Published online Feb 7, 2017. doi: 10.3748/wjg.v23.i5.800
Peer-review started: July 17, 2016
First decision: August 19, 2016
Revised: September 5, 2016
Accepted: September 14, 2016
Article in press: September 14, 2016
Published online: February 7, 2017
Processing time: 193 Days and 2.5 Hours
To compare the genomic variability and the multiple colonization of Helicobacter pylori (H. pylori) in patients with chronic gastritis from two Colombian populations with contrast in the risk of developing gastric cancer (GC): Túquerres-Nariño (High risk) and Tumaco-Nariño (Low risk).
Four hundred and nine patients from both genders with dyspeptic symptoms were studied. Seventy-two patients were included in whom H. pylori was isolated from three anatomic regions of the gastric mucosa, (31/206) of the high risk population of GC (Túquerres) and (41/203) of the low risk population of GC (Tumaco). The isolates were genotyped by PCR-RAPD. Genetic diversity between the isolates was evaluated by conglomerates analysis and multiple correspondence analyses.
The proportion of virulent genotypes of H. pylori was 99% in Túquerres and 94% in Tumaco. The coefficient of similarity of Nei-Li showed greater genetic diversity among isolates of Túquerres (0.13) than those of Tumaco (0.07). After adjusting by age, gender and type of gastritis, the multiple colonization was 1.7 times more frequent in Túquerres than in Tumaco (P = 0.05).
In Túquerres, high risk of GC there was a greater probability of multiple colonization by H. pylori. From the analysis of the results of the PCR-RAPD, it was found higher genetic variability in the isolates of H. pylori in the population of high risk for the development of GC.
Core tip: Multiple colonization of Helicobacter pylori (H. pylori) occurred more frequently in individuals living in the Colombian population with higher risk of gastric cancer (GC) (Túquerres). In the two populations contrasted in relation to the risk of developing GC. (Túquerres high risk and Tumaco low risk) H. pylori was identified with specific genetic characteristics for each region and with varying stages of genomic variability. The diversity of H. pylori dependent of the anatomic regions of the gastric mucosa, obstructs the eradication of the microorganism. Identifying the multiple colonization and evaluating the genetic diversity of H. pylori individuals may be sifted that require particular schemes of early treatment and prevention of the precursor lesions of GC.
- Citation: Matta AJ, Pazos AJ, Bustamante-Rengifo JA, Bravo LE. Genomic variability of Helicobacter pylori isolates of gastric regions from two Colombian populations. World J Gastroenterol 2017; 23(5): 800-809
- URL: https://www.wjgnet.com/1007-9327/full/v23/i5/800.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i5.800
Helicobacter pylori (H. pylori) is recognized as the principal etiologic agent of chronic gastritis; it is related to the development of peptic ulcer, and is the most important factor in the pathogenesis of gastric adenocarcinoma[1].
The high genetic variability between strains of H. pylori is associated to the population or geographic origin of the individuals of the population[2]. Variations in the genomic DNA of H. pylori allows the identification of individuals infected by multiple strains that colonize different anatomic regions of the gastric mucosa, phenomenon described as multiple colonization[3]. In Colombia, there are regions of high and low risk for the development of gastric cancer (GC), although the prevalence of the infection by H. pylori is similar (> 80%)[4,5]. In Tumaco (low risk of GC) and Túquerres (high risk of GC), strains of H. pylori were identified with different stages of genetic variability for each region. The genetic variability of H. pylori that colonizes the different anatomical regions from the gastric mucosa has not been studied in Colombia and it makes the eradication of the microorganism difficult. Therefore it is important to study the multiple colonization of H. pylori as a strategy to prevent the precursor lesions of GC. The high genetic diversity between strains of H. pylori has been evaluated by several methods of characterization, including polymorphisms in the length of the restriction fragments (RFLP), Restriction fragment polymorphism amplified by PCR (AFLP-PCR), ribotyping and random amplification of polymorphic DNA (RAPD)[6,7].
The technique most used to discriminate isolates of H. pylori, in the genotyping studies, is the RAPD with a high level of discrimination (0.99-1.0), able to reveal a marked genetic diversity between isolated strains of different patients and it is discriminatory in the determination of the similarity of the isolated strains from the same patient in the biopsies taken from different anatomical sites from the gastric mucosa[8,9].
Previous investigations describe diversity of H. pylori strains with differences in their virulence factors such as the gene associated with cagA[10] cytotoxicity and the gene related with the production of vacuolizing toxin vacA[11].
A standard treatment to eradicate the H. pylori doesn’t exist, possibly because the gastric mucosa of the same host may be colonized by multiple strains of the microorganism with differences in the virulence factors and differences in the genomic DNA and eventually antimicrobial susceptible and resistant isolates coexist[12]. This phenomena, not studied in Colombia, makes the eradication of H. pylori difficult and doesn’t allow the establishment of therapeutic efficacy as a strategy for the prevention of precursor lesions of GC.
It is important to genotype the isolated H. pylori from the different locations of the gastric mucosa (antrum and body) to evaluate genetic diversity and determine the multiple colonization, in the two Colombian populations with contrast in the risk of developing GC.
Adult men and women with symptoms of dyspepsia were included (n = 203) from Tumaco and (n = 206) from Túquerres. Four biopsies from the gastric mucosa were obtained from each patient, two from the antrum and two from the gastric body for its histopathologic evaluation. Histological sections cut into microtome at 6 μm, were colored with hematoxylin-eosin and were interpreted according to the Sydney[13] classification system. The presence of H. pylori was determined with Giemsa’s staining.
For cultivation and genotyping of H. pylori three biopsies of gastric mucosa were used, two from the antrum and one from the gastric body, that were preserved in thioglycolate and glycerol at (25%). The biopsies were immediately frizzed in liquid nitrogen and subsequently transported on dry ice at -70 °C until its analysis in the Microbiology and Histopathological Laboratory of the Pathology Department of the Universidad del Valle-Colombia. This research counts with the approval of the Human Ethics Committee (CIREH) of the Universidad del Valle. All the participants signed the informed consent.
Fragments of gastric mucosa were homogenized in 200 μL of sterile saline solution 0.89%. The homogenate was seeded in columbia agar plates (Oxoid, Basingstoke, Hampshire, England) with defibrillated lamb blood to 7% plus selective supplement for H. pylori (Dent) and incubated in microaerophilic conditions (6% O2, 6% CO2, 88% N2 using CampyPak Plus Envelop, BBL, Nashville, TN United States) at 37 °C during 4 to 8 d[4]. The compatible colonies with H. pylori were transferred to columbia agar defibrillated lamb blood to 10%, to purify and identify them by the tests of urease, catalase, oxidase, Gram staining. A PCR of the ureA gene was used to confirm the species[14].
From a pure cultivation in petri dish colonies were transferred to 1.0 mL of PBS 1 × pH = 7.2, and it was centrifuge at 13000 RPM × 2 min. The cell pellet was re-suspended in 300 μL of extraction buffer [Proteinase K 100 μg/mL, sodium dodecyl sulfate (SDS) 0.5%, ethylenediaminetetraacetic acid (EDTA) 5 mmol/L, Tris-HCl 10 mmol/L, pH = 8.0 and 276 μL of distilled water], the cell button was homogenized and taken to the dry block (Labnet®) at 56 °C for 18 h, subsequently the proteinase K was inactivated at 76 °C for 10 min and NaCl 5M was added. It was shaken in vortex for 15 seg, and then it was centrifuge at 13000 RPM for 5 min. To the supernatant only, 2 volumes of absolute ethanol were added. It was centrifuge at 13000 RPM for 20 min at 4 °C. The decanted was precipitated by addition of 2 volumes of absolute ethanol at 70%, it was mixed and centrifuge for 5 min at 13000 RPM and the supernatant was discharged. The DNA pellet dried by tube inversion for 10 min. The precipitate DNA was re-suspended in 100 μL of TE buffer (Tris 10 mmol, EDTA 1 mmol) and it was stored at -20 °C. The yield and purity of the DNA were determined by optical density at 260/280 nm in Gene Quant II® spectrophotometer (Pharmacia Biotech, Piscataway, NJ, United States) according to the manufacturer’s instructions[15].
The molecular identification of H. pylori was performed by PCR amplification of the ureA gene. The following reagents were added to a 0.2 mL tube: 1 × PCR buffer (Buffer green 5 ×, Promega®); MgCl2 1 μmol/L (Promega®); 0.25 mmol/L of dNTPs (deoxyribonucleoside 5’-triphosphates - dATP, dCTP, dGTP and dTTP Promega®); 50 pmol/L of each primer (sense 3’-AAGACATCACTATCAACG-5’/anti-sense 5’-CCCGCTCGCAATGTCTAA-3’); 0.5 U of GoTaq DNA polymerase (Promega®) and 25 ng of genomic DNA from H. pylori in a final volume of 25 μL. Amplification was performed at 95 °C/2 min followed by 35 cycles (95 °C/1 min, 54 °C/1 min and 72 °C/1 min) and a final extension at 72 °C/15 min.
The virulence factors were analyzed by PCR amplification of cagA gene. An amplicon of 183 bp was obtained with specific primers (CagAF and CagAR). The negative isolates for the amplification of cagA gene were confirmed by PCR assay empty-site with primers [ES-F (+) - ES-R (-)]. The vacA gene alleles (s1/s2, m1/m2) were analyzed with the specific primers (VA1F and VA1R) to obtain the sizes of the amplicons; 176 bp and 203 bp for vacA s1 and s2, respectively. PCR was performed for vacA gene alleles m1/m2 with primers (HPMGF- HPMGR) to obtain the sizes of 401 bp amplicons for vacA m1 and 476 bp for vacA m2. PCR was performed under the following conditions: 2 min of pre-incubation at 95 °C, followed by 40 cycles of 1 min at 95 °C, 1 min at 52 °C, a min at 72 °C, and subsequently, a final extension of 72 °C for 5 min. In each experiment, H. pylori reference strain Tx30a (ATCC 51932) was included as a negative control. A positive control, H. pylori reference strain 700392 (ATCC 26695), along with inhibition controls for each sample, were used to rule out false negative results[16].
PCR amplification was performed in separate reactions with the primers, 1281 (5’-AACGCGCAAC-3’) and 1254 (5’-CCGCAGCCAA-3’)[17], under the following amplification conditions: two cycles of denaturation at 94 °C for 5 min; amplification at 36 °C for 1.30 min and extension at 72 °C for 5 min; 40 cycles of denaturation at 94 °C for 1 min; amplification at 36 °C for 1.30 min; extension at 72 °C for 2 min and a final cycle of extension at 72 °C for 10 min. The components of the PCR were the buffer (10 mmol/L Tris pH = 9.0, 50 mmol/L KCl, Triton 100 × 0.1%); MgCl2 3 mmol/L 1 × (Promega®); dNTPs 0.2 mmol/L (5’-deoxyribonucleoside triphosphates - dATP, dCTP, dGTP and dTTP - Promega®); 25 pmol of the primer 1281 and 25 pmol of the primer 1254; 1 μL of genomic bacterial DNA; 0.2 U of enzyme GoTaq DNA polymerase (Promega®) and milliQ water until completing at 12.5 mL. A reaction without DNA was used as a negative control. The amplification was performed in a thermocycler (Swift MiniProTM, Esco).
All the amplicons obtained in each of the previously described PCR reactions were run on agarose gel (SeaKim, FMC Bioloabs) at 2%, stained with ethidium bromide (Invitrogen, Carlsbad, CA, United States) at 0.5 μg/mL, in an electrophoresis chamber (Fotodyne Inc., Hartland, WI, United States). This process was performed by an EC-105 Compact Power Supply (Thermo Fisher Scientific Inc., Asheville, NC, United States) at 75 V for 40 min.
The images obtained through the amplification by RAPD-PCR were analyzed by Gel-Pro Analyzer 4.5 for Windows (Media Cybernetics Inc.). A Binary data matrix was constructed based on the presence (1) and absence (0) of bands observed on electrophoresis gel. Relations between isolates were established by cluster analysis and multiple correspondence analysis (MCA), whereby similarities between individuals were evaluated in terms of their molecular profiles, in which the distance between each pair of individuals is proportional to its molecular differences. The estimation of distances between each pair or group of isolates of H. pylori was calculated with the similarity of Neid, 1973. H. pylori isolates were classified as virulent when cagA (+) gene amplification was detected. Low virulence was established when there was an absence of cag marker by the amplification of empty site cagA. To identify multiple colonization in relation to virulence of H. pylori isolates, patients were sub-grouped according to the following characteristics: gender, age, type of gastritis and cancer risk. Univariate and bivariate analyses were performed using the χ2 test. Subsequently, the risk of presenting multiple colonization was then evaluated according to the characteristics of the populations through Odds Ratio with a 95%CI. Statistical analysis was performed using the SAS statistical package, version 9.0. The statistical significance was accepted with a P value ≤ 0.05.
The prevalence of the infection by H. pylori diagnosed by histopathology was higher in the population of low risk of GC (Tumaco, 88.7%) than in the population of high risk of GC (Túquerres, 86.4%). However, the bacteria was isolated from three anatomical places of the stomach (antrum greater curvature, antrum lesser curvature and body greater curvature) in 41 (19.7%) of the infected patients in Tumaco. Through histological tests it was possible to determine that from 206 patients in the population of Túquerres, 178 (86.4%) were positive for H. pylori; of which 165 (80.1%) were positive for isolation of H. pylori and 31 (15.04%) with positive isolation in the three anatomical places of the gastric mucosa described previously. From the 41 positive patients for isolation of H. pylori of Tumaco in the three anatomical regions of the gastric mucosa, (70.8%) were women, with ages between 19 and 68 years old; (90.24%) had non atrophic gastritis and (9.75%) atrophic gastritis. From the 31 positive patients for isolation of H. pylori in the three anatomical regions of the gastric mucosa of the population of Túquerres, (54.8%) were women, with ages between 19 and 68 years old; (93.5%) had non atrophic gastritis and (6.5%) had atrophic gastritis.
In the population of Tumaco it was found that the prevalence of the virulence marker cagA (+) of H. pylori, in the lesser curvature of the gastric antrum was (85.4%); (100%) in the greater gastric antrum curvature and (100%) in the greater curvature of body, prevalence significantly higher than the marker cagA (-) (P = 0.002) (Table 1). In the population of Túquerres the prevalence of the virulence gene cagA (+) of H. pylori on the lesser curvature of the gastric antrum was 96.4%; 100% in the greater curvature of the antrum and 100% in the greater curvature of body, prevalence significantly higher than the marker cagA (-) (P = 0.036) (Table 1).
| Alleles | Tumaco n = 41 | P value | Túquerres n = 31 | P value | ||||
| Antrum | Body | Antrum | Body | |||||
| Lesser curvature | Greater curvature | Greater curvature | Lesser curvature | Greater curvature | Greater curvature | |||
| cagA | ||||||||
| cagA (+) | 85.4% | 100% | 100% | 0.002 | 96.4% | 100% | 100% | 0.36 |
| cagA (-) | 14.6% | 0% | 0% | 0% | 0% | 0% | ||
| vacA | ||||||||
| m1 | 73.2% | 78.1% | 70.7% | 0.360 | 71.0% | 61.3% | 58.1% | 0.26 |
| m2 | 26.8% | 21.9% | 24.3% | 12.9% | 22.6% | 35.5% | ||
| vacA | ||||||||
| s1 | 80.5% | 78.5% | 80.5% | 0.730 | 74.2% | 80.7% | 64.5% | 0.44 |
| s2 | 19.5% | 19.5% | 19.5% | 19.4% | 19.4% | 25.8% | ||
In the population of Tumaco the prevalence of the allele vacA m1 of H. pylori, in the lesser curvature of the gastric antrum was 73.2%; 78.1% in the greater curvature of the antrum and 70.7% in the greater curvature of body, prevalence higher than the marker vacA m2, without being significant (P = 0.36) (Table 1). The allele s1 from the gene vacA of H. pylori was more frequent than the allele s2. 80.5% in the lesser curvature of the gastric antrum, 78.5% in the greater curvature of the antrum and 80.5% in the greater curvature of body, without the differences being significant (P = 0.73) (Table 1).
In the population of Túquerres the prevalence of the allele m1 of H. pylori, in the lesser curvature of the gastric antrum was 71%; 61.3% in the greater curvature of the antrum and 58.1% in the greater curvature of body, prevalence higher than the marker vacA m2, without being significant (P = 0.257). The allele s1 from the gene vacA was more frequent than the allele s2, 74.2% in the lesser curvature of the gastric antrum, 80.7% in the greater curvature of the antrum and 64.5% in the greater curvature of body, without being significant (P = 0.44) (Table 1).
The study included DNA of H. pylori from 72 patients, 41 from Tumaco and 31 from Túquerres. Through bivariate analysis it was found that the infection with multiple strains wasn’t significant with respect to the age (P = 0.063), being more frequent in patients from 18 to 35 years and 36 to 47 years (36.8%), than in patients between 48 and 68 years. It was observed that the infection with strains of multiple genotype was higher in women than in men (52.6% and 47.4%) respectively, without being this difference statistically significant (P = 0.067). Significant differences were observed of multiple colonization according the risk population of GC, being higher in the population of Túquerres (55.3%) than in the Tumaco (44.7%) (P = 0.027). It was found that the infection with multiple strains wasn’t significant with respect to the type of gastric lesion (P = 0.203) (Table 2).
| Features | Type of colonization | Total n = 72 | P value | |
| Single | Multiple | |||
| n = 34 | n = 38 | |||
| Gender | 0.067 | |||
| Male | 26.5% | 47.4% | 37.5% | |
| Female | 73.5% | 52.6% | 62.5% | |
| Age (yr) | 0.633 | |||
| 18-35 | 41.2% | 36.8% | 38.9% | |
| 36-47 | 26.5% | 36.8% | 31.9% | |
| 48-63 | 32.4% | 26.3% | 29.2% | |
| Cancer risk | 0.027 | |||
| Low (Tumaco) | 70.6% | 44.7% | 56.9% | |
| High (Túquerres) | 29.4% | 55.3% | 43.1% | |
| Type of gastritis | 0.203 | |||
| Non atrophic | 79.4% | 89.5% | 84.7% | |
| MAG | 20.6% | 7.9% | 13.9% | |
The multivariate analysis allowed to determine there aren’t significant differences in the risk of presenting colonization with multiple strains according the population characteristics, OR 0.13. It was observed that the risk of colonization by multiple strains of H. pylori is (2477) higher in men than in women and (1.4) times higher in patients in the age range between 36 and 47 years, than in patients between the two intervals studied. Depending the place of origin the risk of multiple colonization also increases, being (2721) times higher in the population of Túquerres (high risk) than in Tumaco (P = 0.056), while the risk of multiple colonization decreases with respect to the type of gastritis, being lower in atrophic gastritis (Table 3).
| Characteristics | Odds ratio | 95%CI | P > z | |
| Gender | ||||
| Female | 1.000 | |||
| Male | 2.477 | 0.858 | 7.154 | 0.094 |
| Age (yr) | ||||
| 18-35 | 1.000 | |||
| 36-47 | 1.401 | 0.412 | 4.769 | 0.589 |
| 48-63 | 1.160 | 0.330 | 4.076 | 0.816 |
| Cancer risk | ||||
| Low (Tumaco) | 1.000 | |||
| High (Túquerres) | 2.721 | 0.973 | 7.609 | 0.056 |
| Type of gastritis | ||||
| Non atrophic | 1.000 | |||
| MAG | 0.325 | 0.066 | 1.594 | 0.166 |
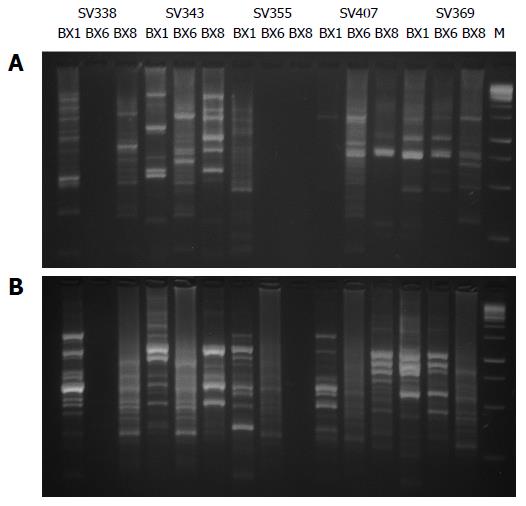
In the 123 isolates from Tumaco and 93 isolates from Túquerres the amplification by PCR-RAPD gave as result well defined DNA bands with each of the primers, 1281, 1254. Isolates with slight variation were found in the banding patterns, which varied between 100 pb and 1000 pb (Figure 1).
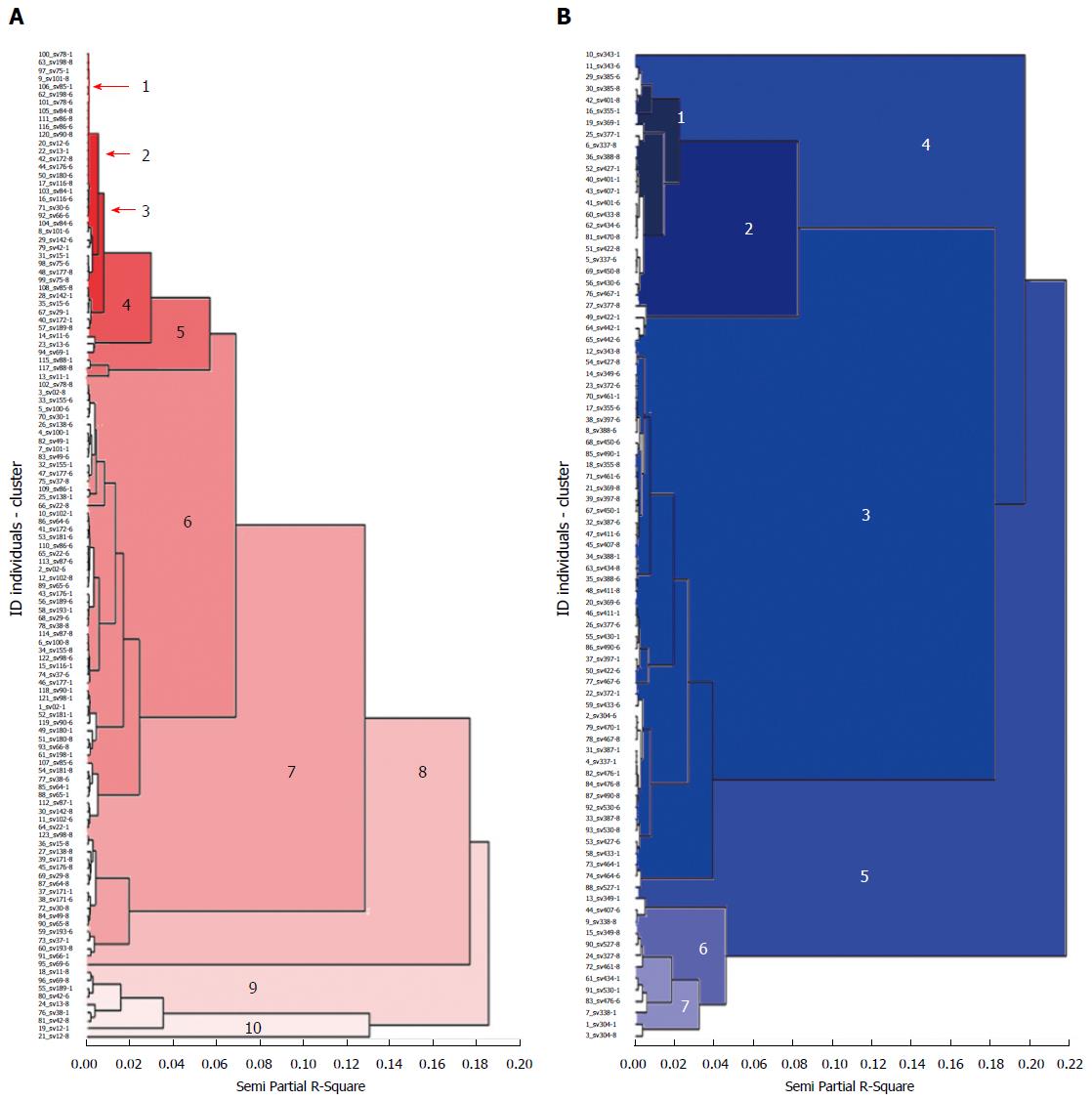
The analysis of the dendogram executed with the UPGMA classification method and the coefficient of similarity of Neid, showed that there is no relation between the groups formed and the characteristics of the population as gender, age and type of gastritis at a level of similarity of 0.07. In the population of Tumaco the dendogram was formed with ten groups (Figure 2). Group one was formed by 35 isolates, from these 33 were from patients with chronic non atrophic gastritis and two patients with chronic atrophic gastritis. Group two was structured with 47 isolates among which, four were from patients with chronic atrophic gastritis and 43 from patients with chronic non atrophic gastritis; group four was formed by 16 isolates, among which one was from a patient with chronic atrophic gastritis and the rest were from patients with non-atrophic gastritis, group five was formed by seven isolates of patients with non-atrophic gastritis; group six was formed by three isolates of patients with atrophic gastritis, group seven was also formed with three isolates from which two were from patients with non-atrophic gastritis and one from a patient with atrophic gastritis, the groups 8, 9, 10 were formed by one isolate of a patient with non-atrophic gastritis (Figure 2).
Through multiple correspondence multivariate analysis (MCA), the three dimensional representation of the group of the isolates of the population of Tumaco, showed that there is no relation between the groups and the characteristics of the population, but a low variation is shown between the isolates of Tumaco by the formation of a relatively homogeneous group.
In the population of Túquerres the dendogram was formed by seven groups. In these there was no relation between the segregation of the isolates in the groups and the characteristics of the population at a similarity level of 0.13.
Group one was formed by 30 isolates, from which 27 were from patients with chronic non-atrophic gastritis and three with chronic atrophic gastritis. Group two joined 16 isolates, among which was found one with chronic atrophic gastritis and 15 with chronic non atrophic gastritis; group three was formed by 13 isolates of patients with chronic non atrophic gastritis and two of patients with chronic atrophic gastritis; group four, with three isolates of patients with chronic non atrophic gastritis; in group five with five isolates, from which one was from a patient with chronic atrophic gastritis; groups six and seven were formed by three and two isolates of patients with chronic non-atrophic gastritis respectively (Figure 2). Through the MCA, it is confirmed that there is no relation between the groups and the characteristics of the population, but a higher variation is showed between the isolates of Túquerres by the formation of a less homogeneous group (Figure 2).
The prevalence of the infection of H. pylori was high and similar in both populations: (88.7%) in Tumaco and (86.4%) in Túquerres[5]. However, there are strong differences in the incidence rates of gastric cancer among these populations, separated by only 200 km, being up to 25 times higher in the population of Túquerres in the Colombian Andes with predominant ancestry Amerindian and European, than in the population of Tumaco, with African ancestors and coming from the pacific coast in Colombia[18].
It was found that the multiple colonization evaluated with respect to genes of virulence of H. pylori, was 52.8% of all the analyzed cases (Table 2), and significantly higher in the population of Túquerres (55.3%), than in Tumaco (44.7%) (P = 0.027) (Table 2). Similar findings, of multiple colonization are described in different geographical regions of the world, including: Korea (60%), México (65%), Chile (32%), Portugal (30%) and Brazil (15%)[18,19].
The genetic similarity was higher between the isolates of H. pylori from the patients of Tumaco than those from Túquerres. Additional to this finding, the MCA showed in the population of Túquerres higher diversity with the formation of a less homogeneous group (Figure 2). People living on the Nariño Pacific Coast (Tumaco), have African ancestors and are infected with H. pylori strains, from African origin that, presumably they acquired when their ancestors brought to the new world during the slave trade and that they have coevolved with his host towards commensalism and in the time adapted, and in consequence are less virulent[18]. In contrast, in Túquerres (Colombian Andes), the phylogenetic origin of H. pylori that colonize the people of the region is predominantly European, and possibly by selective Amerindian strains tend to disappear gradually over time[5]. The disruption of host-bacteria coevolution may favor multiple colonization and determine less favorable biological relations for the host[5].
The H. pylori strains are genetically heterogeneous, with deep variations between patients and in the same patient[20]. An individual may be colonized with one strain or with genetically predominant strains with a same genomic DNA, or by different strains that show high genomic diversity, evident facts in the DNA sequences found in different anatomical places of patients[21]. This genomic diversity reflects a process of persistent accumulation of mutations in the strains as a result of horizontal transfer and DNA incorporation by genetic transformation or spontaneous mutations. The persistent accumulation of mutations, principally in the virulence genes, provides a set of variants to H. pylori that can be selected to colonize particular gastric niches[11]. When all the individuals in a microbial population are identical, the concept of establishment in the host is relatively simple. However, the genetic variability of the microorganism that colonizes different regions in the gastric mucosa of a same host carries a more complex relationship[22].
The host responses to genomic variations of the bacteria leads immune local mechanisms for the anatomical place of the gastric mucosa where the microorganism colonizes. These variations in the local response towards the microorganisms represents an environmental pressure for the strains of H. pylori, living in the gastric mucosa where the microorganism colonizes. These variations in the local response towards the microorganisms represent an environmental pressure for the strains of H. pylori, living in the gastric mucosa. Such selective pressures will affect the relative proportions of the bacterial strains in the population or may involve changes in the alleles of important genes and cause the displacement of the balance point, that favors the percentage of new genetic variants of the bacteria. The continuous rate of mutation assures that the dynamic equilibrium of the strains of H. pylori is maintained present in the population[23].
A more complex condition occurs when two or more strains of H. pylori colonizes one same gastric mucosa, since there is a space to promote genetic recombination which gives rise to new more resistant genotypes to the environmental conditions and they compete with their precursor strains. Each bacterial cell may create independent signals in the host and trigger immune specific answers. However, a strong localized response of the host may affect all the bacterial cells from the environment. The selective pressure results as negative influence in the development of most of the autochthonous bacterial cells, but the new recombinants survive because they may selectively adapt to hostile environments, they can form population structures more organized and optimize the use of resources from the host[24].
The results from the genotype comparison by RAPD-PCR between isolates collected from different patients and the same patient in different anatomical places of the gastric mucosa, is a secure method for the study of the genomic variability of H. pylori.
In this investigation the stage of genomic diversity between the clinical isolates obtained with the primers 1281 and 1254, based in a clear distinction of the patterns with multiple differences in the bands, reveal that 4 of 41 patients shelter one unique strain of H. pylori, while 37 showed heterogeneous traces of DNA that suggests infection by multiple strains in different anatomical places of the gastric mucosa in the same patient in the population of Tumaco (Figure 2). In the population of Túquerres 30 patients shelter different strains between the regions of the gastric mucosa of the same patient (Figure 2). Cellini et al[25], reported similar results in a population with different strains of H. pylori that showed genotypic variation, in the same patient. The genetic diversity of the H. pylori may also be due to multiple genotypes worldwide. A great number of variants H. pylori coexist as product of natural transformation and re genetic arrangements or alterations during the adaptation process and colonization in the host[26]. Nevertheless, the evidence of the high variety of strains of H. pylori may be the result of a continuous evolution, which occurs when the stomach of a person gets infected, since nucleotide mutation may occur, excisions in the isle cag PAI, transposition and insertion of elements cag PAI, recombination with DNA of surrounding strains that aren’t involved in the disease and horizontal transmission of new genes[26]. It is proposed that the high diversity is due to the limited direct competition between most of the strains, even if they are residents in different people in the same community. The diversity emphasizes by the geographical breakup of many towns of the world and in consequence the phyllogeographic origin of H. pylori plays a preponderant role in the high diversity and in the pathogenicity of the bacteria[27,28]. The divergence between strains of H. pylori increases even more, by differences between the characteristics of the people and the individual features of the strains[28]. Some features include types of Lewis antigens for the adhesion of H. pylori, the specificity and intensity of the inflammatory response and the complex regulation of gastric acid secretion.
In conclusion, the population of Túquerres located in the Colombian Andes (high risk of gastric cancer), are more likely to have a multiple colonization of H. pylori, with greater genetic diversity than the infectious strains from the patients of Tumaco (low risk of GC), which should be taken into account when formulating eradication programs of the microorganisms as a strategy for the primary prevention of gastric cancer in these populations of Colombia.
This work was made possible thanks to the support of the Registro Poblacional de Cáncer de Cali Research group (RPCC), Universidad del Valle.
Infection by virulent strains of Helicobacter pylori (H. pylori) is one of the most important risk factors for the development of precursor lesions of gastric cancer. Ideal treatment is not yet possible because the gastric mucosa of a host can be colonized by multiple strains of H. pylori (virulent and avirulent). The strains of H. pylori can coexist with susceptible bacterial isolates and resist antibiotics. This phenomenon makes eradication of the H. pylori difficult as a strategy in treatment regimens and prevention of gastric cancer precursor lesions.
The eradication of H. pylori from the gastric mucosa is the cornerstone in the treatment of diseases such as chronic gastritis, peptic ulcer, metaplasia and dysplasia; it is necessary to have genotyping isolates of H. pylori obtained from different gastric locations (antrum and body) to evaluate multiple strain colonization of H. pylori by analysis of genetic diversity in patients with gastritis.
In this research, the degree of variability of genomic DNA from clinical isolates, obtained with primers 1281 and 1254, based on a clear distinction of patterns with multiple differences in bands, shows that 4 out of 41 patients harbor a single strain of H. pylori, while 37 showed heterogeneous DNA traces that indicate infection by multiple strains in different anatomical sites in the same patient in the population of Tumaco. In the population of Túquerres, 30 patients harbor different strains within their gastric mucosa. While the difference in risk is associated with the virulence of the infecting strain, it is considered that the multiple colonization of gastric mucosa and its interrelation with environmental agents is a condition that could boost the development of precancerous lesions and eventually lead to the development of gastric carcinoma. The eradication of H. pylori is a valid strategy for the prevention of gastric carcinoma. However, treatment failure is inherent to multiple colonization by multiple strains of H. pylori (virulent and avirulent) and the coexistence of susceptible bacteria with bacteria that are resistant to antibiotics.
These in vitro findings are especially important because of their possible implications for the treatment and evolution of gastro-duodenal diseases caused by H. pylori, suggesting that in most of the cases, polycolonization by virulent strains of H. pylori is one of the most important risk factors for the development of precursor lesions of GC and gastric carcinoma. Our results suggest differences in genetic characteristics of the circulating strains, with different degrees of greater genetic diversity between anatomical regions in the same patient, showing multiple colonization. Eradication of H. pylori from the gastric mucosa is the only valid strategy for GC prevention. However, therapeutic failure of polymicrobial treatment is inherent and can be attributed to the colonization of the gastric mucosa by multiple strains of H. pylori (virulent and avirulent).
The phenomenon known as multiple colonization refers to the colonization of the gastric mucosa by multiple strains of H. pylori with different characteristics at the genetic level with varying degrees of pathogenicity (virulent and avirulent) between anatomical regions in a single patient. In Colombia, the risk of gastric cancer varies according to geographical areas with high and low risk regions for developing GC. This contrast of risk between nearby towns with a prevalence of similar infection by H. pylori is explained by the high genetic diversity of bacteria, with varying levels of pathogenicity.
Personalized medicine has important role in treatment. Authors should list comparison of the genomic variability of H. pylori isolates of gastric regions from Columbian population.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Colombia
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Safaei HG, Yamaoka Y S- Editor: Qi Y L- Editor: Filipodia E- Editor: Wang CH
| 1. | Taylor NS, Fox JG, Akopyants NS, Berg DE, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter FM. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918-923. [PubMed] |
| 2. | Norazah A, Rasinah WZ, Zaili Z, Aminuddin A, Ramelah M. Analysis of PCR-RAPD DNA and antibiotic susceptibility profiles of antrum and corpus isolates of Helicobacter pylori from Malaysian patients. Malays J Pathol. 2009;31:29-34. [PubMed] |
| 3. | Kim JW, Kim JG, Chae SL, Cha YJ, Park SM. High prevalence of multiple strain colonization of Helicobacter pylori in Korean patients: DNA diversity among clinical isolates from the gastric corpus, antrum and duodenum. Korean J Intern Med. 2004;19:1-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Figueroa M, Cortés A, Pazos Á, Bravo LE. Antimicrobial susceptibility of Helicobacter pylori with chronic gastritis. Biomedica. 2012;32:32-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 5. | Pazos A. Ancestros filogeográficos, susceptibilidad antimicrobial y marcadores de virulencia de. Helicobacter pylori en pacientes con gastritis crónica. Universidad del Valle. 2012; Available from: http://bibliotecadigital.univalle.edu.co/bitstream/10893/7793/1/CB-0516176.pdf. |
| 6. | Berg DE, Akopyants NS, Kersulyte D. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol. Cell Biol. 1994;5:13-24 Available from: https://www.researchgate.net/publication/279554115. |
| 7. | Foxall PA, Hu LT, Mobley HL. Use of polymerase chain reaction-amplified Helicobacter pylori urease structural genes for differentiation of isolates. J Clin Microbiol. 1992;30:739-741. [PubMed] |
| 8. | Carroll IM, Ahmed N, Beesley SM, Khan AA, Ghousunnissa S, Moráin CA, Habibullah CM, Smyth CJ. Microevolution between paired antral and paired antrum and corpus Helicobacter pylori isolates recovered from individual patients. J Med Microbiol. 2004;53:669-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Han FC, Ng HC, Ho B. Stability of randomly amplified polymorphic DNA fingerprinting in genotyping clinical isolates of Helicobacter pylori. World J Gastroenterol. 2003;9:2021-2024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 10. | Tomasini ML, Zanussi S, Sozzi M, Tedeschi R, Basaglia G, De Paoli P. Heterogeneity of cag genotypes in Helicobacter pylori isolates from human biopsy specimens. J Clin Microbiol. 2003;41:976-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Atherton JC, Cao P, Peek RM, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771-17777. [PubMed] |
| 12. | van der Ende A, van Doorn LJ, Rooijakkers S, Feller M, Tytgat GN, Dankert J. Clarithromycin-susceptible and -resistant Helicobacter pylori isolates with identical randomly amplified polymorphic DNA-PCR genotypes cultured from single gastric biopsy specimens prior to antibiotic therapy. J Clin Microbiol. 2001;39:2648-2651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161-1181. [PubMed] |
| 14. | Pruszkowski J, Ziółkowski G, Gonciarz Z, Besser P. [Isolation of Helicobacter pylori from gastric mucosa depending on the growth medium used]. Med Dosw Mikrobiol. 1994;46:305-311. [PubMed] |
| 15. | Ho SA, Hoyle JA, Lewis FA, Secker AD, Cross D, Mapstone NP, Dixon MF, Wyatt JI, Tompkins DS, Taylor GR. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J Clin Microbiol. 1991;29:2543-2549. [PubMed] |
| 16. | Lopes AI, Vale FF, Oleastro M. Helicobacter pylori infection - recent developments in diagnosis. World J Gastroenterol. 2014;20:9299-9313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 51] [Reference Citation Analysis (0)] |
| 17. | Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137-5142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 564] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 18. | Kodaman N, Pazos A, Schneider BG, Piazuelo MB, Mera R, Sobota RS, Sicinschi LA, Shaffer CL, Romero-Gallo J, de Sablet T. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci USA. 2014;111:1455-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 178] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 19. | Figueiredo C, Van Doorn LJ, Nogueira C, Soares JM, Pinho C, Figueira P, Quint WG, Carneiro F. Helicobacter pylori genotypes are associated with clinical outcome in Portuguese patients and show a high prevalence of infections with multiple strains. Scand J Gastroenterol. 2001;36:128-135. [PubMed] |
| 20. | Go MF, Chan KY, Versalovic J, Koeuth T, Graham DY, Lupski JR. Cluster analysis of Helicobacter pylori genomic DNA fingerprints suggests gastroduodenal disease-specific associations. Scand J Gastroenterol. 1995;30:640-646. [PubMed] |
| 21. | Makristathis A, Barousch W, Pasching E, Binder C, Kuderna C, Apfalter P, Rotter ML, Hirschl AM. Two enzyme immunoassays and PCR for detection of Helicobacter pylori in stool specimens from pediatric patients before and after eradication therapy. J Clin Microbiol. 2000;38:3710-3714. [PubMed] |
| 22. | Kienesberger S, Cox LM, Livanos A, Zhang XS, Chung J, Perez-Perez GI, Gorkiewicz G, Zechner EL, Blaser MJ. Gastric Helicobacter pylori Infection Affects Local and Distant Microbial Populations and Host Responses. Cell Rep. 2016;14:1395-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 118] [Article Influence: 13.1] [Reference Citation Analysis (1)] |
| 23. | Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 24. | Bauer B, Meyer TF. The human gastric pathogen Helicobacter pylori and its association with gastric cancer and ulcer disease. Ulcers. 2011;11:1-23. |
| 25. | Cellini L, Grande R, Di Campli E, Di Bartolomeo S, Capodicasa S, Marzio L. Analysis of genetic variability, antimicrobial susceptibility and virulence markers in Helicobacter pylori identified in Central Italy. Scand J Gastroenterol. 2006;41:280-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Achtman M, Azuma T, Berg DE, Ito Y, Morelli G, Pan ZJ, Suerbaum S, Thompson SA, van der Ende A, van Doorn LJ. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459-470. [PubMed] |
| 27. | Kersulyte D, Mukhopadhyay AK, Velapatiño B, Su W, Pan Z, Garcia C, Hernandez V, Valdez Y, Mistry RS, Gilman RH. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210-3218. [PubMed] |
| 28. | Dubois A, Berg DE, Incecik ET, Fiala N, Heman-Ackah LM, Del Valle J, Yang M, Wirth HP, Perez-Perez GI, Blaser MJ. Host specificity of Helicobacter pylori strains and host responses in experimentally challenged nonhuman primates. Gastroenterology. 1999;116:90-96. [PubMed] |