Published online Jun 14, 2016. doi: 10.3748/wjg.v22.i22.5254
Peer-review started: March 3, 2016
First decision: March 31, 2016
Revised: April 22, 2016
Accepted: May 4, 2016
Article in press: May 4, 2016
Published online: June 14, 2016
Processing time: 81 Days and 3.4 Hours
AIM: To detect the expression of the long noncoding RNA HOTAIR in colon cancer and analyze its relationship with clinicopathological parameters of colon cancer.
METHODS: Total RNA was extracted from 80 colon cancer tissues and matched tumor-adjacent normal colon tissues and reverse transcribed. Quantitative polymerase chain reaction was used to detect the expression of HOTAIR. The relationship between the expression of HOTAIR and clinicopathological parameters of colon cancer was analyzed.
RESULTS: The expression of HOTAIR was significantly higher in colon cancer tissues than in matched tumor-adjacent normal colon tissues (P < 0.05). HOTAIR expression was significantly higher in cases with lymph node metastasis than in those without metastasis; in lowly differentiated and undifferentiated cases than in highly and moderately differentiated cases; and in stages III + IV cases than in stages I + II cases (P < 0.05).
CONCLUSION: HOTAIR expression is upregulated in colon cancer, suggesting that HOTAIR plays an important role in the tumorigenesis, development and metastasis of colon cancer. HOTAIR may act as an oncogene and represents a new molecular target for the treatment of colon cancer.
Core tip: This study aimed to detect the expression of HOTAIR in colon cancer and analyze its relationship with clinicopathological parameters of colon cancer. Total RNA was extracted from 80 colon cancer tissues and matched tumor-adjacent normal colon tissues and reverse transcribed. HOTAIR expression was upregulated in colon cancer, suggesting that it may play an important role in the tumorigenesis, development and metastasis of colon cancer. HOTAIR might acts as an oncogene and could be a new molecular target for the treatment of colon cancer.
- Citation: Luo ZF, Zhao D, Li XQ, Cui YX, Ma N, Lu CX, Liu MY, Zhou Y. Clinical significance of HOTAIR expression in colon cancer. World J Gastroenterol 2016; 22(22): 5254-5259
- URL: https://www.wjgnet.com/1007-9327/full/v22/i22/5254.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i22.5254
Colon cancer is a clinically common, highly malignant tumor of the digestive tract. Although drugs targeting epidermal growth factor receptor (EGFR) and KRAS mutations have significantly extended the survival of some colon cancer patients[1-3], only a small number of patients can benefit from these drugs because of the complex etiology of this malignancy. Overall, the effects of current therapies for colon cancer are not satisfactory[4,5].
Long noncoding RNAs (lncRNAs) are non-protein coding transcripts of around 200 nucleotides, which exist widely in the genome and can regulate gene expression[6]. HOTAIR is one of the extensively studied lncRNAs in recent years. Many studies have indicated that HOTAIR plays an important role in breast cancer, pancreatic cancer, liver cancer, gastric cancer, esophageal cancer and non-small cell lung cancer[7-10]. Studies in colon cancer suggest that HOTAIR is an important oncogene that affects the biological behavior of colon cancer[11] and can serve as an independent risk factor[12]. The latest research suggests that the expression of HOTAIR is associated with tumor metastasis[13].
In the present study, we detected the expression of HOTAIR in 80 colon cancer tissue samples by quantitative polymerase chain reaction (qPCR). Based on the clinical and pathological parameters of colon cancer patients, we analyzed the possible role of HOTAIR in colon cancer development, metastasis and sensitivity to treatment, with an emphasis on the role of HOTAIR in colon cancer treatment. The findings will provide a theoretical basis for developing a new, targeted therapy for colon cancer.
Eighty patients with pathologically proven colon cancer who underwent surgery at our hospital from September 2011 to September 2013, and had complete clinical records, were included. All patients provided written informed consent, and the study protocol was approved by the Medical Ethics Committee of Zhengzhou University. The mean age of the patients was 64 ± 16 years. There were 46 patients with stage I or II disease, and 34 patients with stage III or IV disease. Forty-one patients had well or moderately differentiated tumors, and 34 patients had poorly differentiated or undifferentiated tumors. No patients had undergone radiotherapy or chemotherapy before surgery. Tumor tissues and normal colon tissues at least 7 cm away from the tumor were taken, frozen in liquid nitrogen within 30 min and preserved for further use.
Trizol was purchased from Invitrogen. The reverse transcription kit and DNA ladder were purchased from Takara. Primers for qPCR were designed and synthesized by Shanghai GenePharma. The qPCR kit was purchased from Thermo.
Tissue samples preserved in liquid nitrogen were put into an RNase-free mortar with liquid nitrogen and pulverized. For each 100 mg of tissue, 1 mL of Trizol was added. RNA preparation was then performed following the manufacturer’s instructions. The obtained RNA was dissolved in DEPC-treated water, and the RNA concentration was measured using a micro UV-Vis fluorescence spectrophotometer (e-spect, Malcom, Japan). The obtained RNA was preserved at -80 °C for further use.
RNA reverse transcription was performed using a reverse transcription kit in a 20-μL system, containing 11 μL of DEPC-treated water, 1 μL of total RNA, 4 μL of 5 × buffer, 1 μL of RNase inhibitor, 2 μL of dNTPs, and 1 μL of reverse transcriptase. Reaction parameters were 42 °C for 60 min and 95 °C for 5 min. The obtained cDNA was preserved at -80 °C for further use.
qPCR: qPCR was performed in a 20-μL system containing 1 μL of cDNA, 10 μL of 2 × Master Mix with 0.03 × ROX added, 1 μL of forward primer (final concentration of 0.5 μmol/L), 1 μL of reverse primer, and 8 μL of DEPC-treated water on a Mx3005p cycler. PCR amplification was performed in triplicate. Cycling parameters were 95 °C for 7 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s.
The expression levels of HOTAIR in tissues are expressed as mean ± SD and were compared using a two-sample t-test. Statistical analyses were performed using SPSS13.0. P-values < 0.05 were considered statistically significant.
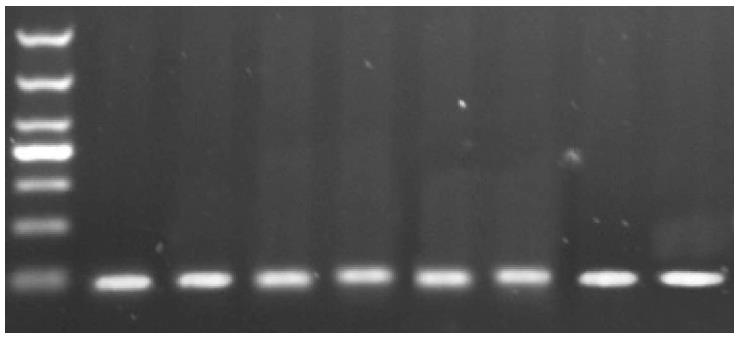
The length of the expected PCR product for HOTAIR is 91 bp, and agarose gel electrophoresis showed that qPCR yielded PCR products of expected size (Figure 1).
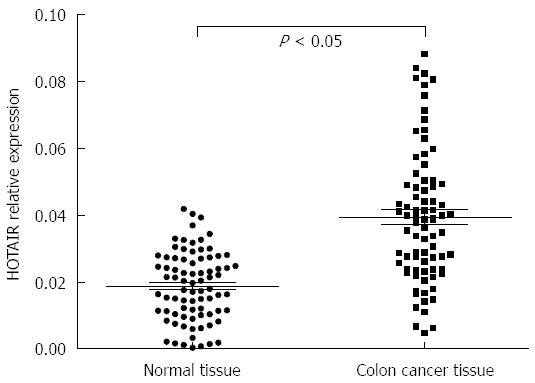
QPCR analysis showed that, although the expression of GAPDH showed no significant differences, the Ct value of HOTAIR was significantly lower in colon cancer tissues than in tumor-adjacent normal colonic tissues, suggesting that HOTAIR expression is upregulated in colon cancer. When the relative expression level is expressed as N (N = 2-∆Ct, ∆Ct = CtHOTAIR - CtGAPDH[14]), the relative expression level of HOTAIR was significantly higher in colon cancer tissues than in tumor-adjacent normal colonic tissues (P < 0.05, Figure 2).
HOTAIR expression was significantly correlated with lymph node metastasis, tumor differentiation and TNM stage (P < 0.05). HOTAIR expression was significantly higher in cases with lymph node metastasis than in those without metastasis, in lowly differentiated and undifferentiated cases than in highly and moderately differentiated cases, and in stages III + IV cases than in stages I + II cases. By contrast, HOTAIR expression had no significant correlation with patient gender, age or tumor size (P > 0.05) (Tables 1-3).
| Primer | Sequence |
| HOTAIR | |
| Forward | 5'-CAGTGGGGAACTCTGACTCG-3' |
| Reverse | 5'-GTGCCTGGTGCTCTCTTACC-3' |
| GAPDH | |
| Forward | 5'-GTCAACGGATTTGGTCTGTATT-3' |
| Reverse | 5'-AGTCTTCTGGGTGGCAGTGAT-3' |
| Clinicopathological parameter | No. of cases | HOTAIR expression | P value |
| Age (yr) | |||
| < 6 | 49 | 3.69 ± 1.94 | 0.188 |
| ≥ 60 | 31 | 3.45 ± 1.55 | |
| Gender | |||
| Male | 43 | 3.91 ± 1.85 | 0.761 |
| Female | 37 | 3.23 ± 1.68 | |
| Tumor size (cm) | |||
| < 7 | 38 | 3.59 ± 1.59 | 0.599 |
| ≥ 7 | 42 | 3.60 ± 1.81 | |
| Lymph node metastasis | |||
| Yes | 48 | 4.27 ± 1.54 | 0.024 |
| No | 32 | 3.11 ± 1.92 | |
| Tumor differentiation | |||
| High and moderate | 41 | 2.93 ± 1.62 | 0.019 |
| Low and undifferentiated | 39 | 4.35 ± 1.82 | |
| TNM stage | |||
| I + II | 46 | 3.17 ± 1.77 | 0.034 |
| III + IV | 34 | 3.87 ± 1.66 |
| Variable | Regression coefficient | SE | χ2 | P value | OR | 95%CI for OR | |
| Lower | Upper | ||||||
| TNM stage | 0.732 | 0.345 | 4.489 | 0.034 | 2.0790 | 1.056 | 4.090 |
| Lymph node metastasis | -2.512 | 1.088 | 5.325 | 0.021 | 0.0081 | 0.010 | 0.685 |
| HOTAIR expression | -2.048 | 0.785 | 6.806 | 0.090 | 0.1290 | 0.028 | 0.601 |
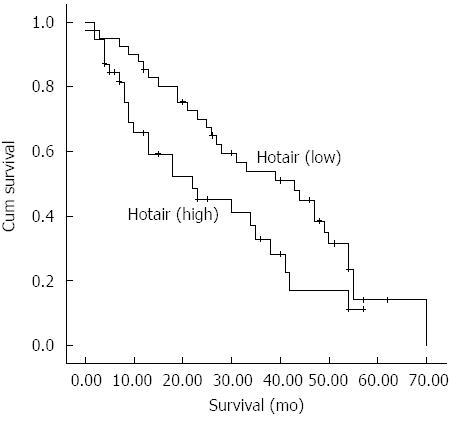
The Kaplan-Meier method was used to assess the impact of HOTAIR expression on survival of patients with colon cancer. The cumulative survival rate was significantly higher in patients with low HOTAIR expression than in those with high HOTAIR expression (P < 0.05) (Figure 3).
Using prognosis of colon cancer patients as the dependent variable and factors possibly influencing the prognosis as independent variables, Cox multiple regression analysis was performed. The results showed that TNM stage, lymph node metastasis and HOTAIR expression were independent risk factors for prognosis of colon cancer patients.
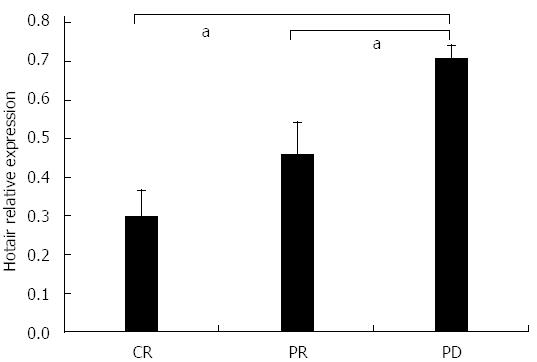
The relationship between prognosis of colon cancer patients after chemotherapy and HOTAIR expression was analyzed. The results showed that high HOTAIR expression was associated with poorer prognosis (Figure 4).
Colon cancer is a common malignancy[15,16]. With the development of diagnostic technology, advances in endoscopy and imaging techniques, as well as the clinical application of the carcinoembryonic antigen assay, have greatly improved the early diagnosis and treatment effect of colon cancer[17,18]. Patients with early colon cancer have localized lesions, and surgery with adjuvant radiochemotherapy is the preferred treatment, which is often associated with a good prognosis. However, because of the unbalanced regional development in China, many patients with colon cancer, especially those in rural regions, are diagnosed at advanced stages, and some patients even present with metastases as the first manifestation. Although drugs targeting EGFR and KRAS mutations have been effective in some patients with colon cancer[1-3], molecular targeted drugs, which often target only one or several molecules, are not suitable for all patients because of the complexity etiology of colon cancer. Therefore, there is an urgent need to find new therapeutic targets.
LncRNAs are non-protein coding transcripts of around 200 nucleotides that are widely distributed in the genome. Many lncRNAs can bind to DNA binding proteins and alter the chromosome state to participate in the regulation of many genes[6,19]. HOTAIR is an lncRNA located in the HOXC locus, and it can interact with polycomb repressive complex 2 and mediate the histone H3 lysine 27 methylation and lysine 4 demethylation in the HOXD locus, in which EZH2 also plays an important role[9,20,21]. HOTAIR can alter the state of chromosomes, thus affecting the expression of many genes. Researchers have found that HOTAIR expression is upregulated in cancer tissue samples from patients with breast cancer, pancreatic cancer, liver cancer, gastric cancer, or non-small cell lung cancer, and the expression is even higher in metastatic tissue. Both in vivo and in vitro studies have confirmed that upregulated expression of HOTAIR enhances the ability of tumors to invade and metastasize[7-9]. The aim of this study was to detect the expression of HOTAIR in tissue samples from patients with colon cancer, analyze the relationship between HOTAIR expression and clinicopathological parameters and explore the role of HOTAIR in colon cancer development and metastasis.
The results showed that the expression of HOTAIR is upregulated in colon cancer, suggesting that HOTAIR may act as an oncogene in the development of colon cancer. We also discovered that HOTAIR expression was significantly higher in lowly differentiated and undifferentiated cases compared with highly and moderately differentiated cases; in stages III + IV cases compared with stages I + II cases; and in cases with lymph node metastasis compared with those without. These results are similar to the findings of a previous study[22]; however, that study found that the expression of HOTAIR did not differ significantly between cases with and without lymph node metastasis, but was significantly higher in cases with liver metastasis compared with those without. The present study did not compare the HOTAIR expression between cases with and without liver metastasis. Low differentiation, late stage or lymph node metastasis in colon cancer are often associated with poor prognosis; therefore, our findings need to be validated by studies with a larger sample size.
Although HOTAIR might affect response to therapy in some tumors; for example, HOTAIR is associated with resistance to chemotherapy in ovarian cancer and sarcoma[23,24], there have been no reports in colon cancer. Our study, together with previous research, found that HOTAIR has an impact on the biological behavior of colon cancer[13], and detecting the level HOTAIR in blood could be used to predict prognosis of colon cancer. We speculated that this finding may be related to the role of HOTAIR in chemotherapy resistance, and this, therefore, was the focus of this study. We found that tumors with high expression of HOTAIR tended to develop resistance to chemotherapy, which may be the reason that high expression of HOTAIR is associated with a poor prognosis.
This finding also suggested that it is essential to explore the relationship between HOTAIR and resistance to chemotherapy in vitro, as well as the impact of HOTAIR on the biological behavior of tumor cells. Several studies have revealed that HOTAIR has an important role in tumor metastasis. On the basis of these findings, our subsequent follow-up study will expand the sample size to conduct prognostic and survival analyses to further define the relationship between HOTAIR and tumor metastasis in colon cancer, to explore the mechanism of pathogenesis of this malignancy and provide new targets for molecular therapy for colon cancer patients.
Long noncoding RNAs are non-protein coding transcripts of around 200 nucleotides that are widely distributed in the genome.
The expression of HOTAIR, a long noncoding RNA, is upregulated in cancer tissue samples from patients with breast cancer, pancreatic cancer, liver cancer, gastric cancer, or non-small cell lung cancer, and the expression is even higher in metastatic tissue.
High expression of HOTAIR tended to develop resistance to chemotherapy, which may be the reason that high expression of HOTAIR is associated with a poor prognosis.
By exploration of the mechanism of HOTAIR expression in colon cancer, the authors might identify new targets for molecular therapy for colon cancer patients.
HOTAIR might acts as an oncogene and represents a new molecular target for the treatment of colon cancer.
It is a very good and interesting study. The authors found that the expression of HOTAIR was significantly higher in colon cancer tissues than in matched tumor-adjacent normal colon tissues. HOTAIR expression was significantly higher in cases with lymph node metastasis than in those without metastasis, in lowly differentiated and undifferentiated cases than in highly and moderately differentiated cases. HOTAIR expression is upregulated in colon cancer, which may plays an important role in tumorigenesis, development and metastasis of colon cancer.
P- Reviewer: Ong J, Osuga T, Roman ID S- Editor: Ma YJ L- Editor: Stewart G E- Editor: Ma S
| 1. | Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknæs M, Hektoen M, Lind GE, Lothe RA. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 525] [Cited by in RCA: 674] [Article Influence: 56.2] [Reference Citation Analysis (0)] |
| 2. | Corso G, Pascale V, Flauti G, Ferrara F, Marrelli D, Roviello F. Oncogenic mutations and microsatellite instability phenotype predict specific anatomical subsite in colorectal cancer patients. Eur J Hum Genet. 2013;21:1383-1388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Mouradov D, Domingo E, Gibbs P, Jorissen RN, Li S, Soo PY, Lipton L, Desai J, Danielsen HE, Oukrif D. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am J Gastroenterol. 2013;108:1785-1793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 4. | Cekaite L, Eide PW, Lind GE, Skotheim RI, Lothe RA. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget. 2016;7:6476-6505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 5. | Esin E, Yalcin S. Maintenance strategy in metastatic colorectal cancer: A systematic review. Cancer Treat Rev. 2016;42:82-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 6. | Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4004] [Cited by in RCA: 3939] [Article Influence: 246.2] [Reference Citation Analysis (0)] |
| 7. | Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 203] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 8. | Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616-1625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 631] [Cited by in RCA: 685] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 9. | Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-1076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4202] [Cited by in RCA: 4229] [Article Influence: 281.9] [Reference Citation Analysis (0)] |
| 10. | Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 305] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 11. | Xue Y, Ma G, Gu D, Zhu L, Hua Q, Du M, Chu H, Tong N, Chen J, Zhang Z. Genome-wide analysis of long noncoding RNA signature in human colorectal cancer. Gene. 2015;556:227-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H, Tong N, Chen J, Zhang Z, Wang M. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30:303-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 13. | Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, Qiu GQ, Peng ZH, Yan DW. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 173] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 14. | Chikamatsu K, Aono A, Kato T, Takaki A, Yamada H, Sasaki Y, Izumi K, Yi L, Mitarai S. COBAS® TaqMan® MTB, smear positivity grade and MGIT culture; correlation analyses of three methods for bacillary quantification. J Infect Chemother. 2016;22:19-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 15. | Shahbazian H, Nasuri Y, Hosseini SM, Arvandi S, Razzaghi S. A report of the frequency of colorectal carcinoma and involved lymph nodes in South-West Iran. Indian J Med Paediatr Oncol. 2016;37:38-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 16. | Lien GS, Lin CH, Yang YL, Wu MS, Chen BC. Ghrelin induces colon cancer cell proliferation through the GHS-R, Ras, PI3K, Akt, and mTOR signaling pathways. Eur J Pharmacol. 2016;776:124-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 17. | Lloyd JM, McIver CM, Stephenson SA, Hewett PJ, Rieger N, Hardingham JE. Identification of early-stage colorectal cancer patients at risk of relapse post-resection by immunobead reverse transcription-PCR analysis of peritoneal lavage fluid for malignant cells. Clin Cancer Res. 2006;12:417-423. [PubMed] |
| 18. | Zaenker P, Ziman MR. Serologic autoantibodies as diagnostic cancer biomarkers--a review. Cancer Epidemiol Biomarkers Prev. 2013;22:2161-2181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 19. | Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667-11672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2182] [Cited by in RCA: 2350] [Article Influence: 146.9] [Reference Citation Analysis (0)] |
| 20. | Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311-1323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3533] [Cited by in RCA: 3389] [Article Influence: 188.3] [Reference Citation Analysis (0)] |
| 21. | Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2754] [Cited by in RCA: 2649] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 22. | Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320-6326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 1039] [Article Influence: 74.2] [Reference Citation Analysis (0)] |
| 23. | Milhem MM, Knutson T, Yang S, Zhu D, Wang X, Leslie KK, Meng X. Correlation of MTDH/AEG-1 and HOTAIR Expression with Metastasis and Response to Treatment in Sarcoma Patients. J Cancer Sci Ther. 2011;S5:pii 004. [PubMed] |
| 24. | Teschendorff AE, Lee SH, Jones A, Fiegl H, Kalwa M, Wagner W, Chindera K, Evans I, Dubeau L, Orjalo A. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 2015;7:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 127] [Article Influence: 12.7] [Reference Citation Analysis (0)] |