs3811463 polymorphism is located in the 3’-untranslated region (UTR) of LIN28A, which is also near the target region of let-7. The C allele of rs3811463 could attenuate the let-7-induced repression of LIN28A mRNA, resulting in the increased production of LIN28A protein, which could in turn downregulate the level of mature let-7 via an LIN28A/let-7 double-negative feedback loop and alter breast cancer risk[13]. In GC, however, the related studies are limited. This study was aimed to determine the role of let-7 related SNPs in the susceptibility to and prognosis of GC.
This study was approved by the Ethics Committee of the First Hospital of Jilin University (2013-005). All participants provided written informed consent prior to joining the study.
GC patients who were hospitalized for potential tumor resection were invited to participate in the study from July 2008 to December 2013 at the First Hospital of Jilin University. A total of 898 patients who had not undergone chemotherapy or radiotherapy before surgery were recruited to the study, and all of them were histologically diagnosed with GC. Demographical and clinicopathological information was collected. The tumor histological type was evaluated by the World Health Organization criteria, and clinical stages were classified according to the 7th edition of the TNM staging system of the Union for International Cancer Control/American Joint Committee on Cancer (2010) based on postoperative pathologic examination.
During the same period, tumor-free controls were recruited from the Physical Examination Center of the same hospital. Controls were frequency-matched with cases by sex and age (± 5 years), and 992 controls were included in the study.
GC patients were followed periodically after tumorectomy. Follow-ups were performed at 3 mo, 6 mo, and 1 year after surgery and annually afterwards. Information on general status and postoperative chemotherapy was collected during each follow-up. If the patients had died, the date of death and potential cause were recorded. Survival time was defined as the duration from the date of surgery to the date of death. If the patient was alive, survival time was defined as the duration from the date of surgery to the date of the last follow-up. If the patient was lost to follow-up, survival time was defined as the duration from the date of surgery to the date of the last successful interview. Survival time was right-censored in the latter two cases. Patients were excluded from the survival analysis if they were lost to follow-up at the first interview or if they died of postoperative complications such as uncontrollable bleeding during the perioperative period.
All subjects donated blood samples. Genomic DNA was isolated using blood genomic DNA extraction kits (Axygen Biosciences, Union City, CA, United States). Serum immunoglobulin G antibodies to Helicobacter pylori (H. pylori) were assessed using an enzyme-linked immunosorbent assay (Biohit, Helsinki, Finland). Titers higher than 30 enzyme immunounits were defined as positive for H. pylori according to the manufacturer’s instructions.
Let-7-related SNPs that were reportedly associated with cancer were selected and genotyped. Six SNPs are in the let-7 gene region (rs13293512, rs562052, rs547008, rs1143770, rs629367, and rs10877887) and four SNPs are in the 3’-UTR of potential target genes of let-7 (rs3811463 in LIN28A, rs10889677 in IL23R, rs7963551 in RAD52, and rs712 in KRAS). Genotypes were determined with the MassARRAY system (Sequenom Inc, CA, United States). The call rates were all greater than 95% (13 subjects for rs13293512, 10 for rs562052, 10 for rs547008, 26 for rs1143770, 2 for rs629367, 53 for rs10877887, 7 for rs10889677, 6 for rs7963551, and 31 for rs712, respectively, failed to genotype). Ten randomly selected samples were simultaneously genotyped twice, and the concordance rates were 100% for all loci.
Continuous variables are shown as the mean ± standard deviation (SD) and compared by Student’s t-test. Categorical variables are presented as frequencies with percentages and were compared with the χ2 test or Fisher’s exact test when appropriate. Multivariate logistic regression analysis was employed to select the independent loci associated with the development of GC, and odds ratios (ORs) with their 95% confidence intervals (CIs) were calculated. Survival curves within each stratification of variables were plotted by the Kaplan-Meier method and compared by log-rank test. The multivariate Cox proportional hazard model was used to evaluate the prognostic role of polymorphisms, and hazard ratios (HRs) with their 95% CIs were calculated. All analyses were performed using SAS 9.2 software (SAS Institute Inc, United States). A two-tailed P-value < 0.05 indicated statistical significance.
From 2008 to 2013, 898 GC cases and 992 tumor-free controls were enrolled in the study. Gender distribution was uniform between cases and controls (P = 0.886). The mean age of GC patients was 2 years older than that of the controls (61.1 years vs 59.2 years, respectively; P = 0.0002). Six hundred and three GC cases were positive for H. pylori, and the H. pylori positive rate (67.5%) was much higher in GC patients than in the control group (487/992 = 49.1%, P < 0.001) (Table 1).
| Cases | Controls | P value | |
| n | 898 | 992 | |
| Gender | |||
| Male | 649 (72.3) | 714 (72.0) | 0.886 |
| Female | 249 (27.7) | 278 (28.0) | |
| Age (yr) | 61.1 ± 11.4 | 59.2 ± 10.0 | < 0.001 |
| ≤ 65 | 582 (64.8) | 703 (70.9) | 0.005 |
| > 65 | 316 (35.2) | 289 (29.1) | |
| Helicobacter pylori | |||
| Negative | 290 (32.5) | 505 (50.9) | < 0.001 |
| Positive | 603 (67.5) | 487 (49.1) |
Ten SNPs were genotyped in the study. All ten loci were in agreement with the Hardy-Weinberg equilibrium in the control group (P > 0.05). The distribution of the genotypes is presented in Table 2. The rs3811463 polymorphism in the 3’-UTR of LIN28A was distributed differently between GC cases and controls (P = 0.024). The proportions of T/C and C/C genotypes were lower in the GC group than in the control group (T/C: 22.6% vs 26.7%, respectively; C/C: 2.6% vs 3.8%, respectively). When compared to the population with the T/T genotype, individuals with the heterozygous T/C genotype had a decreased risk of GC (OR = 0.76, 95%CI: 0.61-0.94), and the risk was even lower for individuals with the C/C genotype (OR = 0.59, 95%CI: 0.34-1.01) after adjusting for age, gender, and H. pylori status. When the dominant model was used, the C allele (C/T + C/C) was associated with a reduced risk of GC (OR = 0.74, 95%CI: 0.60-0.91, P = 0.004). In stepwise logistic regression analysis, which included all loci, age, gender, and H. pylori status, rs3811463 was the only SNP site in the final model, and the risk of GC was lower by 26% with each increase of the number of C alleles (OR = 0.74, 95%CI: 0.61-0.88, P = 0.001), in addition to H. pylori status (OR = 2.20, 95%CI: 1.81-2.67, P < 0.001) and age (> 65 years vs ≤ 65 years, OR = 1.31, 95%CI: 1.07-1.61, P = 0.010) (Table 3). The other nine SNPs, however, were not associated with GC risk.
| Gene | SNP | Genotypes | Cases | Controls | P valuea | OR (95% CI)b |
| let-7a-1 | rs13293512 | T/T | 270 (30.3) | 310 (31.5) | 0.593 | 1.00 |
| T/C | 442 (49.6) | 494 (50.2) | 1.03 (0.83-1.28) | |||
| C/C | 180 (20.2) | 181 (18.4) | 1.13 (0.86-1.48) | |||
| let-7a-2 | rs562052 | G/G | 382 (43.0) | 439 (44.3) | 0.814 | 1.00 |
| G/A | 413 (46.5) | 446 (45.0) | 1.06 (0.87-1.29) | |||
| A/A | 94 (10.6) | 106 (10.7) | 0.96 (0.70-1.32) | |||
| let-7a-2 | rs547008 | C/C | 529 (59.2) | 575 (58.3) | 0.875 | 1.00 |
| C/T | 321 (35.9) | 358 (36.3) | 0.96 (0.79-1.17) | |||
| T/T | 44 (4.9) | 53 (5.4) | 0.85 (0.56-1.30) | |||
| let-7a-2 | rs1143770 | T/T | 255 (28.9) | 282 (28.7) | 0.262 | 1.00 |
| T/C | 456 (51.8) | 482 (49.0) | 1.04 (0.83-1.29) | |||
| C/C | 170 (19.3) | 219 (22.3) | 0.83 (0.64-1.09) | |||
| let-7a-2 | rs629367 | A/A | 532 (59.2) | 597 (60.3) | 0.583 | 1.00 |
| A/C | 323 (36.0) | 355 (35.9) | 1.01 (0.83-1.22) | |||
| C/C | 43 (4.8) | 38 (3.8) | 1.21 (0.76-1.92) | |||
| let-7i | rs10877887 | T/T | 370 (42.3) | 406 (42.2) | 0.777 | 1.00 |
| T/C | 399 (45.6) | 449 (46.7) | 0.96 (0.78-1.17) | |||
| C/C | 106 (12.1) | 107 (11.1) | 1.04 (0.76-1.42) | |||
| LIN28A | rs3811463 | T/T | 672 (74.8) | 689 (69.5) | 0.024 | 1.00 |
| T/C | 203 (22.6) | 265 (26.7) | 0.76 (0.61-0.94) | |||
| C/C | 23 (2.6) | 38 (3.8) | 0.59 (0.34-1.01) | |||
| Dominant | T/T | 672 (74.8) | 689 (69.5) | 0.009 | 1.00 | |
| T/C+C/C | 226 (25.2) | 303 (30.5) | 0.74 (0.60-0.91) | |||
| IL23R | rs10889677 | A/A | 476 (53.2) | 545 (55.2) | 0.685 | 1.00 |
| A/C | 356 (39.8) | 375 (38.0) | 1.10 (0.91-1.34) | |||
| C/C | 63 (7.0) | 68 (6.9) | 1.07 (0.74-1.55) | |||
| RAD52 | rs7963551 | T/T | 597 (66.6) | 672 (68.0) | 0.666 | 1.00 |
| T/G | 267 (29.8) | 287 (29.0) | 1.06 (0.87-1.31) | |||
| G/G | 32 (3.6) | 29 (2.9) | 1.18 (0.69-2.01) | |||
| KRAS | rs712 | G/G | 540 (61.0) | 589 (60.5) | 0.646 | 1.00 |
| G/T | 296 (33.4) | 339 (34.8) | 0.94 (0.77-1.15) | |||
| T/T | 49 (5.5) | 46 (4.7) | 1.14 (0.74-1.75) |
| Factor | OR | 95%CI | P value |
| rs3811463 (each increase in C allele) | 0.74 | 0.61-0.88 | 0.001 |
| Helicobacter pylori (positive vs negative) | 2.20 | 1.81-2.67 | < 0.001 |
| Age (> 65 vs ≤ 65 yr) | 1.31 | 1.07-1.61 | 0.010 |
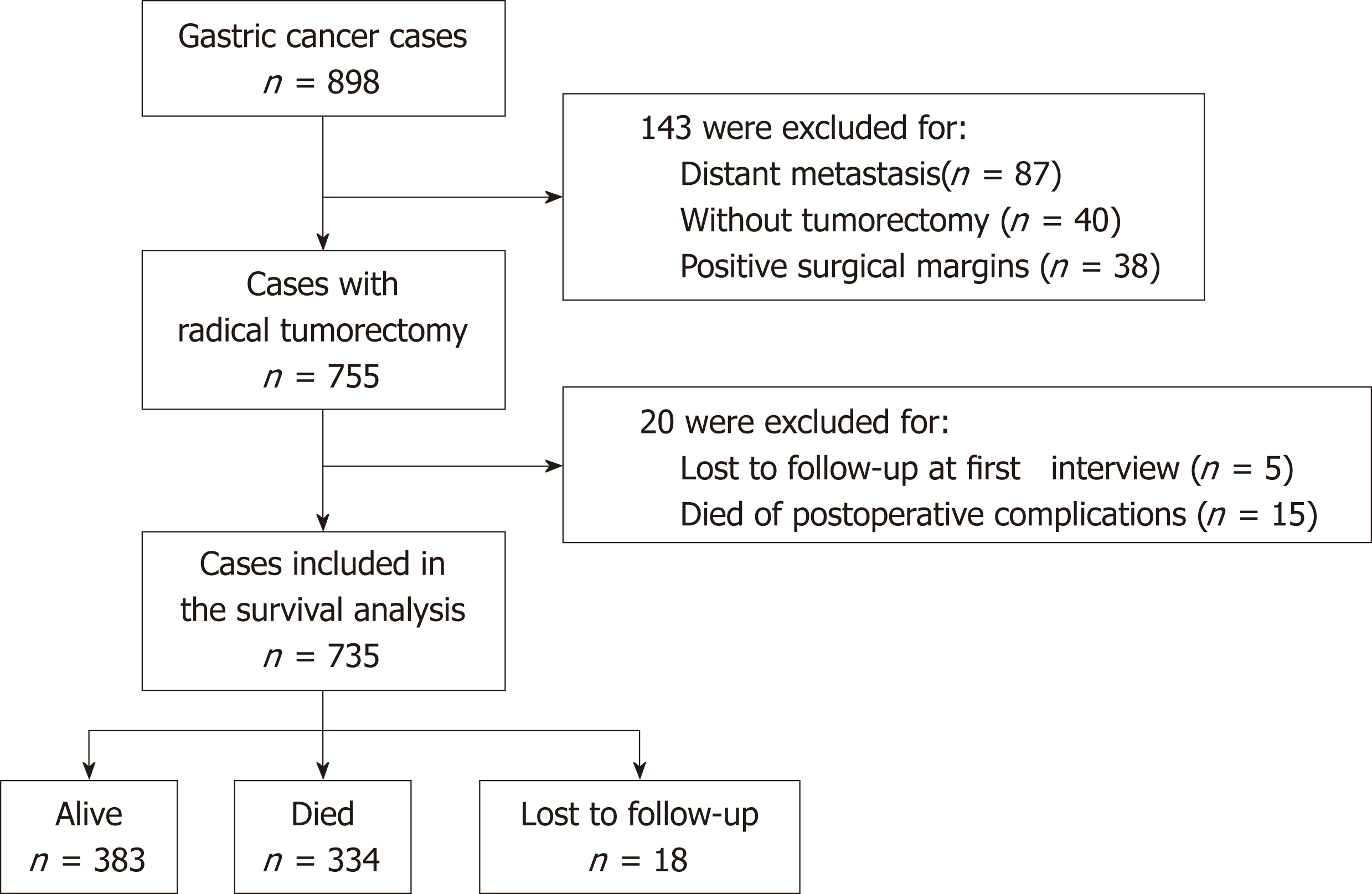
Given the prognostic value of surgical tumorectomy, survival analysis was performed in patients who had undergone radical tumorectomy (Figure 1). Among the 898 GC cases included in this study, 143 were excluded for one of the following reasons: (1) distant metastasis; (2) not receiving curative tumorectomy; or (3) positive surgical margins. Of the remaining 755 patients, 5 were lost to follow-up at the first interview, and 15 died of postoperative complications within one month of surgery. These 20 patients were also excluded from the survival analysis. Thus, 735 patients were included in the final survival analysis (Figure 1). The median follow-up time until September 2017 for these 735 patients was 59.8 mo. During the follow-up period, 334 patients (45.4%) died, 383 (52.1%) survived, and 18 (2.4%) were lost to follow-up. The 5-year survival rate was 53.9% (95% CI 50.1%-57.6%) and the median survival time was estimated to be 77.7 mo.
We tested the association between SNP genotypes and the overall survival of GC patients with radical resection. We observed that patients with different rs10889677 genotypes had different survival curves (log-rank P = 0.049, Figure 2). More precisely, patients had worse survival with each increase in the number of C alleles of rs10889677. By the end of the study, 41.7% of patients with no rs10889677 C allele (A/A genotype) had died, while 48.5% of patients with one C allele (A/C genotype) and 56.6% of patients with two C alleles (C/C genotype) had died (Table 4). The five-year survival rate for A/A genotype patients was 57.7%, which was higher than that of patients with the A/C genotype (50.3%) or C/C genotype (46.0%). We further transformed the rs10889677 genotypes to a quantifiable trait (the number of C alleles) and observed that the death HR increased by 23% with each additional C allele (HR = 1.23, 95%CI: 1.04-1.45, P = 0.015). The other nine SNP loci were not significantly associated with the overall survival of GC patients (Table 4).
| SNP | Genotype | Total | Death (%) | 5-yr survival rate | Median survival time | P valuea |
| rs13293512 | T/T | 213 | 100 (46.9) | 51.6% | 85.2 | 0.508 |
| T/C | 370 | 172 (46.5) | 53.3% | 75.8 | ||
| C/C | 148 | 60 (40.5) | 59.0% | -b | ||
| rs562052 | G/G | 313 | 150 (47.9) | 51.5% | 76.2 | 0.253 |
| G/A | 337 | 141 (41.8) | 56.9% | -b | ||
| A/A | 77 | 40 (51.9) | 50.7% | 65.4 | ||
| rs547008 | C/C | 434 | 200 (46.1) | 53.0% | 85.2 | 0.148 |
| C/T | 264 | 112 (42.4) | 56.2% | -b | ||
| T/T | 34 | 21 (61.8) | 46.7% | 33.8 | ||
| rs1143770 | T/T | 215 | 98 (45.6) | 54.3% | -b | 0.535 |
| T/C | 370 | 164 (44.3) | 54.4% | -b | ||
| C/C | 138 | 68 (49.3) | 51.3% | 65.4 | ||
| rs629367 | A/A | 435 | 205 (47.1) | 52.8% | 75.8 | 0.834 |
| A/C | 264 | 113 (42.8) | 55.6% | -b | ||
| C/C | 36 | 16 (44.4) | 55.8% | 85.2 | ||
| rs10877887 | T/T | 301 | 147 (48.8) | 50.4% | 64.7 | 0.235 |
| T/C | 330 | 141 (42.7) | 57.2% | -b | ||
| C/C | 88 | 39 (44.3) | 53.7% | -b | ||
| rs3811463 | T/T | 555 | 251 (45.2) | 53.7% | 85.2 | 0.971 |
| T/C | 162 | 75 (46.3) | 54.9% | 77.7 | ||
| C/C | 18 | 8 (44.4) | 52.0% | -b | ||
| rs10889677 | A/A | 379 | 158 (41.7) | 57.7% | -b | 0.049 |
| A/C | 301 | 146 (48.5) | 50.3% | 65.4 | ||
| C/C | 53 | 30 (56.6) | 46.0% | 41.0 | ||
| rs7963551 | T/T | 508 | 236 (46.5) | 52.56% | 85.2 | 0.144 |
| T/G | 199 | 82 (41.2) | 59.35% | -b | ||
| G/G | 27 | 16 (59.3) | 38.22% | 41.3 | ||
| rs712 | G/G | 452 | 215 (47.6) | 51.51% | 73.7 | 0.227 |
| G/T | 235 | 95 (40.4) | 58.64% | -b | ||
| T/T | 36 | 17 (47.2) | 55.01% | 77.7 |
Age (P < 0.001), tumor differentiation level (P = 0.010), depth of tumor invasion (T stage, P < 0.001), local lymph node metastasis (N stage, P < 0.001), TNM stage (P < 0.001), lymphovascular invasion (P < 0.001), and neural invasion (P < 0.001) were associated with the long-term survival of GC patients in univariate survival analysis (Table 5). Postoperative chemotherapy was marginally related to the outcome (P = 0.054). H. pylori status, which was previously reported to be related to GC prognosis[14], was not associated with the long-term OS in our study (P = 0.364, Table 5).
| Variable | Classification | na | Death (%) | 5-yr survival (%) | P valueb |
| Age (yr) | ≤ 65 | 494 | 197 (39.9) | 59.7 (55.0-64.0) | < 0.001 |
| > 65 | 241 | 137 (56.8) | 42.3 (35.6-48.8) | ||
| Gender | Female | 195 | 82 (42.1) | 56.8 (49.2-63.7) | 0.261 |
| Male | 540 | 252 (46.7) | 52.9 (48.4-57.2) | ||
| Differentiation | Poor | 508 | 241 (47.4) | 50.9 (46.2-55.3) | 0.010 |
| Moderate and high | 209 | 85 (40.7) | 61.6 (54.4-68.0) | ||
| WHO type | Tubular adenocarcinoma | 614 | 275 (44.8) | 54.8 (50.6-58.8) | 0.205 |
| Signet ring cell | 72 | 32 (44.4) | 52.8 (39.8-64.2) | ||
| Other | 45 | 25 (55.6) | 44.5 (29.2-58.6) | ||
| T stage | T1 | 100 | 13 (13.0) | 85.8 (76.5-91.6) | < 0.001 |
| T2 | 102 | 16 (15.7) | 84.4 (74.8-90.6) | ||
| T3 | 423 | 220 (52.0) | 48.1 (43.1-52.9) | ||
| T4 | 103 | 78 (75.7) | 17.4 (9.2-27.8) | ||
| N stage | N0 | 219 | 38 (17.4) | 84.2 (78.4-88.5) | < 0.001 |
| N1 | 184 | 63 (34.2) | 66.0 (58.2-72.6) | ||
| N2 | 150 | 87 (58.0) | 38.7 (30.3-47.1) | ||
| N3 | 175 | 139 (79.4) | 17.3 (11.7-23.9) | ||
| TNM | I | 137 | 12 (8.8) | 90.6 (83.9-94.6) | < 0.001 |
| II | 278 | 92 (33.1) | 68.3 (62.1-73.6) | ||
| III | 313 | 223 (71.2) | 25.8 (20.7-31.2) | ||
| Lymphovascular invasion | Negative | 213 | 38 (17.8) | 83.0 (77.0-87.6) | < 0.001 |
| Positive | 511 | 287 (56.2) | 42.6 (38.1-47.0) | ||
| Neural invasion | Negative | 328 | 100 (30.5) | 70.2 (64.6-75.0) | < 0.001 |
| Positive | 396 | 225 (56.8) | 41.1 (36.0-46.2) | ||
| Chemotherapy | No | 488 | 218 (44.7) | 54.8 (50.0-59.3) | 0.054 |
| FOLFOX-4 | 136 | 63 (46.3) | 52.1 (43.2-60.3) | ||
| XELOX | 73 | 28 (38.4) | 61.7 (49.1-72.0) | ||
| Other | 38 | 25 (65.8) | 34.0 (19.1-49.4) | ||
| Helicobacter pylori | Negative | 228 | 109 (47.8) | 49.8 (42.7-56.4) | 0.364 |
| Positive | 503 | 222 (44.1) | 56.1 (51.4-60.4) |
Further multivariate analysis showed that the rs10889677 polymorphism was an independent prognostic predictor and that the death HR increased by 23% with each increase in the number of C alleles (HR = 1.25, 95%CI: 1.04-1.45, P = 0.012) (Table 6). In addition, old age (> 65 years, HR = 1.39, 95%CI: 1.10-1.75), high TNM stage [stage II (HR = 3.49, 95%CI: 1.88-6.48) or III (HR = 11.12, 95%CI: 6.0-20.61)], and positive lymphovascular invasion (HR = 1.89, 95%CI: 1.32-2.72) were also independently associated with decreased long-term survival. Additionally, chemotherapy after radical tumorectomy reduced the death hazard (HR = 0.75, 95%CI: 0.59-0.96, P = 0.021) (Table 6).
| Variable | Comparison | HR | 95%CI | P value |
| rs10889677 | each increase in C allele | 1.25 | 1.05-1.49 | 0.012 |
| Age (yr) | > 65 vs ≤ 65 | 1.39 | 1.10-1.75 | 0.005 |
| TNM stage | II vs I | 3.49 | 1.88-6.48 | < 0.001 |
| III vs I | 11.12 | 6.00-20.61 | < 0.001 | |
| Lymphovascular invasion | Positive vs Negative | 1.89 | 1.32-2.72 | 0.001 |
| Chemotherapy | Yes vs No | 0.74 | 0.59-0.95 | 0.016 |
The let-7 family members play an important role in several hallmarks of cancer, including repressing cellular proliferation, inducing cell apoptosis, suppressing aerobic glycolysis, inhibiting invasion and metastasis, and regulating tumor innate immune reactions by interacting with their targets[8]. Let-7-related SNPs, which are located in the let-7 gene region or in the target gene region, were reported to be associated with the development or prognosis of tumors. In this study, we explored the association of let-7-related SNPs with the susceptibility to and long-term overall survival of GC in a Chinese population. We observed that the rs3811463 in the 3’-UTR of LIN28A was associated with the susceptibility of GC and that rs10889677 in the 3’-UTR of IL23R was associated with the OS of resectable GC.
The let-7 family and LIN28A compose a double-negative feedback loop. LIN28A represses let-7 by inhibiting the biogenesis and inducing the degradation of the let-7 family; meanwhile, let-7 inhibits the expression and function of LIN28A via binding to complementary sites at the 3’-UTR of LIN28A mRNA[8]. In contrast to the role of let-7 in tumors, LIN28A promotes the proliferation of cancer cells and is related to the poor prognosis[15]. Rs3811463 is located in the 3’-UTR of LIN28A, near the binding region of let-7. Chen et al[13] reported that the C allele of rs3811463 corresponded to an increased risk of breast cancer by attenuating the let-7-induced LIN28A repression, which in turn downregulates the mature let-7 via the LIN28A/let-7 double-negative feedback loop. A subsequent study on oral cancer did not report any association between oral cancer risk and rs3811463 polymorphism in 384 cases and 731 controls[16]. In our study, however, the C allele of rs3811463 was found to be associated with a reduced risk of GC. Moreover, this association demonstrated a dose-response manner in which the risk was lower with each increase in the number of C alleles compared to that of the patients without the C allele (i.e., genotype T/T). The online bioinformatics tool SNPinfo (http://snpinfo.niehs.nih.gov/) predicts that the change from T to C might make LIN28A gain the target of miR-490, which would reduce the level of LIN28A, thus associating with a lower risk of GC. However, further studies are needed to verify this hypothesis.
Interleukin-23 receptor (IL23R), a crucial subunit of the IL-23 receptor complex, is involved in innate and adaptive immune processes by interacting with IL-23, which plays a crucial role in many human diseases including autoimmune diseases and tumors[17,18]. Because IL23R presents tumor-promoting effects, knockdown of IL23R restricts tumor growth and decreases cancer metastasis[17,18]. The rs10889677 polymorphism, located in the 3’-UTR of the IL23R gene, has been reported to be associated with cancer risk, including the risk of breast cancer[19], esophageal cancer[20], bladder cancer[21], ovarian cancer[22], oral cancer[23], and GC[24,25]. The results of these studies, however, are controversial[26]. In GC, Chen et al showed that the presence of the C allele of rs10889677 predicted lower GC risk (A/C: OR = 0.81, 95%CI: 0.66–0.99; C/C: OR = 0.47, 95%CI: 0.31–0.71)[24], while another study by Dong et al[25] observed an opposite relationship; the presence of the C allele increased the GC risk by 24% (Callelevs Aallele: OR = 1.24, 95%CI: 1.02–1.52). In our study, however, we did not find any significant association between rs10889677 and GC risk (P > 0.05). Nonetheless, we observed that the C allele was related to decreased survival of GC patients after curative tumorectomy in a dose-dependent manner, and the death HR increased by 25% (HR = 1.25, 95%CI: 1.05-1.49) with each increment in the number of the C alleles. This is the first study reporting that the rs10889677 of IL23R is related to the prognosis of GC, as most studies only focused on cancer susceptibility. Zwiers et al[27] showed that the C allele of rs10889677 was corresponded to reduced IL23R levels possibly through the target gain of let-7. However, this could not explain what we had observed, and another unknown mechanism might be involved.
Studies have reported a significant correlation between SNPs in the let-7 gene region and the susceptibility to or prognosis of malignant tumors, for example, rs629367 in let-7a and the risk of GC[10], rs10877887 in let-7i and the risk of papillary thyroid carcinoma[28] and survival of hepatocellular carcinoma[12], and rs1143770 in let-7a and the overall survival and disease-free survival of surgically resected non-small cell lung cancer[29]. Other studies, however, observed no such associations[30]. In our study of a relatively large number of GC patients, we did not find that the six polymorphisms in the let-7 gene region (rs13293512, rs562052, rs547008, rs1143770, rs629367, and rs10877887) were associated with the risk of development and the overall survival of GC. Several factors might contribute to the discrepancies among studies, such as different origins of the tumors studied and study populations from different ethnic groups. Therefore, more studies with a large sample size, including populations from different ethnic groups and tumors from multiple origins, are warranted in the future.
Two limitations should be noted in our study. First, although we observed that two SNPs were associated with GC, we could not verify the underlying mechanism for these associations because of our study design. Second, GC patients and controls from only one hospital were included in the study. The generalization of these results to other populations should be cautious because there has been no external validation. Therefore, more studies are needed to verify the results.
In summary, this study provides evidence that polymorphisms represent a genetic factor that modifies the susceptibility to and prognosis of GC. Individuals carrying the C allele of rs3811463 in the 3’-UTR of LIN28A have a lower risk of GC, and GC patients with the C allele of rs10889677 in the 3’-UTR of IL23R have a shorter lifespan than patients without it.
Gastric cancer (GC) is one of the most common malignancies worldwide. Despite the advances in diagnosis and treatment of GC in recent decades, prognosis of GC patients is still poor. It is of great importance to identify biomarkers that could be helpful in the improvement of screening of high-risk individuals, early diagnosis, and predicting outcome for the individualized therapy.
The microRNA lethal-7 (let-7) often exhibits tumor-suppressor functions in tumorigenesis. Single nucleotide polymorphisms (SNPs) in the let-7 gene region or let-7 target genes have been reported to modulate the risk of several cancers including breast cancer and lung cancer. In GC, the related studies are limited
By including a relatively large number of GC patients, this study aimed to determine the role of let-7-related polymorphisms in GC in a Chinese population.
From 2008 to 2013, 898 consecutive GC patients and 992 tumor-free controls were recruited into the study. GC patients were followed periodically to determine their prognosis. Ten SNPs in the let-7 gene region or their target mRNAs were genotyped using MassArray system and the associations with the risk or overall survival of GC were analyzed.
Two SNPs in let-7 target genes were associated with GC in a dose-dependent manner. Rs3811463 in the 3’-UTR of LIN28A was associated with lower risk of GC and the risk was reduced by 26% with each increase of the C allele of rs3811463. The rs10889677 in the 3’-UTR of IL23R was corresponded to the prognosis of GC, and the death risk increased by 25% with each increment of the C allele of rs10889677, after controlling for clinicopathological parameters.
Let-7-related SNPs were related to GC. The rs3811463 in LIN28A is associated with the susceptibility to and rs10889677 in IL23R is associated with the prognosis of GC.
Our research adds evidence that polymorphisms represent a genetic factor to modify the susceptibility to and prognosis of GC. The underlying mechanisms of the associations should be elucidated in future studies.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Aoyagi K, Nardone G, Park WS S- Editor: Yan JP L- Editor: Wang TQ E- Editor: Yin SY
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55742] [Article Influence: 7963.1] [Reference Citation Analysis (132)] |
| 2. | Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, Xia C, Sun K, Yang Z, Li H, Wang N, Han R, Liu S, Li H, Mu H, He Y, Xu Y, Fu Z, Zhou Y, Jiang J, Yang Y, Chen J, Wei K, Fan D, Wang J, Fu F, Zhao D, Song G, Chen J, Jiang C, Zhou X, Gu X, Jin F, Li Q, Li Y, Wu T, Yan C, Dong J, Hua Z, Baade P, Bray F, Jemal A, Yu XQ, He J. Changing cancer survival in China during 2003-15: A pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6:e555-e567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 795] [Cited by in RCA: 964] [Article Influence: 137.7] [Reference Citation Analysis (2)] |
| 3. | Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1159] [Cited by in RCA: 1323] [Article Influence: 120.3] [Reference Citation Analysis (0)] |
| 4. | Stenholm L, Stoehlmacher-Williams J, Al-Batran SE, Heussen N, Akin S, Pauligk C, Lehmann S, Senff T, Hofheinz RD, Ehninger G, Kramer M, Goekkurt E. Prognostic role of microRNA polymorphisms in advanced gastric cancer: A translational study of the Arbeitsgemeinschaft Internistische Onkologie (AIO). Ann Oncol. 2013;24:2581-2588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 5. | Espinosa-Parrilla Y, Muñoz X, Bonet C, Garcia N, Venceslá A, Yiannakouris N, Naccarati A, Sieri S, Panico S, Huerta JM, Barricarte A, Menéndez V, Sánchez-Cantalejo E, Dorronsoro M, Brennan P, Duarte-Salles T, B As Bueno-de-Mesquita H, Weiderpass E, Lund E, Clavel-Chapelon F, Boutron-Ruault MC, Racine A, Numans ME, Tumino R, Canzian F, Campa D, Sund M, Johansson M, Ohlsson B, Lindkvist B, Overvad K, Tjønneland A, Palli D, Travis RC, Khaw KT, Wareham N, Boeing H, Nesi G, Riboli E, Gonzalez CA, Sala N. Genetic association of gastric cancer with miRNA clusters including the cancer-related genes MIR29, MIR25, MIR93 and MIR106: Results from the EPIC-EURGAST study. Int J Cancer. 2014;135:2065-2076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 6. | Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163-6169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 343] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 7. | Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170-6175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 275] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 8. | Wang T, Wang G, Hao D, Liu X, Wang D, Ning N, Li X. Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol Cancer. 2015;14:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 148] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 9. | Sun X, Liu J, Xu C, Tang SC, Ren H. The insights of Let-7 miRNAs in oncogenesis and stem cell potency. J Cell Mol Med. 2016;20:1779-1788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Xu Q, Dong Q, He C, Liu W, Sun L, Liu J, Xing C, Li X, Wang B, Yuan Y. A new polymorphism biomarker rs629367 associated with increased risk and poor survival of gastric cancer in chinese by up-regulated miRNA-let-7a expression. PLoS One. 2014;9:e95249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Jiang Y, Qin Z, Hu Z, Guan X, Wang Y, He Y, Xue J, Liu X, Chen J, Dai J, Jin G, Ma H, Wang S, Shen H. Genetic variation in a hsa-let-7 binding site in RAD52 is associated with breast cancer susceptibility. Carcinogenesis. 2013;34:689-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Xie K, Liu J, Zhu L, Liu Y, Pan Y, Wen J, Ma H, Zhai X, Hu Z. A potentially functional polymorphism in the promoter region of let-7 family is associated with survival of hepatocellular carcinoma. Cancer Epidemiol. 2013;37:998-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Chen AX, Yu KD, Fan L, Li JY, Yang C, Huang AJ, Shao ZM. Germline genetic variants disturbing the Let-7/LIN28 double-negative feedback loop alter breast cancer susceptibility. PLoS Genet. 2011;7:e1002259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Tsai KF, Liou JM, Chen MJ, Chen CC, Kuo SH, Lai IR, Yeh KH, Lin MT, Wang HP, Cheng AL, Lin JT, Shun CT, Wu MS; Taiwan Gastrointestinal Disease and Helicobacter Consortium. Distinct Clinicopathological Features and Prognosis of Helicobacter pylori Negative Gastric Cancer. PLoS One. 2017;12:e0170942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 15. | Xu C, Shen J, Xie S, Jiang Z, Huang L, Wang L. Positive expression of Lin28 is correlated with poor survival in gastric carcinoma. Med Oncol. 2013;30:382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Zhang Y, Zhu L, Wang R, Miao L, Jiang H, Yuan H, Ma H, Chen N. Genetic variants in let-7/Lin28 modulate the risk of oral cavity cancer in a Chinese Han population. Sci Rep. 2014;4:7434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | Yan J, Allen S, Vijayan D, Li XY, Harjunpää H, Takeda K, Liu J, Cua DJ, Smyth MJ, Teng MWL. Experimental Lung Metastases in Mice Are More Effectively Inhibited by Blockade of IL23R than IL23. Cancer Immunol Res. 2018;6:978-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, Basham B, McClanahan T, Kastelein RA, Oft M. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 739] [Cited by in RCA: 772] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 19. | Zheng J, Jiang L, Zhang L, Yang L, Deng J, You Y, Li N, Wu H, Li W, Lu J, Zhou Y. Functional genetic variations in the IL-23 receptor gene are associated with risk of breast, lung and nasopharyngeal cancer in Chinese populations. Carcinogenesis. 2012;33:2409-2416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 20. | Ni B, Chen S, Xie H, Ma H. Functional polymorphisms in interleukin-23 receptor and susceptibility to esophageal squamous cell carcinoma in Chinese population. PLoS One. 2014;9:e89111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 21. | Tang T, Xue H, Cui S, Gong Z, Fei Z, Cheng S, Gui C. Association of interleukin-23 receptor gene polymorphisms with risk of bladder cancer in Chinese. Fam Cancer. 2014;13:619-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 22. | Zhang Z, Zhou B, Zhang J, Chen Y, Lai T, Yan L, Liang A, Li Y, Wang Y, Chen Y, Zhang L, Xi MR. Association of interleukin-23 receptor gene polymorphisms with risk of ovarian cancer. Cancer Genet Cytogenet. 2010;196:146-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Chien MH, Hsin CH, Chou LS, Chung TT, Lin CH, Weng MS, Chou MY, Chen MK, Yang SF. Interleukin-23 receptor polymorphism as a risk factor for oral cancer susceptibility. Head Neck. 2012;34:551-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Chen B, Zeng Z, Xu L, Wu X, Yu J, Xue L, Hao Y, Wang Y, Sung JJ, Chen M, Hu P. IL23R +2199A/C polymorphism is associated with decreased risk of certain subtypes of gastric cancer in Chinese: a case-control study. Cancer Epidemiol. 2011;35:165-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Dong K, Xu Y, Yang Q, Shi J, Jiang J, Chen Y, Song C, Wang K. Associations of Functional MicroRNA Binding Site Polymorphisms in IL23/Th17 Inflammatory Pathway Genes with Gastric Cancer Risk. Mediators Inflamm. 2017;2017:6974696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Liu XH, Dai ZM, Kang HF, Lin S, Ma XB, Wang M, Liu K, Dai C, Wang XJ, Dai ZJ. Association of IL-23R Polymorphisms (rs6682925, rs10889677, rs1884444) With Cancer Risk: A PRISMA-Compliant Meta-Analysis. Medicine (Baltimore). 2015;94:e2361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Zwiers A, Kraal L, van de Pouw Kraan TC, Wurdinger T, Bouma G, Kraal G. Cutting edge: A variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. J Immunol. 2012;188:1573-1577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 28. | Wang Y, Wei T, Xiong J, Chen P, Wang X, Zhang L, Gao L, Zhu J. Association Between Genetic Polymorphisms in the Promoter Regions of Let-7 and Risk of Papillary Thyroid Carcinoma: A Case-Control Study. Medicine (Baltimore). 2015;94:e1879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Shin KM, Jung DK, Hong MJ, Kang HJ, Lee WK, Yoo SS, Lee SY, Cha SI, Lee J, Kim CH, Seok Y, Cho S, Son JW, Lee EB, Jheon S, Kim YT, Park JY. The pri-let-7a-2 rs1143770C>T is associated with prognosis of surgically resected non-small cell lung cancer. Gene. 2016;577:148-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 30. | Huang F, Hu LM, Liu JB, Zhang YX, Hu ZB. [Relationship between genetic polymorphism of promoter region let-7 and genetic susceptibility to hepatocellular carcinoma]. Zhonghua Yu Fang Yi Xue Za Zhi. 2011;45:1093-1098. [PubMed] |