Published online Feb 14, 2016. doi: 10.3748/wjg.v22.i6.1953
Peer-review started: August 26, 2015
First decision: September 29, 2015
Revised: October 19, 2015
Accepted: December 19, 2015
Article in press: December 21, 2015
Published online: February 14, 2016
Processing time: 150 Days and 19.3 Hours
Hepatitis C virus (HCV) infects over 150 million people worldwide. In most cases, HCV infection becomes chronic causing liver disease ranging from fibrosis to cirrhosis and hepatocellular carcinoma. Viral persistence and pathogenesis are due to the ability of HCV to deregulate specific host processes, mainly lipid metabolism and innate immunity. In particular, HCV exploits the lipoprotein machineries for almost all steps of its life cycle. The aim of this review is to summarize current knowledge concerning the interplay between HCV and lipoprotein metabolism. We discuss the role played by members of lipoproteins in HCV entry, replication and virion production.
Core tip: The aim of the review is to summarize current knowledge concerning the interplay between hepatitis C virus (HCV) and lipoprotein metabolism. In particular, the manuscript discusses the role played by members of lipoproteins family in all steps of HCV life cycle.
- Citation: Grassi G, Di Caprio G, Fimia GM, Ippolito G, Tripodi M, Alonzi T. Hepatitis C virus relies on lipoproteins for its life cycle. World J Gastroenterol 2016; 22(6): 1953-1965
- URL: https://www.wjgnet.com/1007-9327/full/v22/i6/1953.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i6.1953
Hepatitis C virus (HCV) infection is one of the main causes of chronic liver disease worldwide. It has been estimated that 130-170 million people are chronically infected with HCV[1-3] with a prevalence, in selected countries, ranging from 0.4% to 12.3%[4].
Acute infection is spontaneously cleared only in 15%-30% of individuals and the majority of patients develop chronic infection. HCV infection is generally a slowly progressive disease characterized by different liver damages that can progress to life-threatening diseases, such as cirrhosis and hepatocellular carcinoma[5,6].
The recent advent of highly potent direct-acting antiviral drugs (DAAs), employed in interferon-containing and interferon-free combinations, has led to virus elimination in more than 90% of treated patients[7]. However, it is yet unclear whether, and how, the virus-induced liver damages are reversible; therefore, it is important to fully elucidate the mechanism of HCV-induced pathogenesis.
HCV does not cause a direct cytopathic effect on host cells and most of the related liver dysfunctions are likely due to the virus-mediated alteration of host processes such as immune responses and several metabolic pathways[8-10]. In particular, HCV interferes with the host lipid metabolism and cholesterol homeostasis. Several lipids abnormalities have been associated to HCV chronic infection, such as liver steatosis, particularly evident in patients infected with the genotype 3 of the virus, hypobetalipoproteinemia and hypocholesterolemia[11].
The relationship existing between the virus and the lipid metabolism is very intimate, every step of the viral life cycle relies at least on one member involved in lipid pathways[12,13].
HCV is an enveloped positive-strand RNA virus, a member of the genus Hepacivirus within the family of Flaviviridae. HCV enters the cell by receptor-mediated endocytosis involving multiple cell surface molecules (as recently reviewed by Ding and coauthors[14]). After pH-dependent fusion and uncoating, the 9.6 kb single-stranded RNA genome is translated at the rough endoplasmic reticulum (ER). The resulting polyprotein precursor is processed by cellular and viral proteases into ten mature proteins; core and envelope glycoproteins E1 and E2 are the main constituents of the virus particle, p7 and nonstructural protein (NS) 2 participate in virus assembly, while NS3, NS4A, NS4B, NS5A, and NS5B are sufficient for viral RNA replication and are involved in virus assembly[15]. Replication takes place in ER-derived membrane spherules called membranous web, which formation and architecture remain to be fully elucidated. Progeny RNA is then packaged into virus particles that exit the cell via the secretory pathway[16].
Lipoproteins are responsible for lipids packaging and transport through the bloodstream and for their delivery to target tissues. The transported lipids, which are the core of the lipoproteins, are cholesteryl esters (CE) and triglycerides (TG), derived either from the diet or from liver neo-synthesis. They are enveloped by a layer of phospholipids, free cholesterol and proteins (mainly apolipoproteins), which control lipoproteins assembly, transport and metabolism by mediating interactions with receptors, enzymes and lipid transport proteins[17,18]. Lipoproteins vary in the content of lipids and proteins. Their classification and isolation procedures are commonly based on their density, which reflect their different content of CE, TG, free cholesterol and apolipoproteins. The main lipoproteins particles generated by the liver are the very low-density lipoproteins (VLDL), intermediate-density lipoproteins (IDL), low-density lipoproteins (LDL), in which apolipoprotein(apo)B-100 (apoB) is the main structural component, and high-density lipoproteins (HDL), where apoA-I is the main structural component[17,18].
In the endogenous transport pathway, the liver releases TG and CE in the circulation mainly through the generation of VLDL particles[19]. Mobilized lipid storage pool in the liver, as well as de novo synthesis of fatty acids and phospholipids, contribute to hepatic VLDL synthesis. Lipoprotein lipase (LPL) hydrolyses the TG present in the core of circulating VLDL, releasing free-fatty acids (FFA) to the target tissues. A large proportion of the resulting particles (IDL) is efficiently removed from plasma by the hepatocytes through the LDL receptor (LDLR). The remaining part is converted to LDL by a reaction catalyzed by hepatic lipase, which further reduces the amount of TG. Once formed, CE-rich LDL delivers cholesterol to peripheral tissues where are taken up by the LDLR and internalized via a clathrin-dependent pathway. Following endocytosis, LDL are degraded into lysosomes and free cholesterol is released and either accumulated within the cells or incorporated into new lipoproteins.
In the reverse transport system, HDL carry the excess of cholesterol from extrahepatic cells to the liver. HDL biosynthesis and maturation are complex multistep processes that involve the secretion of proteins-rich and lipid-poor particles (nascent HDL) and a massive extracellular lipid acquisition of mainly phospholipids and cholesterol. The major lipid components of HDL are CE and phospholipids, while apoA-I and apoA-II are the main apolipoproteins required for normal HDL biosynthesis[17,18].
Several steps of HCV life cycle are strictly linked to host lipoprotein metabolism, the aim of this review is to describe this close relationship (summarized in Table 1).
| Name | Lipoprotein association | Main function | HCV life cycle step | Function | Ref. |
| GAGs | Lipoproteins adsorption | Entry | LVP adsorption | [33-36,40] | |
| LDLR | apoE- and apoB-containing lipoproteins receptor | Entry | LVP binding | [37-44] | |
| RNA replication | Cellular distribution of lipids | [42] | |||
| ApoB | VLDL and LDL | Structural protein of VLDL/LDL | Virion production | ? | [102-105,110] |
| ApoE | VLDL, LDL and HDL | Ligand of LDL receptor | Entry | GAGs/LDLR binding | [35,36,51] |
| Virion production | ? | [106-111] | |||
| ApoA-I | HDL >> VLDL | Structural protein of HDL | RNA replication | ? | [82,83] |
| Virion production | ? | [82,110,111] | |||
| ApoH | VLDL >> HDL | Cholesterol efflux | RNA replication | ? | [85] |
| ApoA-II | HDL >> VLDL | Structural protein of HDL | Virion production | ? | [110,111] |
| ApoC-I | VLDL, LDL, HDL | Inhibitor of CETP activity | Entry | Viral and cellular membranes fusion | [58,59] |
| Virion production | ? | [110,111] | |||
| ApoC-II | VLDL, LDL, HDL | Activator of LPL | Virion production | ? | [110,111] |
| ApoC-III | VLDL, LDL, HDL | Inhibitor of LPL and HL | Virion production | ? | [110,111] |
| ACSL3 | Phosphatidylcholine synthesis for apoB | Virion production | ? | [105] |
Viral particles purified from infected patients sera revealed that HCV has a spherical morphology of different sizes (range 40-70 nm diameter), with an enveloped membrane displaying the two surface viral glycoproteins E1 and E2[20,21]. HCV exists as a mixture of infectious and noninfectious particles, both in vivo and in vitro, and, very interestingly, virions found with a very low buoyant density (range 1.10-1.14 g/mL) displayed the highest infectivity[22-26].
The high buoyant density is mainly due to the association with apoB-containing lipoproteins (VLDL/LDL) to form the so-called lipoviral particles (LVP). Different lipoproteins components, such as cholesterol, TG, apoB, apoE, apoA-I and apoCs, have been found in the LVP of HCV infected patients[27,28]. In vitro produced HCV particles have confirmed these associations[26,29,30].
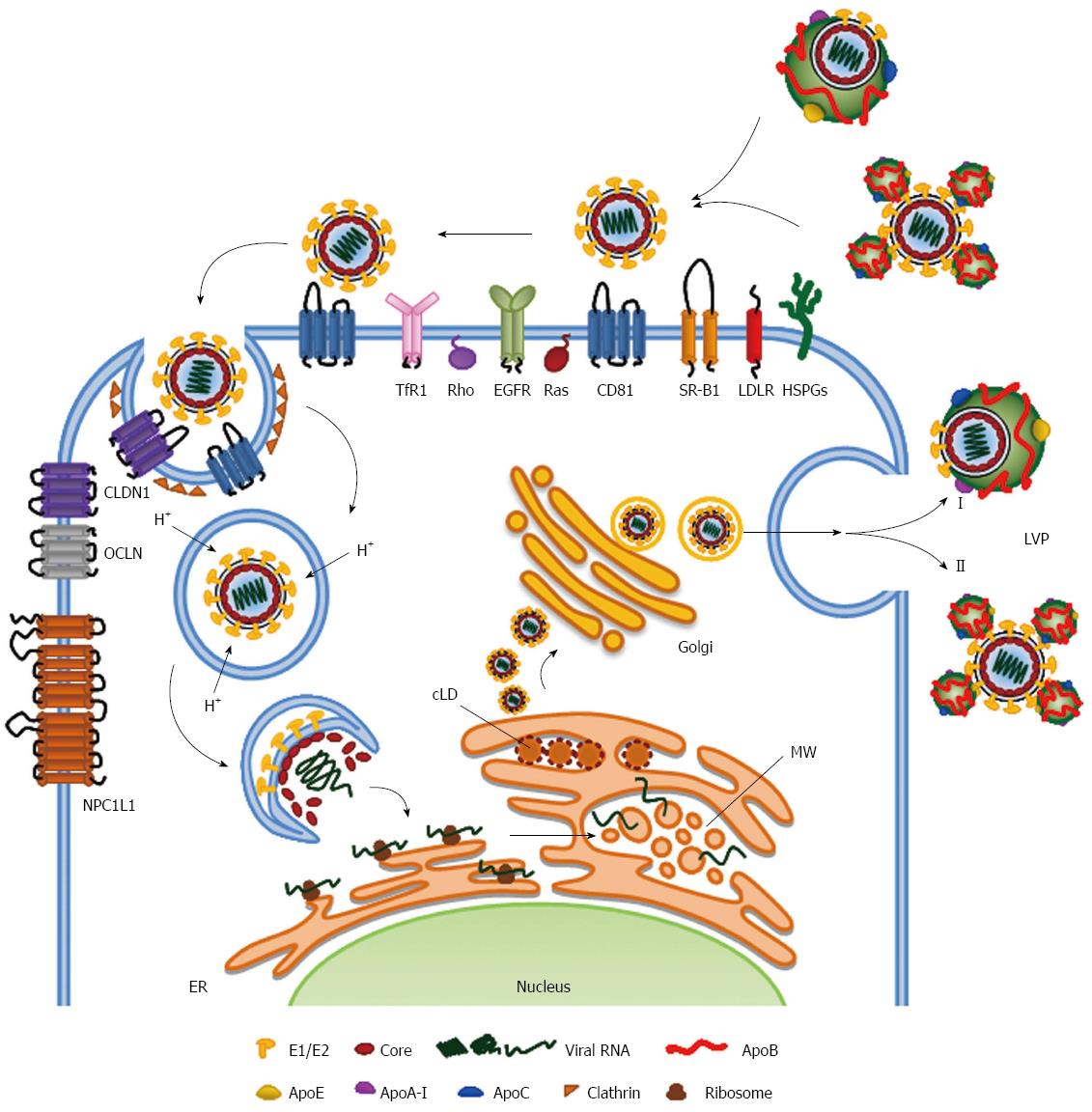
HCV LVP enter into the cells via a multi-step endocytosis that requires a growing number of receptors, co-receptors and host factors, which probably are responsible for the hepatotropism of the virus. The long list of these host proteins includes two binding factors glycosaminoglycans (GAGs) and low-density lipoprotein receptor (LDLR), four receptors CD81, scavenger receptor class B type 1 (SR-BI), claudin-1 (CLDN1), occludin (OCLN), and several co-receptors epidermal growth factor receptor (EGFR), ephrin receptor A2 (EphA2), the cholesterol transporter Niemann-Pick C1-like 1 (NPC1L1), transferrin receptor 1 (TfR1) and the cell-death-inducing DFFA-like effector b (CIDEB). For an exhaustive description of all known HCV receptors and their involvement in HCV cell entry, we refer the reader to recent reviews and references therein[14,31]. Here we will focus on molecules involved in lipoprotein metabolism.
HCV entry into the cells is a process that requires spatial and temporal control of these cellular cofactors. The putative mechanism consists of three main steps (1) viral attachment to the hepatocyte; (2) receptor-mediated endocytosis; and (3) endosomal fusion. Members of the lipoprotein metabolism seem to be involved in all these steps.
Attachment of the virus to the host cell is firstly obtained by the interactions with GAGs and LDLR present on the surface of the cells, which are known to mediate lipoprotein metabolism[32]. It has been demonstrated that HCV binds to the GAGs present on heparan sulfate (HS) proteoglycans (HSPGs) and syndecan 1 and syndecan 4 have been reported to be involved in this process[33,34]. The minimum HS oligosaccharide length required for HCV infection is a decasaccharide with the N- and 6-O-sulfation. Very interestingly, it has been reported that apoE is responsible for this process, while viral glycoproteins, although capable of binding to GAGs, are not involved[35,36].
The LDLR, which transports the cholesterol-rich LDL intracellularly via clathrin-mediated endocytosis[37], is involved in this first step of HCV entry. However, its exact role in HCV infection still remains controversial; it is not clear whether it acts as a HCV receptor or as a facilitator of initial attachment to hepatocyte surface or for other steps of the virus life cycle, such as viral replication[38-44]. It is worth to note that the internalization of infectious particles and lipoproteins mediated by LDLR displays different kinetics, thus suggesting distinct uptake steps/pathways for HCV and lipoproteins[42].
The capture of viral particles is mediated by other members of the lipoprotein metabolism. In fact, after the initial interaction with GAGs and LDLR, HCV utilizes SR-BI, a major receptor of high-density lipoproteins (HDL) that can bind also apoB-containing lipoproteins (VLDL/LDL) and oxidized forms of LDL[45,46]. SR-BI is a glycoprotein with two N- and C-terminal cytoplasmic domains separated by a large extracellular domain, which is involved in lipoprotein metabolism, mediating the uptake and the intracellular delivery of the cholesterol esters (CE). Interestingly, the SR-BI-mediated intracellular lipid transportation is different from that of the LDLR. In fact, it binds lipoproteins on the cell surface and delivers CE intracellularly without internalization of the intact lipoprotein particle (as reviewed by Shen and coauthors[47]). This could partially explained the aforementioned distinct internalization pathways described for HCV and lipoproteins[42].
The interaction between HCV and SR-BI is thought to mediate the dissociation of lipoproteins from LVP, likely through the SR-BI-mediated cholesterol transfer activity, and to induce conformational changes in the E2 glycoprotein, exposing its CD81 interaction domains[48,49]. Interestingly, it has been reported that HDL increases HCV entry into the cells by accelerating their endocytosis through SR-BI activation, with the consequence of decreasing the neutralizing effect of the anti-HCV antibodies[50]. On the other hand, apoB-containing lipoproteins competed and effectively inhibited the interaction between HCV and hepatocytes and, as reported for LDLR, the binding to SR-BI is not mediated by E2 but mainly, although not exclusively, by apoE[51].
Together, these studies suggest that the first step of HCV entry is regulated by the complex interactions occurring between lipoproteins components, lipoproteins receptors (i.e., GAGs, LDLR and SR-BI) and HCV envelope glycoproteins.
After the attachment to the cells, HCV binds its receptors, the tetraspanin CD81 and the tight junction proteins CLDN1 and OCLN, leading to cellular internalization of the virus through a clathrin-dependent endocytosis process. After the binding to HCV, CD81 moves toward the tight junctions and interacts with CLDN1. This movement depends on the activation of several transduction pathways such as EGFR, Ras and Rho GTPases, which trigger the actin-mediated lateral membrane diffusion of HCV-CD81 complexes[52-54]. It is reported that also TfR1 is engaged at a post-CD81 binding step in HCV entry and it is involved in the viral entry. Although the exact mechanism of action is unknown, the TfR1 inhibition decreases significantly the infection of HCV derived from cell culture (HCVcc) and HCV pseudoparticles (HCVpp), thus suggesting that it is involved in viral internalization. Interestingly, the cell-to-cell spread is less dependent on this molecule[55]. Similarly, OCLN is required for HCV entry and it acts after the GAGs and SR-BI post binding step and prior to endosomal acidification, thus suggesting that the tight junction region is the last to be encountered by the virion before cellular internalization[56].
The interaction of HCV-CD81 complexed with CLDN1 induces clathrin-mediated endocytosis. Following uptake, HCV co-receptor complexes are trafficked to RAB5A containing endosomes for HCV fusion[57]. Interestingly, receptor-specific antibodies and HCV particles increased CD81 and CLDN1 endocytosis, thus supporting a model wherein HCV stimulates receptor trafficking to promote viral particle internalization.
After viral internalization, the interaction between E2 and CD81 induces a yet unknown fusion mechanism between the viral glycoproteins and the endosomal membrane in a low-pH environment. Interestingly, this process is favoured by apoC-I, an exchangeable apolipoprotein that predominantly resides in HDL[58,59]. ApoC-I specifically enhances the infectivity of HCVcc and HCVpp as well as of HCV isolated from viremic chimpanzees. ApoC-I increases the infectivity via a direct interaction with the HCV glycoproteins. Interestingly, the hypervariable region 1 (HVR1), located at the N-terminus of the HCV E2 glycoprotein, is the essential viral component for the apoC-I-mediated activity[58]. ApoC-I activity does not rely on SR-BI or CD81, thus not influencing the binding or the internalization steps of HCV entry. ApoC-I instead enhances the pH-dependent fusion rates between viral and target membranes, as measured by a HCVpp/liposome fusion assay[58].
Following fusion, HCV genomic RNA is released into the cytosol, where it is directly translated to produce viral proteins and initiate viral replication.
HCV RNA replication is a multi-step process regulated by several viral and cellular proteins[60]. It is well established that the minimal viral proteins necessary and sufficient for viral replication are NS3-4A, NS4B, NS5A and NS5B, together with 5’ and 3’ untranslated regions[15]. HCV replication occurs within a dense cluster of rearranged intracellular membranes referred to as membranous web. It is composed by double-membrane vesicle structures, most likely derived from the ER[61]. This membranous matrix is the center of HCV replication. In fact, it contains all the non-structural viral proteins necessary for replication as well as the newly synthesized viral RNA[62]. It is worth to note that, although the membranous web is induced by all HCV replicase factors (NS3-5B)[61], the sole expression of NS4B and NS5A induces very similar membrane alterations[63,64]. HCV, through NS5A, hijacked the isomerase activity of cyclophilin A (CypA)[65,66] and the membrane-deforming ability of proline-serine-threonine phosphatase interacting protein 2 (PSTPIP2)[67] for remodeling intracellular membranes and, thus, enhancing HCV replication.
The replication step of HCV life cycle is highly linked to the host lipid metabolism processes. RNA replication occurs in membranes rich in cholesterol and sphingolipids, two lipids not abundant in the ER membrane[68]. Since HCV hijacks host lipid metabolism at different levels, it is likely that these lipids are transported to the membranous web, rather than that the replication complexes take place in specific area of the ER enriched in cholesterol and sphingolipids. In fact, HCV is able to alter the lipid composition of membranes affecting the subcellular localization of the lipid kinase phosphatidylinositol-4-kinase IIIα (PI4KIIIα), which leads to a different distribution of its product phosphatidylinositol-4-phosphate (PI4P) from the Golgi compartment and plasma membrane to the cytoplasm[69,70]. HCV utilizes the redistribution of PI4P to alter the lipid composition of the membranous web, attracting sphingolipids and cholesterol through the recruitment of the PI4P-interacting lipid transfer proteins four-phosphate adaptor protein 2 (FAPP2), which is also a glycosphingolipid-binding protein, and oxysterol-binding protein (OSBP), respectively[71,72].
In addition to these structural and molecular alteration of membranes, HCV also induces de novo lipid and membrane biosynthesis modulating the expression of a number of genes involved in lipid metabolism. This is likely mediated by the transcriptional activity of the sterol regulatory element-binding protein (SREBP) pathway[73,74]. In this regards, it has been well described how HCV infection is able to alter the lipidomic profile of hepatocytes[75].
While the lipid metabolism involvement in HCV replication has been well demonstrated, the role of members of the lipoprotein machineries is far to be fully proved. It has been reported that LDLR is necessary for viral RNA replication. The use of a neutralizing antibody against LDLR after HCV RNA electroporation into Huh7 cells induced a decreased RNA production. The treatment with this antibody induced a changed in the cellular lipid profile, with an increase of CE level and a change in phospholipids [i.e., increased phosphatidylethanolamine (PE) and a lower phosphatidylcholine (PC) content]. Therefore, it is likely that LDLR is necessary to the virus to have the adequate amount and variety of lipids at the membranous web[42].
Although how LDLR contributes to HCV life cycle, in general, and to the viral entry step, in particular, is still controversial, its importance is undisputed. The role of this receptor in viral life cycle is further emphasized by the direct or indirect ability of HCV to modulate LDLR expression by both increasing its gene transcription and inhibiting its PCSK9-mediated protein degradation[44]. Interestingly, the HCV-mediated PCSK9 regulation has been reported to be different in patients infected with the genotype 3 compare to those with genotype 1, thus suggesting that HCV can affect lipoprotein metabolism in a genotype specific-manner[76].
This hypothesis is indirectly supported by the observation that hepatoma cells transfected with core proteins of the different HCV genotypes displayed a genotype-specific intracellular TG accumulation[77]. This accumulation is probably caused by the virus-mediated inhibition of the microsomal triglyceride transfer protein (MTTP), a key enzyme in the apoB-containing lipoproteins assembly pathway[78]. It has been also found that the HCV genotype could also affect the circulating levels of apolipoproteins. In chronic patients the infection with genotype 1b was found to be an independent factor significantly associated with higher levels of apoA-II and apoE, and lower levels of apoC-II and apoC-III, while genotype 2 infection was associated only with lower levels of apoC-II and apoC-III[79]. Moreover, the HCV genotypes influence the levels of LDL-cholesterol differently in patients with different IL28B polymorphisms, as well as the lipid-related genes expression in cultured cells[80]. These different effects on lipoproteins machinery, together with other pathogenic effects[81] could explain, at least partially, the HCV genotype-specific steatogenic effects.
Another connection between the lipoprotein metabolism and the RNA replication step of the HCV life cycle was discovered in our laboratory. We found that apoA-I is involved in the replication step of HCV. In fact, the downregulation of apoA-I induces significant decrease of viral RNA levels in either replicon carrying cells and in the HCVcc infected cells[82].
Although apoA-I is the major structural protein of the HDL, we focused our attention on this exchangeable apolipoprotein because a decreased association of apoA-I to the circulating LDL of HCV infected patients was found by a proteomic analysis. This result suggests that the function of apoA-I necessary for HCV replication could be linked to a its putative role in the biogenesis of apoB-containing lipoproteins rather than that of HDL. This is indirectly supported by the finding that the siRNA-mediated downregulation of apoA-I induces a significant reduction of HCV RNA only at later time point (day 4-6 post-silencing), when a decreased levels of apoB secretion was observed (unpublished results).
The apoA-I involvement in HCV replication were confirmed by others using different replicon system and siRNAs[83], thus lowering a possible off-targets effect, as recently underlined by a work on MOBKL1B by Rice’s group[84]. Interestingly, Saito’s laboratory found that one of the transcriptional effect of a histone deacetylase inhibitor treatment (SAHA) was the down-regulation of apoA-I and also the up-regulation of osteopontin (OPN), which are per se sufficient to induced the inhibition of HCV RNA replication[83].
Another apolipoprotein that has been described to influence HCV replication is apoH (also known as b2-glycoprotein I), which was able to limit RNA replication using human liver slices as a HCV infection model[85]. Although, the authors have not proved a direct effect on HCV replication, treatment with apoH reduced HCV production while not affecting HCV entry[85]. A negative effect of apoH on the virus is also supported by the positive correlation between high plasma levels of apoH and viral clearance, both in spontaneous remission and in response to pegylated-interferon/ribavirin therapy in HCV patients[86]. Interestingly, patients carrying the favorable IL28B rs12979860 CC SNP correlated with high plasma concentration of apoH, thus unveiling this apolipoprotein as a quantitative trait associated with IL28B[86].
Remarkably, apoH is part of the LDL[87] and it is known to influence the size and the lipid composition of LDL[88-90]. This further reinforces the idea that proteins affecting the generation of apoB-containing lipoproteins could have also an effect on HCV replication. However, since the inhibition of either apoB expression or MTTP function does not affect viral replication, this suggests that the distribution or the quality of lipids related to the lipoprotein machinery could influence HCV RNA production.
The last step of the intracellular HCV life cycle is the formation of viral particle. The dynamic of virus assembly is challenging to track, suggesting that this process is either rare or rapid. However, it is now well recognized that the HCV virion biogenesis strictly relies mainly on lipid droplets (LD), the storage sites for neutral lipids in cells, and on the apoB-containing lipoprotein machinery[91-93].
HCV particles production is a coordinated and complex process regulated spatially and temporally by all viral proteins and host factors. Virion assembly is coordinated between the synthesis of new positive RNA strands, its encapsidation and the acquisition of envelop, likely via budding into the ER[94].
The RNA replication site and the formation of nucleocapsid has to be separated to avoid competition for the binding of viral RNA. This is obtained by the localization of the core protein on the surface of cytosolic LD, probably through the presence of two amphipathic helices and a palmitoylated conserved cysteine residue. The MAPK-regulated cytosolic phospholipase A2, group IVA (PLA2G4A) is important for the core recruitment at the LD and for the specific cleavages of lipids with arachidonic acid, which is essential for the production of highly infectious viral particles[95]. LD localization of core may be also enhanced by the diacylglycerol acyltransferase-1 (DGAT-1)[96], an enzyme involved in LD morphogenesis that is also known to influence VLDL biogenesis[97]. The LD localization of core is an essential step. In fact, it regulates the recruitments of the other viral components and cellular factors, which regulate the transfer of both the newly replicated viral genomes from the membranous web and the HCV glycoproteins E1 and E2 from the ER, to the assembly site[94,98,99]. Although this step is not fully elucidated, it is likely regulated through protein-protein interactions between multiple viral proteins and a yet not fully unveil list of cellular proteins, as recently reviewed elsewhere[94,100].
Nascent virions, following maturation and acquisition of the typical low density, exit the cell via the secretory pathway. Although, the exact mechanisms are still poorly understood, it is now evident that assembly and secretion of HCV particles are associated with the VLDL/LDL pathway. There are different experimental evidences supporting this model.
Notably, in humanized livers of transplanted SCID/Alb-uPA mice, HCV infection occurs only when the engraftment of human hepatocytes is sufficient to obtain a human-like lipoproteins profile while it is not correlated to the number of human hepatocytes[101].
In Huh7 cells, the isolation of membrane vesicles in which HCV replicates lead to the enrichment in different members of the apoB-containing lipoproteins such as apoB, apoE and MTTP[102]. Interestingly, the impairment of VLDL/LDL production through the downregulation of apoB or the inhibition of either MTTP, which stabilizes apoB by transferring lipids during its translation, or long chain acyl-coenzyme A synthetase 3 (ACSL3), which mediates the phosphatidylcholine synthesis required for apoB secretion, lead to a decreased HCV particles production[102-105].
However, the direct requirement of apoB is controversial. Other researchers did not find a dependency of HCV production on apoB but rather on the activity of apoE[106-108]. Moreover, it has been reported that MTTP inhibitors at low doses, which are effective for apoB secretion inhibition, are ineffective for HCV production, while at higher concentrations those inhibitors blocked apoE expression and secretion and, consequently, suppressed the generation of viral particles[106]. The ectopic expression of apoE, but not apoB or MTTP, was found necessary also for the production of infectious HCV trans-complemented particles in human non-liver cells[108]. Moreover, apoE but not apoB was found necessary also for the cell-to-cell transmission of the virus. In fact, either the silencing of apoE in donor cells, but not in acceptor cells, inhibited the cell-to-cell viral spread[109].
A recent work of Matsuura’s lab smooth out the controversy. They showed that apoB and apoE redundantly participate in the formation of infectious HCV particles[110]. They generated Huh7 cells knock out for either or both apoB and apoE by zinc finger nucleases and found that the single knock out cells have a slightly reduction of HCV virions, while the apoB/apoE double knock out severely impaired the formation of infectious viral particles. More interestingly, they showed that the overexpression of different exchangeable apolipoproteins (i.e., apoA-I, apoA-II, apoC-I, apoC-II, apoC-III, apoE but not apoH) in the double knock out Huh7 cells rescued the capability of producing viral particles[110].
These results were independently confirmed in complemented HCV virus production experiments using the non-permissive 293T/miR-122 cells transfected with HCV, in which the expression of cDNAs encoding for all members of the apoA and apoC family, but not apoD, complemented HCV virus production, although at lower levels compared with apoE[111].
These results support the data obtained by different labs in which a requirement of other apolipoproteins for HCV production has been reported.
For instance, apoJ, a small heat shock protein that prevents unfolded secretory protein aggregation identified as a VLDL-associated protein[112], was found involved in efficient infectious HCV virion production[113]. ApoJ silencing decreased the HCV virion production, without affecting the HCV RNA replication, which could be restored upon reconstitution with a siRNA-resistant apoJ. Most likely, apoJ is involved at the step of virion assembly, since it was found to interact with core and NS5A, stabilizing the dual protein complex. Interestingly, immunofluorescence analysis showed that HCV infection induces a cellular redistribution of apoJ from Golgi to LD at the ER-Golgi membrane contact site[113].
Our lab found that the silencing of apoA-I induced a significant decrease in HCVcc production[82], although at lower levels respect of that of apoE (unpublished data). As described for other cellular components also the distribution of apoA-I was affected by HCV. We found that the apoA-I specific association to LDL was reduced in the circulating lipoproteins of HCV infected patients. Although it is known that the exchange of apolipoproteins among lipoprotein particles and interconversion of particles occurs in the plasma compartment, we found that the HCV-induced decreased association of apoA-I with LDL has a cellular origin. The HCV-associated LDL-specific reduction in apoA-I was also observed in the different HCV cellular models (i.e., HCVcc, genomic and subgenomic replicons). The results obtained with subgenomic replicon-carrying Huh7 cells, which recapitulate only the viral RNA replication step, indicate that the sole NS viral proteins are sufficient to impair the apoA-I/LDL association, thus reinforcing the hypothesis that VLDL/LDL biogenesis and viral replication could share common sites and bridging molecules. However, although apoA-I is known to be associated to the circulating VLDL and LDL[114,115], its role in the physiology of these lipoproteins is yet unknown. A possible role of apoA-I in the viral life cycle could be to drive HCV components in the right cellular compartments, and its binding activity to NS5A seems to support this hypothesis[116,117].
Moreover, as above mentioned, Fukuhara and colleagues found that different apolipoproteins are sufficient for HCV viral production in culture. More specifically, they found that the amphipathic α-helices present in the different apolipoproteins possess a redundant role in the assembly of HCV, through a direct interaction with the viral particles at the post-envelopment step[110].
Interestingly, similar results were found for the replication step of HCV life cycle. The N-terminal amphipathic helix of NS5A, which motif is very similar to that of apolipoproteins, bound specifically to PI 4,5-bisphosphate [PI(4,5)P2], inducing a conformational change that stabilizes the interaction between NS5A and TBC1D20, which is required for HCV replication[118].
Since phosphoinositides bind and regulate localization of proteins via a variety of structural motifs, these results support the hypothesis that the requirement of the different apolipoproteins is related to the proper cellular localisation of viral components, which is necessary for the exploitation of lipid, in general, and of the lipoprotein metabolism, in particular. In other words, HCV may modulate the production of lipoproteins in the host cells to make this pathway more appropriate for viral maturation.
This point of view is supported by the observation that there is no correlation between the ability to generate VLDL and the production of LVP. In fact, the VLDL-producing HepG2 cells generated LVP similar, for both density and apolipoproteins content, to those generated by the VLDL-deficient Huh7.5 cells[119].
However, it is important to note that dissimilarities exist in the molecular composition of LVP found in HCV patients or generated in culture; for instance serum LVP could be immunoprecipitated by anti-apoB antibodies while the interaction with HCVcc is less efficient[29].
These discrepancies should be kept in mind because may have major implications for our understanding of HCV assembly and secretion. In fact, the exact model for the LVP composition is not yet defined. It has been proposed either a single-particle model, in which HCV exists as a hybrid particle fused to a VLDL/LDL, or as a two-particles structure, in which the virion is surrounded by lipoproteins (Figure 1).
HCV interacts with and hijacks host cell machineries and pathways to generate a chronic and productive infection. It is now well established that HCV has an intimate relationship with the host lipid metabolism, which has a role in all the steps of the viral infectious cycle. In particular, HCV modulates the production of lipoproteins in the host cells to make it more effective for viral production, propagation and persistence.
The virus circulates in the bloodstream as a highly lipidated LVP, although it is not yet known whether fused with or surrounded by lipoproteins (Figure 1). HCV utilizes the lipoproteins pathways for cell entry, virus assembly and, possibly also for RNA replication. Moreover, it is becoming recognized that its strict resemblance with VLDL/LDL may contribute to the viral immune evasion strategies, such as masking viral epitopes or escaping from anti-HCV neutralizing antibodies directed against either viral proteins or the entry factors involved in lipoproteins pathways (i.e., CD81 and SR-BI as reviewed by Vercauteren and coauthors[120]).
What has not been investigated yet is the possible involvement of lipid/lipoprotein metabolism in the dysregulation of the immune response mediated by HCV, as recently reported for the hepatitis B virus[121]. Recent evidences showing that LVP affect dendritic cells maturation[122], ApoE3 blocks the antiviral effect of ficolin-2[123] and the VLDL and LDL of chronic infected patients induce an altered intracellular lipid production[124] suggest that the HCV-induced modification of lipoprotein metabolism could be involved in the regulation of the immune response.
Overall, decoding the multiple interactions that HCV establishes with the lipoproteins pathways is mandatory to obtain a deeper knowledge of HCV biology and pathogenesis, a step necessary to understand and to manage the reversibility of liver damages upon DAAs-mediated viral clearance.
We sincerely apologize to all those colleagues whose important work was not cited due to space constraints.
P- Reviewer: Otsuka M, Pazienza V S- Editor: Gong ZM L- Editor: A E- Editor: Ma S
| 1. | Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol. 2013;10:553-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 704] [Cited by in RCA: 741] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 2. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1770] [Cited by in RCA: 1847] [Article Influence: 153.9] [Reference Citation Analysis (3)] |
| 3. | Ansaldi F, Orsi A, Sticchi L, Bruzzone B, Icardi G. Hepatitis C virus in the new era: perspectives in epidemiology, prevention, diagnostics and predictors of response to therapy. World J Gastroenterol. 2014;20:9633-9652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 113] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (1)] |
| 4. | Bruggmann P, Berg T, Øvrehus AL, Moreno C, Brandão Mello CE, Roudot-Thoraval F, Marinho RT, Sherman M, Ryder SD, Sperl J. Historical epidemiology of hepatitis C virus (HCV) in selected countries. J Viral Hepat. 2014;21 Suppl 1:5-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 183] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 5. | European Association for Study of Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2014;60:392-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 655] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 6. | Westbrook RH, Dusheiko G. Natural history of hepatitis C. J Hepatol. 2014;61:S58-S68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 615] [Article Influence: 55.9] [Reference Citation Analysis (0)] |
| 7. | Pawlotsky JM, Flisiak R, Sarin SK, Rasenack J, Piratvisuth T, Chuang WL, Peng CY, Foster GR, Shah S, Wedemeyer H. Alisporivir plus ribavirin, interferon free or in combination with pegylated interferon, for hepatitis C virus genotype 2 or 3 infection. Hepatology. 2015;62:1013-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 8. | Heim MH, Thimme R. Innate and adaptive immune responses in HCV infections. J Hepatol. 2014;61:S14-S25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 224] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 9. | Thimme R, Binder M, Bartenschlager R. Failure of innate and adaptive immune responses in controlling hepatitis C virus infection. FEMS Microbiol Rev. 2012;36:663-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 10. | Macaluso FS, Maida M, Minissale MG, Li Vigni T, Attardo S, Orlando E, Petta S. Metabolic factors and chronic hepatitis C: a complex interplay. Biomed Res Int. 2013;2013:564645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Goossens N, Negro F. Is genotype 3 of the hepatitis C virus the new villain? Hepatology. 2014;59:2403-2412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 12. | Popescu CI, Riva L, Vlaicu O, Farhat R, Rouillé Y, Dubuisson J. Hepatitis C virus life cycle and lipid metabolism. Biology (Basel). 2014;3:892-921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 13. | Schaefer EA, Chung RT. HCV and host lipids: an intimate connection. Semin Liver Dis. 2013;33:358-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | Ding Q, von Schaewen M, Ploss A. The impact of hepatitis C virus entry on viral tropism. Cell Host Microbe. 2014;16:562-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 15. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [PubMed] |
| 16. | Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 423] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 17. | Alonzi T, Mancone C, Amicone L, Tripodi M. Elucidation of lipoprotein particles structure by proteomic analysis. Expert Rev Proteomics. 2008;5:91-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Ramasamy I. Recent advances in physiological lipoprotein metabolism. Clin Chem Lab Med. 2014;52:1695-1727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 146] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 19. | Sundaram M, Yao Z. Recent progress in understanding protein and lipid factors affecting hepatic VLDL assembly and secretion. Nutr Metab (Lond). 2010;7:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 127] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 20. | Kaito M, Watanabe S, Tsukiyama-Kohara K, Yamaguchi K, Kobayashi Y, Konishi M, Yokoi M, Ishida S, Suzuki S, Kohara M. Hepatitis C virus particle detected by immunoelectron microscopic study. J Gen Virol. 1994;75:1755-1760. [PubMed] |
| 21. | Li X, Jeffers LJ, Shao L, Reddy KR, de Medina M, Scheffel J, Moore B, Schiff ER. Identification of hepatitis C virus by immunoelectron microscopy. J Viral Hepat. 1995;2:227-234. [PubMed] |
| 22. | Hijikata M, Shimizu YK, Kato H, Iwamoto A, Shih JW, Alter HJ, Purcell RH, Yoshikura H. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol. 1993;67:1953-1958. [PubMed] |
| 23. | Kanto T, Hayashi N, Takehara T, Hagiwara H, Mita E, Naito M, Kasahara A, Fusamoto H, Kamada T. Buoyant density of hepatitis C virus recovered from infected hosts: two different features in sucrose equilibrium density-gradient centrifugation related to degree of liver inflammation. Hepatology. 1994;19:296-302. [PubMed] |
| 24. | Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294-9299. [PubMed] |
| 25. | Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol. 2006;80:2418-2428. [PubMed] |
| 26. | Gastaminza P, Dryden KA, Boyd B, Wood MR, Law M, Yeager M, Chisari FV. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J Virol. 2010;84:10999-11009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 27. | André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919-6928. [PubMed] |
| 28. | Nielsen SU, Bassendine MF, Martin C, Lowther D, Purcell PJ, King BJ, Neely D, Toms GL. Characterization of hepatitis C RNA-containing particles from human liver by density and size. J Gen Virol. 2008;89:2507-2517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 29. | Merz A, Long G, Hiet MS, Brügger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286:3018-3032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 284] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 30. | Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, Silvestry M, Kuhn RJ, Rice CM. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci USA. 2013;110:9505-9510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 210] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 31. | Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol. 2013;11:688-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 282] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 32. | Karangelis DE, Kanakis I, Asimakopoulou AP, Karousou E, Passi A, Theocharis AD, Triposkiadis F, Tsilimingas NB, Karamanos NK. Glycosaminoglycans as key molecules in atherosclerosis: the role of versican and hyaluronan. Curr Med Chem. 2010;17:4018-4026. [PubMed] |
| 33. | Shi Q, Jiang J, Luo G. Syndecan-1 serves as the major receptor for attachment of hepatitis C virus to the surfaces of hepatocytes. J Virol. 2013;87:6866-6875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 34. | Lefèvre M, Felmlee DJ, Parnot M, Baumert TF, Schuster C. Syndecan 4 is involved in mediating HCV entry through interaction with lipoviral particle-associated apolipoprotein E. PLoS One. 2014;9:e95550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 35. | Jiang J, Wu X, Tang H, Luo G. Apolipoprotein E mediates attachment of clinical hepatitis C virus to hepatocytes by binding to cell surface heparan sulfate proteoglycan receptors. PLoS One. 2013;8:e67982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 36. | Xu Y, Martinez P, Séron K, Luo G, Allain F, Dubuisson J, Belouzard S. Characterization of hepatitis C virus interaction with heparan sulfate proteoglycans. J Virol. 2015;89:3846-3858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 37. | Sorrentino V, Nelson JK, Maspero E, Marques AR, Scheer L, Polo S, Zelcer N. The LXR-IDOL axis defines a clathrin-, caveolae-, and dynamin-independent endocytic route for LDLR internalization and lysosomal degradation. J Lipid Res. 2013;54:2174-2184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 38. | Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766-12771. [PubMed] |
| 39. | Monazahian M, Böhme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J Med Virol. 1999;57:223-229. [PubMed] |
| 40. | Germi R, Crance JM, Garin D, Guimet J, Lortat-Jacob H, Ruigrok RW, Zarski JP, Drouet E. Cellular glycosaminoglycans and low density lipoprotein receptor are involved in hepatitis C virus adsorption. J Med Virol. 2002;68:206-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 115] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 41. | Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411-419. [PubMed] |
| 42. | Albecka A, Belouzard S, Op de Beeck A, Descamps V, Goueslain L, Bertrand-Michel J, Tercé F, Duverlie G, Rouillé Y, Dubuisson J. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology. 2012;55:998-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 133] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 43. | Prentoe J, Serre SB, Ramirez S, Nicosia A, Gottwein JM, Bukh J. Hypervariable region 1 deletion and required adaptive envelope mutations confer decreased dependency on scavenger receptor class B type I and low-density lipoprotein receptor for hepatitis C virus. J Virol. 2014;88:1725-1739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 44. | Syed GH, Tang H, Khan M, Hassanein T, Liu J, Siddiqui A. Hepatitis C virus stimulates low-density lipoprotein receptor expression to facilitate viral propagation. J Virol. 2014;88:2519-2529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 45. | Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017-5025. [PubMed] |
| 46. | Dreux M, Dao Thi VL, Fresquet J, Guérin M, Julia Z, Verney G, Durantel D, Zoulim F, Lavillette D, Cosset FL. Receptor complementation and mutagenesis reveal SR-BI as an essential HCV entry factor and functionally imply its intra- and extra-cellular domains. PLoS Pathog. 2009;5:e1000310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 101] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 47. | Shen WJ, Hu J, Hu Z, Kraemer FB, Azhar S. Scavenger receptor class B type I (SR-BI): a versatile receptor with multiple functions and actions. Metabolism. 2014;63:875-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 48. | Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, Pawlotsky JM, Lavillette D, Cosset FL. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J Virol. 2005;79:8217-8229. [PubMed] |
| 49. | Zahid MN, Turek M, Xiao F, Thi VL, Guérin M, Fofana I, Bachellier P, Thompson J, Delang L, Neyts J. The postbinding activity of scavenger receptor class B type I mediates initiation of hepatitis C virus infection and viral dissemination. Hepatology. 2013;57:492-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 50. | Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, Keck Z, Foung S, Vu-Dac N, Dubuisson J. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem. 2006;281:18285-18295. [PubMed] |
| 51. | Maillard P, Huby T, Andréo U, Moreau M, Chapman J, Budkowska A. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/Cla1 is mediated by ApoB-containing lipoproteins. FASEB J. 2006;20:735-737. [PubMed] |
| 52. | Brazzoli M, Bianchi A, Filippini S, Weiner A, Zhu Q, Pizza M, Crotta S. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol. 2008;82:8316-8329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 144] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 53. | Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 569] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 54. | Zona L, Lupberger J, Sidahmed-Adrar N, Thumann C, Harris HJ, Barnes A, Florentin J, Tawar RG, Xiao F, Turek M. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe. 2013;13:302-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 55. | Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci USA. 2013;110:10777-10782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 166] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 56. | Sourisseau M, Michta ML, Zony C, Israelow B, Hopcraft SE, Narbus CM, Parra Martín A, Evans MJ. Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies. PLoS Pathog. 2013;9:e1003244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 57. | Farquhar MJ, Hu K, Harris HJ, Davis C, Brimacombe CL, Fletcher SJ, Baumert TF, Rappoport JZ, Balfe P, McKeating JA. Hepatitis C virus induces CD81 and claudin-1 endocytosis. J Virol. 2012;86:4305-4316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 58. | Dreux M, Boson B, Ricard-Blum S, Molle J, Lavillette D, Bartosch B, Pécheur EI, Cosset FL. The exchangeable apolipoprotein ApoC-I promotes membrane fusion of hepatitis C virus. J Biol Chem. 2007;282:32357-32369. [PubMed] |
| 59. | Meunier JC, Russell RS, Engle RE, Faulk KN, Purcell RH, Emerson SU. Apolipoprotein c1 association with hepatitis C virus. J Virol. 2008;82:9647-9656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 60. | Lohmann V. Hepatitis C virus RNA replication. Curr Top Microbiol Immunol. 2013;369:167-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 61. | Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 383] [Cited by in RCA: 402] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 62. | Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487-5492. [PubMed] |
| 63. | Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974-5984. [PubMed] |
| 64. | Gouttenoire J, Castet V, Montserret R, Arora N, Raussens V, Ruysschaert JM, Diesis E, Blum HE, Penin F, Moradpour D. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J Virol. 2009;83:6257-6268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 71] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 65. | Liu Z, Yang F, Robotham JM, Tang H. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J Virol. 2009;83:6554-6565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 66. | Madan V, Paul D, Lohmann V, Bartenschlager R. Inhibition of HCV replication by cyclophilin antagonists is linked to replication fitness and occurs by inhibition of membranous web formation. Gastroenterology. 2014;146:1361-72.e1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 67. | Chao TC, Su WC, Huang JY, Chen YC, Jeng KS, Wang HD, Lai MM. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), a host membrane-deforming protein, is critical for membranous web formation in hepatitis C virus replication. J Virol. 2012;86:1739-1749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 68. | Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol. 2003;77:4160-4168. [PubMed] |
| 69. | Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 426] [Cited by in RCA: 406] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 70. | Bianco A, Reghellin V, Donnici L, Fenu S, Alvarez R, Baruffa C, Peri F, Pagani M, Abrignani S, Neddermann P. Metabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog. 2012;8:e1002576. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 71. | Khan I, Katikaneni DS, Han Q, Sanchez-Felipe L, Hanada K, Ambrose RL, Mackenzie JM, Konan KV. Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2. J Virol. 2014;88:12276-12295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 72. | Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373-85.e1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 73. | Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122-8130. [PubMed] |
| 74. | Olmstead AD, Knecht W, Lazarov I, Dixit SB, Jean F. Human subtilase SKI-1/S1P is a master regulator of the HCV Lifecycle and a potential host cell target for developing indirect-acting antiviral agents. PLoS Pathog. 2012;8:e1002468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 75. | Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 331] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 76. | Bridge SH, Sheridan DA, Felmlee DJ, Crossey MM, Fenwick FI, Lanyon CV, Dubuc G, Seidah NG, Davignon J, Thomas HC. PCSK9, apolipoprotein E and lipoviral particles in chronic hepatitis C genotype 3: evidence for genotype-specific regulation of lipoprotein metabolism. J Hepatol. 2015;62:763-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 77. | Abid K, Pazienza V, de Gottardi A, Rubbia-Brandt L, Conne B, Pugnale P, Rossi C, Mangia A, Negro F. An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol. 2005;42:744-751. [PubMed] |
| 78. | Perlemuter G, Sabile A, Letteron P, Vona G, Topilco A, Chrétien Y, Koike K, Pessayre D, Chapman J, Barba G. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 2002;16:185-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 428] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 79. | Seki N, Sugita T, Aida Y, Itagaki M, Ishiguro H, Sutoh S, Abe H, Tsubota A, Matsushima M, Aizawa Y. Assessment of the features of serum apolipoprotein profiles in chronic HCV infection: difference between HCV genotypes 1b and 2. Hepatol Int. 2014;8:550-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 80. | Rojas Á, del Campo JA, Maraver M, Aparcero R, García-Valdecasas M, Diago M, Carmona I, Andrade RJ, Solà R, Romero-Gómez M. Hepatitis C virus infection alters lipid metabolism depending on IL28B polymorphism and viral genotype and modulates gene expression in vivo and in vitro. J Viral Hepat. 2014;21:19-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 81. | Ripoli M, Pazienza V. Impact of HCV genetic differences on pathobiology of disease. Expert Rev Anti Infect Ther. 2011;9:747-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 82. | Mancone C, Steindler C, Santangelo L, Simonte G, Vlassi C, Longo MA, D’Offizi G, Di Giacomo C, Pucillo LP, Amicone L. Hepatitis C virus production requires apolipoprotein A-I and affects its association with nascent low-density lipoproteins. Gut. 2011;60:378-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 83. | Sato A, Saito Y, Sugiyama K, Sakasegawa N, Muramatsu T, Fukuda S, Yoneya M, Kimura M, Ebinuma H, Hibi T. Suppressive effect of the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) on hepatitis C virus replication. J Cell Biochem. 2013;114:1987-1996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 84. | Chung HY, Gu M, Buehler E, MacDonald MR, Rice CM. Seed sequence-matched controls reveal limitations of small interfering RNA knockdown in functional and structural studies of hepatitis C virus NS5A-MOBKL1B interaction. J Virol. 2014;88:11022-11033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 85. | Sultanik P, Mallet V, Lagaye S, Casrouge A, Dorival C, Barthe Y, Fontaine H, Hézode C, Mottez E, Bronowicki JP. Plasma apolipoprotein H limits HCV replication and associates with response to NS3 protease inhibitors-based therapy. Liver Int. 2015;35:1833-1844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 86. | Laird ME, Mohsen A, Duffy D, Mamdouh R, LeFouler L, Casrouge A, El-Daly M, Rafik M, Abdel-Hamid M, Soulier A. Apolipoprotein H expression is associated with IL28B genotype and viral clearance in hepatitis C virus infection. J Hepatol. 2014;61:770-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 87. | Diffenderfer MR, Schaefer EJ. The composition and metabolism of large and small LDL. Curr Opin Lipidol. 2014;25:221-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 88. | Takada D, Ezura Y, Ono S, Iino Y, Katayama Y, Xin Y, Wu LL, Larringa-Shum S, Stephenson SH, Hunt SC. Apolipoprotein H variant modifies plasma triglyceride phenotype in familial hypercholesterolemia: a molecular study in an eight-generation hyperlipidemic family. J Atheroscler Thromb. 2003;10:79-84. [PubMed] |
| 89. | Bossé Y, Feitosa MF, Després JP, Lamarche B, Rice T, Rao DC, Bouchard C, Pérusse L, Vohl MC. Detection of a major gene effect for LDL peak particle diameter and association with apolipoprotein H gene haplotype. Atherosclerosis. 2005;182:231-239. [PubMed] |
| 90. | Foulkes AS, Matthews GJ, Das U, Ferguson JF, Lin R, Reilly MP. Mixed modeling of meta-analysis P-values (MixMAP) suggests multiple novel gene loci for low density lipoprotein cholesterol. PLoS One. 2013;8:e54812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 91. | Alvisi G, Madan V, Bartenschlager R. Hepatitis C virus and host cell lipids: an intimate connection. RNA Biol. 2011;8:258-269. [PubMed] |
| 92. | Suzuki T. Morphogenesis of infectious hepatitis C virus particles. Front Microbiol. 2012;3:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 93. | Filipe A, McLauchlan J. Hepatitis C virus and lipid droplets: finding a niche. Trends Mol Med. 2015;21:34-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 94. | Lindenbach BD. Virion assembly and release. Curr Top Microbiol Immunol. 2013;369:199-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 95. | Menzel N, Fischl W, Hueging K, Bankwitz D, Frentzen A, Haid S, Gentzsch J, Kaderali L, Bartenschlager R, Pietschmann T. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 2012;8:e1002829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 96. | Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV, Ott M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16:1295-1298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 270] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 97. | Yamazaki T, Sasaki E, Kakinuma C, Yano T, Miura S, Ezaki O. Increased very low density lipoprotein secretion and gonadal fat mass in mice overexpressing liver DGAT1. J Biol Chem. 2005;280:21506-21514. [PubMed] |
| 98. | Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman MJ, Miyamura T. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200-1205. [PubMed] |
| 99. | Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089-1097. [PubMed] |
| 100. | Dubuisson J, Cosset FL. Virology and cell biology of the hepatitis C virus life cycle: an update. J Hepatol. 2014;61:S3-S13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 101. | Steenbergen RH, Joyce MA, Lund G, Lewis J, Chen R, Barsby N, Douglas D, Zhu LF, Tyrrell DL, Kneteman NM. Lipoprotein profiles in SCID/uPA mice transplanted with human hepatocytes become human-like and correlate with HCV infection success. Am J Physiol Gastrointest Liver Physiol. 2010;299:G844-G854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 102. | Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci USA. 2007;104:5848-5853. [PubMed] |
| 103. | Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120-2129. [PubMed] |
| 104. | Nahmias Y, Goldwasser J, Casali M, van Poll D, Wakita T, Chung RT, Yarmush ML. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology. 2008;47:1437-1445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 190] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 105. | Yao H, Ye J. Long chain acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 cells. J Biol Chem. 2008;283:849-854. [PubMed] |
| 106. | Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783-13793. [PubMed] |
| 107. | Jiang J, Luo G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J Virol. 2009;83:12680-12691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 108. | Hueging K, Doepke M, Vieyres G, Bankwitz D, Frentzen A, Doerrbecker J, Gumz F, Haid S, Wölk B, Kaderali L. Apolipoprotein E codetermines tissue tropism of hepatitis C virus and is crucial for viral cell-to-cell transmission by contributing to a postenvelopment step of assembly. J Virol. 2014;88:1433-1446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 109. | Gondar V, Molina-Jiménez F, Hishiki T, García-Buey L, Koutsoudakis G, Shimotohno K, Benedicto I, Majano PL. Apolipoprotein E, but Not Apolipoprotein B, Is Essential for Efficient Cell-to-Cell Transmission of Hepatitis C Virus. J Virol. 2015;89:9962-9973. [PubMed] |
| 110. | Fukuhara T, Wada M, Nakamura S, Ono C, Shiokawa M, Yamamoto S, Motomura T, Okamoto T, Okuzaki D, Yamamoto M. Amphipathic α-helices in apolipoproteins are crucial to the formation of infectious hepatitis C virus particles. PLoS Pathog. 2014;10:e1004534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 111. | Hueging K, Weller R, Doepke M, Vieyres G, Todt D, Wölk B, Vondran FW, Geffers R, Lauber C, Kaderali L. Several Human Liver Cell Expressed Apolipoproteins Complement HCV Virus Production with Varying Efficacy Conferring Differential Specific Infectivity to Released Viruses. PLoS One. 2015;10:e0134529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 112. | Sun HY, Chen SF, Lai MD, Chang TT, Chen TL, Li PY, Shieh DB, Young KC. Comparative proteomic profiling of plasma very-low-density and low-density lipoproteins. Clin Chim Acta. 2010;411:336-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 113. | Lin CC, Tsai P, Sun HY, Hsu MC, Lee JC, Wu IC, Tsao CW, Chang TT, Young KC. Apolipoprotein J, a glucose-upregulated molecular chaperone, stabilizes core and NS5A to promote infectious hepatitis C virus virion production. J Hepatol. 2014;61:984-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 114. | Mancone C, Amicone L, Fimia GM, Bravo E, Piacentini M, Tripodi M, Alonzi T. Proteomic analysis of human very low-density lipoprotein by two-dimensional gel electrophoresis and MALDI-TOF/TOF. Proteomics. 2007;7:143-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 115. | Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics I: mapping of proteins in low-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005;5:551-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 108] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 116. | Shi ST, Polyak SJ, Tu H, Taylor DR, Gretch DR, Lai MM. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology. 2002;292:198-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 239] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 117. | Wang AG, Lee DS, Moon HB, Kim JM, Cho KH, Choi SH, Ha HL, Han YH, Kim DG, Hwang SB. Non-structural 5A protein of hepatitis C virus induces a range of liver pathology in transgenic mice. J Pathol. 2009;219:253-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 118. | Cho NJ, Lee C, Pang PS, Pham EA, Fram B, Nguyen K, Xiong A, Sklan EH, Elazar M, Koytak ES. Phosphatidylinositol 4,5-bisphosphate is an HCV NS5A ligand and mediates replication of the viral genome. Gastroenterology. 2015;148:616-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 119. | Jammart B, Michelet M, Pécheur EI, Parent R, Bartosch B, Zoulim F, Durantel D. Very-low-density lipoprotein (VLDL)-producing and hepatitis C virus-replicating HepG2 cells secrete no more lipoviroparticles than VLDL-deficient Huh7.5 cells. J Virol. 2013;87:5065-5080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 120. | Vercauteren K, Mesalam AA, Leroux-Roels G, Meuleman P. Impact of lipids and lipoproteins on hepatitis C virus infection and virus neutralization. World J Gastroenterol. 2014;20:15975-15991. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 121. | Zeissig S, Murata K, Sweet L, Publicover J, Hu Z, Kaser A, Bosse E, Iqbal J, Hussain MM, Balschun K. Hepatitis B virus-induced lipid alterations contribute to natural killer T cell-dependent protective immunity. Nat Med. 2012;18:1060-1068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 189] [Article Influence: 14.5] [Reference Citation Analysis (2)] |
| 122. | Perrin-Cocon L, Diaz O, André P, Lotteau V. Modified lipoproteins provide lipids that modulate dendritic cell immune function. Biochimie. 2013;95:103-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 123. | Zhao Y, Ren Y, Zhang X, Zhao P, Tao W, Zhong J, Li Q, Zhang XL. Ficolin-2 inhibits hepatitis C virus infection, whereas apolipoprotein E3 mediates viral immune escape. J Immunol. 2014;193:783-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 124. | Napolitano M, Giuliani A, Alonzi T, Mancone C, D’Offizi G, Tripodi M, Bravo E. Very low density lipoprotein and low density lipoprotein isolated from patients with hepatitis C infection induce altered cellular lipid metabolism. J Med Virol. 2007;79:254-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |