Published online Jul 21, 2015. doi: 10.3748/wjg.v21.i27.8382
Peer-review started: January 27, 2015
First decision: March 10, 2015
Revised: March 19, 2015
Accepted: May 4, 2015
Article in press: May 4, 2015
Published online: July 21, 2015
Processing time: 176 Days and 17.9 Hours
AIM: To evaluate tumor necrosis factor-α converting enzyme (TACE) methylation status in patients with chronic hepatitis B (CHB).
METHODS: Eighty patients with hepatitis B e antigen (HBeAg)-positive CHB, 80 with HBeAg-negative CHB, and 40 healthy controls (HCs) were randomly enrolled in this study. Genomic DNA was extracted from peripheral blood mononuclear cells and methylation status of TACE promoter was determined by methylation-specific polymerase chain reaction. The clinical and laboratory parameters were collected.
RESULTS: One hundred and thirty of 160 patients with CHB (81.25%) and 38 of 40 HCs (95%) displayed TACE promoter methylation. The difference was significant (χ2 = 4.501, P < 0.05). TACE promoter methylation frequency in HBeAg-positive CHB (58/80, 72.5%) was significantly lower than that in HBeAg-negative CHB (72/80, 90%; χ2 = 8.041, P < 0.01) and HCs (χ2 = 8.438, P < 0.01). However, no significant difference was observed in the methylation frequency between HBeAg-negative CHB and HCs (χ2 = 0.873, P > 0.05). In the HBeAg-positive group, TACE methylation frequency was significantly negatively correlated with HBeAg (r = -0.602, P < 0.01), alanine aminotransferase (r = -0.461, P < 0.01) and aspartate aminotransferase (r = -0.329, P < 0.01).
CONCLUSION: Patients with HBeAg-positive CHB have aberrant demethylation of the TACE promoter, which may potentially serve as a biomarker for HBeAg seroconversion.
Core tip: We retrospectively recruited 80 patients with hepatitis B e antigen (HBeAg)-positive chronic hepatitis B (CHB), 80 with HBeAg-negative CHB, and 40 healthy controls. We evaluated tumor necrosis factor-α converting enzyme (TACE) promoter methylation status in peripheral blood mononuclear cells and analyzed the association between TACE methylation status and clinical features. Aberrant demethylation of the TACE promoter in HBeAg-positive CHB was associated with high HBeAg, alanine aminotransferase and aspartate aminotransferase levels. These findings imply that demethylation of the TACE promoter may potentially serve as a biomarker for HBeAg seroconversion.
- Citation: Wang ZL, Gao S, Li XY, Sun FK, Li F, Fan YC, Wang K. Demethylation of tumor necrosis factor-α converting enzyme promoter associated with high hepatitis B e antigen level in chronic hepatitis B. World J Gastroenterol 2015; 21(27): 8382-8388
- URL: https://www.wjgnet.com/1007-9327/full/v21/i27/8382.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i27.8382
Hepatitis B virus (HBV) has chronically infected > 350 million people throughout the world and results in 500000 to 700000 deaths annually[1,2]. China has an estimated 120 million people chronically infected with HBV[3]. Nearly 15%-40% of HBV carriers will progress to life-threatening complications including liver cirrhosis[4], decompensated liver disease[5], or hepatocellular carcinoma (HCC)[6,7]. Hepatitis B e antigen (HBeAg) seroconversion is a crucial step during the progression of HBV infection. However, there has been no sensitive and effective model for predicting the occurrence of HBeAg seroconversion until now.
DNA methylation is one of the most important epigenetic mechanisms, which denotes the addition of a methyl group to DNA. It is widespread in the human genome and mainly occurs at cytosine adjacent to guanine (CpG dinucleotides)[8]. DNA methylation in gene promoter regions often results in long-term silencing of gene expression[9]. Meanwhile, aberrant promoter demethylation is usually associated with overexpression of genes that might participate in pathogenesis of many diseases[10]. Previous studies demonstrated that aberrant demethylation in the promoter region of genes occurs in many diseases and may be used as a biomarker[11,12].
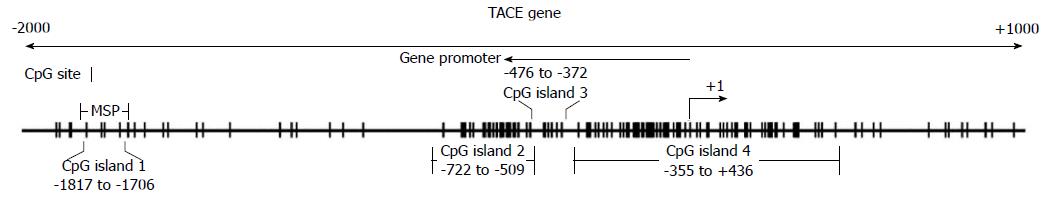
Tumor necrosis factor (TNF)-α-converting enzyme (TACE) is a modular transmembrane protein with a zinc-dependent catalytic domain[13]. The main function of TACE is to mediate cleavage of substrates such as TNF-α, TNF receptor type I and type II[13-15]. It is well documented that the overexpression of TACE is associated with several human diseases, including liver cancer. The human TACE promoter has not been definitely characterized to date. In the present study, we recognized the region within 2000 bp upstream of the transcriptional initiation site as a presumptive promoter region for the TACE gene, as reported previously[16-18]. GenBank indicates that there are four CpG islands located on the TACE promoter sequence (Figure 1). Therefore, aberrant demethylation of the TACE gene promoter may occur in patients with chronic hepatitis B (CHB) and may be associated with disease progression.
In this study, we determined the methylation status of the TACE promoter in patients with HBeAg-positive CHB, HBeAg-negative CHB and healthy controls (HCs), and evaluated the relationship between TACE promoter methylation status and clinical features.
Eighty patients with HBeAg-positive CHB, 80 with HBeAg-negative CHB and 40 HCs were randomly enrolled from July 2012 to June 2014 at the Department of Hepatology, Qilu Hospital of Shandong University, China. Chronic HBV infection was defined as the presence of hepatitis B surface antigen (HBsAg) for > 6 mo prior to the beginning of this study[2]. The HCs were recruited from among eligible blood donors, who had no history of liver diseases. Patients were excluded if they met any of the following criteria: (1) co-infection with hepatitis C virus or HIV; (2) other liver diseases such as autoimmune hepatitis and alcoholic hepatitis; (3) receiving antioxidant agent or interferon therapy; (4) pregnancy; (5) decompensated liver disease or HCC. Prior to sample collection, informed consent was obtained from each participant and the study was approved by the local Ethics Committee of Qilu Hospital of Shandong University.
Peripheral blood mononuclear cells (PBMCs) were isolated using gradient centrifugation via Ficoll-Paque (Pharmacia Diagnostics, Uppsala, Sweden) and stored at -20 °C until use. Genomic DNA was extracted from PBMCs using QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, United States) and stored at -20 °C.
Extracted DNA was treated with sodium bisulfite using an EZ DNA methylation kit (Zymo Research, Orange, CA, United States) and then used to perform methylation-specific polymerase chain reaction (MSP). Methylated and unmethylated primers specific for TACE promoters were designed using MethPrimer (Table 1)[19]. There were four CpG islands in the promoter region of the TACE gene. The reasons for choosing this area included: (1) the primers contained at least one CpG site at the 3’-end; (2) the primers in the M pair and U pair contained the same CpG sites within their sequence; and (3) two sets of primers had similar product Tm values, which were 68.1 °C for the M pair and 67.3 °C for the U pair[18]. The selected primer sets were used to amplify the bisulfite-modified DNA in our study. The M pair primers amplified the -1831 to -1686 site of the 5’-UTR of the TACE gene (+1 for the transcriptional start site). Meanwhile, the U pair primers amplified the -1832 to -1686 site of the 5’-UTR of the TACE gene (Figure 1). MSP was performed in a total volume of 25 μL containing 1 μL bisulfite-treated DNA, 0.5 μL of each primer (10 μmol/L), 10.5 μL nuclease-free water, and 12.5 μL Premix Taq (Zymo Research, Irvine, CA, United States), which consisted of Taq DNA polymerase, reaction buffer, and dNTP mixture. The PCR protocol was composed of an initial denaturation at 95 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C for 40 s, and primer extension at 72 °C for 40 s; followed by final extension at 72 °C for 10 min. Water blank without DNA was used in each round of PCR. PCR products were electrophoresed on a 2% agarose gel, stained with ethidium bromide, and visualized under UV illumination.
| Primer name | Primer sequence (5’-3’) | Product size (bp) | Annealing temperature (°C) |
| M | F: GGAGTTTGAGATTAGTTTGGTTAAC | 146 | 56 |
| R: TAAACACCTCCTAAATTTAAACGAT | |||
| U | F: AGGAGTTTGAGATTAGTTTGGTTAATG | 147 | 56 |
| R: TAAACACCTCCTAAATTTAAACAAT |
Fasting venous blood was collected from each subject. HBeAg was detected by an automatic analyzer (Cobas 6000; Roche Diagnostics, Basel, Switzerland). The HBV DNA level was measured by a real-time PCR system (ABI 7300; Applied Biosystems, Foster City, CA, United States). The serum biochemical markers (Cobas Integra 800; Roche Diagnostics, Mannheim, Germany) included alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL), albumin, and creatinine. Hemostasis markers (ACL TOP 700, Instrument Laboratory, Lexington, MA, United States) included prothrombin time-international normalized ratio (PT-INR), and prothrombin time activity (PTA). These markers were measured using standard methodologies in the Department of Laboratory Medicine, Qilu Hospital, Shandong University, China.
Quantitative variables are expressed as the mean (24th-75th percentiles). Categorical variables are expressed as number (percentage). Statistical analyses were performed with SPSS version 16.0 (SPSS Inc., Chicago, IL, United States). Categorical variables were compared by χ2 test. Quantitative variables were compared by Student’s t test or Mann-Whitney U test. Spearman correlation coefficients were calculated to evaluate correlations between TACE methylation status and clinicopathological parameters. All statistical analyses were two-sided and P < 0.05 was considered statistically significant. The statistical methods of this study were reviewed by Dr. Shu-Mei Wang from Department of Epidemiology and Biostatistics, School of Public Health, Shandong University.
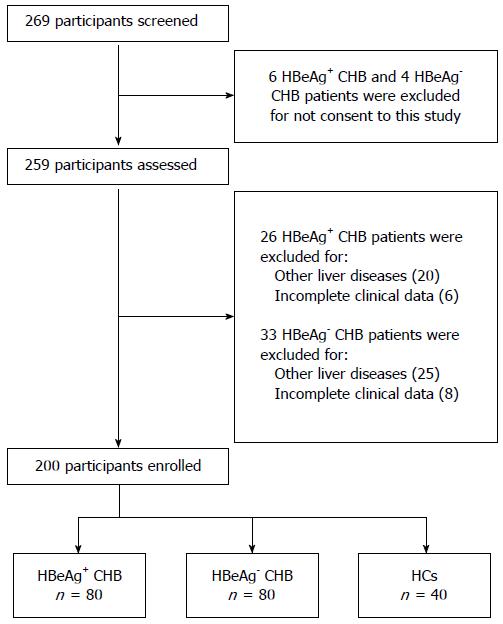
From June 2012 to July 2014, 269 participants (112 HBeAg-positive CHB patients, 117 HBeAg-negative CHB patients, and 40 HCs) were screened at the Department of Hepatology, Qilu Hospital. Six HBeAg-positive CHB patients and four HBeAg-negative patients were excluded for not giving consent to the study. Twenty-six HBeAg-positive CHB patients were excluded because of other liver diseases (n = 20) or incomplete clinical data (n = 6). Thirty-three HBeAg-negative CHB patients were excluded because of other liver diseases (n = 25) or incomplete clinical data (n = 8), leaving 200 participants. In the enrolled participants, there were 80 patients in the HBeAg-positive group, 80 in the HBeAg-negative group, and 40 in the HC group (Figure 2). The basic characteristics of the enrolled subjects are summarized in Table 2.
| Variable | HBeAg+ CHB group | HBeAg- CHB group | HC group |
| Number | 80 | 80 | 40 |
| Age (yr) | 41.50 (33.25-51.0) | 45.00 (33.25-52.0) | 36.50 (31.0-44.50) |
| Male sex, n (%) | 57 (71.25) | 53 (66.25) | 28 (70.00) |
| HBeAg | 132.85 (24.30-512.12) | NA | NA |
| Log10 (HBV DNA) | 5.23 (3.90-6.41) | 3.08 (0.0-5.43) | NA |
| ALT (U/L) | 189.50 (78.25-441.75) | 91.50 (44.0-236.50) | NA |
| AST (U/L) | 106.0 (46.0-287.75) | 56.50 (30.0-108.75) | NA |
| TBIL (μmol/L) | 16.95 (11.10-20.0) | 18.75 (15.00-21.18) | NA |
| ALB (g/L) | 44.85 (42.83-48.48) | 45.56 (42.05-49.38) | NA |
| Cr (μmol/L) | 63.0 (53.0-75.50) | 64.00 (56.0-77.75) | NA |
| PT-INR | 1.02 (0.99-1.09) | 1.01 (0.96-1.05) | NA |
| PTA (%) | 81.50 (71.25-93.0) | 87.0 (80.0-102.0) | NA |
| Methylation, n (%) | 58 (72.5) | 72 (90) | 38 (95) |
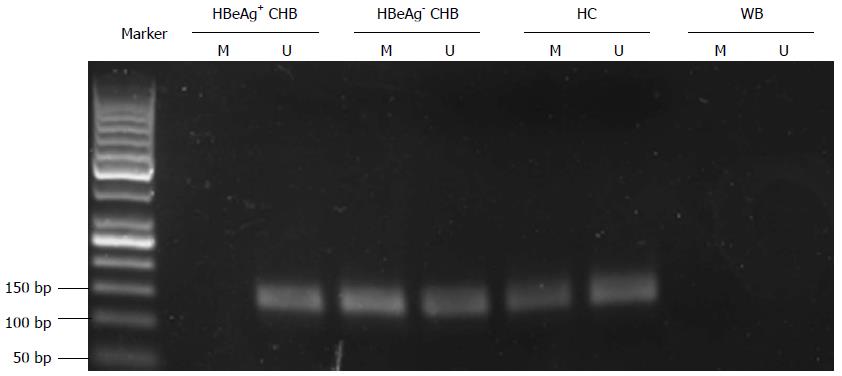
The methylation status of the TACE promoter was determined in PBMCs of all participants (Figure 3). One hundred and thirty of the 160 patients with CHB (81.25%) and 38 of the 40 HCs (95%) displayed aberrant TACE promoter methylation and the difference was significant (χ2 = 4.501, P < 0.05). TACE promoter methylation frequency in the HBeAg-positive CHB group (58/80, 72.5%) was significantly lower than that in the HBeAg-negative CHB (72/80, 90%; χ2 = 8.041, P < 0.01) and HC (χ2 = 8.438, P < 0.01) groups. However, no significant difference could be observed in the methylation frequency between the HBeAg-negative CHB and HC groups (χ2 = 0.873, P > 0.05) groups.
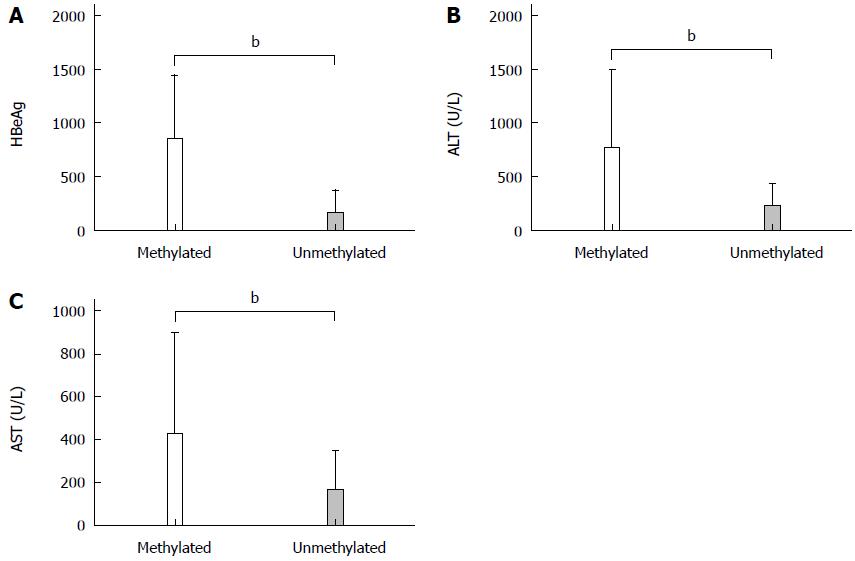
In the HBeAg-positive CHB group, there was a negative correlation between TACE promoter methylation and HBeAg (r = -0.602, P < 0.01), ALT (r = -0.461, P < 0.01) and AST (r = -0.329, P < 0.01) (Figure 4). There was no significant correlation between TACE promoter methylation and sex (r = 0.011, P = 0.922), age (r = 0.119, P = 0.294), log10 HBV DNA (r = 0.203, P = 0.07), albumin (r = -0.045, P = 0.689), TBIL (r = -0.128, P = 0.258), PT-INR (r = 0.027, P = 0.810), PTA (r = -0.138, P = 0.223) and creatinine (r = 0.079, P = 0.484).
In the HBeAg-negative CHB group, there was no significant correlation between TACE promoter methylation and sex (r = -0.009, P = 0.934), age (r = 0.023, P = 0.842), HBeAg (r = 0.076, P = 0.500), log10 HBV DNA (r = 0.060, P = 0.596), ALT (r = -0.111, P = 0.327), AST (r = -0.073, P = 0.519), albumin (r = 0.016, P = 0.886), TBIL (r = -0.144, P = 0.201), PT-INR (r = -0.195, P = 0.084), PTA (r = 0.156, P = 0.167) and creatinine (r = -0.023, P = 0.836).
We investigated the methylation status of the TACE gene in PBMCs of 80 patients with HBeAg-positive CHB, 80 patients with HBeAg-negative CHB, and 40 HCs. TACE promoter methylation frequency in HBeAg-positive CHB patients (58/80, 72.5%) was significantly lower than that in HBeAg-negative CHB patients (72/80, 90%; χ2 = 8.041, P < 0.01) and HCs (χ2 = 8.438, P < 0.01). However, no significant difference could be observed in the methylation frequency between the HBeAg-negative CHB patients and HCs (χ2 = 0.873, P > 0.05). In the HBeAg-positive group, the TACE methylation frequency was significantly negatively correlated with HBeAg (r = -0.602, P < 0.01), ALT (r = -0.461, P < 0.01) and AST (r = -0.329, P < 0.01).
The prevalence of hepatitis B is a major concern worldwide. Approximately one-third of the global population has serological evidence of past or present infection with HBV, and 350-400 million people are chronic HBsAg carriers. Once chronically infected, the covalently closed circular DNA of HBV is hard to eradicate from the nucleus of hepatocytes[20,21]. People with hepatitis B are at an increased risk of developing hepatic decompensation, cirrhosis, and HCC[1]. HBeAg seroconversion is a crucial step during the progression of HBV infection. Until now, no sensitive and effective model for predicting the occurrence of HBeAg seroconversion has been proposed.
DNA methylation and demethylation are important epigenetic mechanisms. Demethylation of a promoter region is often associated with long-term gene overexpression and is linked to many diseases[22-24]. DNA methylation status is usually stable and suitable for use as a biomarker for disease detection and prognosis prediction[25]. The present study revealed that the TACE methylation frequency was significantly lower in HBeAg-positive CHB than in HBeAg-negative CHB and HCs. Also, the TACE methylation frequency was negatively correlated with serum ALT and AST. Aberrant TACE methylation status may have a role in the progression of HBV infection and induce liver damage.
Most importantly, this study revealed that TACE demethylation was significantly associated with the HBeAg level in HBeAg-positive CHB, which indicated that it might possess a potential value for predicting HBeAg seroconversion. Further studies are needed to prove its usefulness. The prediction of HBeAg seroconversion is essential for the management of HBV infection. This finding might help clinicians initiate the correct treatment strategy at an early stage and prevent many patients from developing fatal complications such as hepatic decompensation, cirrhosis, or HCC.
There were several limitations in this study. The sample size was relatively small and we could not confirm our results in a second cohort. Therefore, our findings need further validation with large studies prior to clinical application, and the follow-up of the HBeAg-positive CHB patients should also be performed in a further study. Prospective studies with long-term follow-up would be useful for determining the predictive values for HBeAg seroconversion. This is an exploratory study and further studies are needed to reveal the mechanisms involved.
In conclusion, this study demonstrated demethylation of the TACE promoter in HBeAg-positive CHB, which was associated with HBeAg level. TACE promoter demethylation may potentially serve as a marker for HBeAg seroconversion.
Hepatitis B e antigen (HBeAg) seroconversion is a crucial step during the progression of hepatitis B virus (HBV) infection. Nevertheless, there have been no sensitive and effective markers for predicting the occurrence of HBeAg seroconversion until now. Tumor necrosis factor (TNF)-α converting enzyme (TACE) have been demonstrated to be involved in liver inflammation. GenBank indicates that there are four CpG islands located on the TACE promoter sequence. However, the methylation status of the TACE promoter in chronic hepatitis B (CHB) and its significance for HBeAg seroconversion have not been demonstrated.
HBeAg seroconversion is a crucial step during the progression of HBV infection. The current research hotspot is to find effective biomarkers for predicting the occurrence of HBeAg seroconversion. DNA methylation is one of the most important epigenetic mechanisms. Aberrant methylation or demethylation of several genes has been associated with a number of diseases and many methylation biomarkers have been proposed.
This study demonstrated aberrant demethylation of the TACE promoter in HBeAg-positive CHB, which was associated with high HBeAg level, and indicated that TACE demethylation may potentially serve as a biomarker for HBeAg seroconversion.
Aberrant demethylation of TACE promoter may be used as a biomarker for predicting the occurrence of HBeAg seroconversion.
TACE is a modular transmembrane protein with a zinc-dependent catalytic domain. Its main function is to mediate cleavage of substrates such as TNF-α, TNF receptor I and TNF receptor II. DNA methylation is one of the most important epigenetic mechanisms, which donates a methyl group to DNA. A biomarker is a substance used as an indicator of a biological state.
This manuscript presented some useful data showing that aberrant demethylation of TACE promoter existed in HBeAg-positive CHB, which was associated with high HBeAg level. This might potentially serve as a biomarker for HBeAg seroconversion. The research design, and the data analysis and statistics are reasonable.
P- Reviewer: Celikbilek M, He ST, Kemik O S- Editor: Qi Y L- Editor: Cant MR E- Editor: Liu XM
| 1. | Liaw YF, Chu CM. Hepatitis B virus infection. Lancet. 2009;373:582-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 993] [Article Influence: 62.1] [Reference Citation Analysis (1)] |
| 2. | Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45:507-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1794] [Cited by in RCA: 1778] [Article Influence: 98.8] [Reference Citation Analysis (0)] |
| 3. | Cui Y, Jia J. Update on epidemiology of hepatitis B and C in China. J Gastroenterol Hepatol. 2013;28 Suppl 1:7-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 245] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 4. | Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1764] [Cited by in RCA: 1843] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 5. | Sato S, Suzuki K, Akahane Y, Akamatsu K, Akiyama K, Yunomura K, Tsuda F, Tanaka T, Okamoto H, Miyakawa Y. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann Intern Med. 1995;122:241-248. [PubMed] |
| 6. | Lok AS. Chronic hepatitis B. N Engl J Med. 2002;346:1682-1683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 337] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 7. | Bosch FX, Ribes J, Cléries R, Díaz M. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2005;9:191-211, v. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 625] [Cited by in RCA: 605] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 8. | Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215-219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1914] [Cited by in RCA: 1591] [Article Influence: 93.6] [Reference Citation Analysis (0)] |
| 9. | Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3811] [Cited by in RCA: 4209] [Article Influence: 323.8] [Reference Citation Analysis (0)] |
| 10. | Qian J, Chen Q, Yao DM, Yang L, Yang J, Wen XM, Zhang YY, Chai HY, Ma JC, Deng ZQ. MOK overexpression is associated with promoter hypomethylation in patients with acute myeloid leukemia. Int J Clin Exp Pathol. 2015;8:127-136. [PubMed] |
| 11. | Zhang W, Jiao H, Zhang X, Zhao R, Wang F, He W, Zong H, Fan Q, Wang L. Correlation between the expression of DNMT1, and GSTP1 and APC, and the methylation status of GSTP1 and APC in association with their clinical significance in prostate cancer. Mol Med Rep. 2015;12:141-146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 12. | Søes S, Daugaard IL, Sørensen BS, Carus A, Mattheisen M, Alsner J, Overgaard J, Hager H, Hansen LL, Kristensen LS. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience. 2014;1:367-374. [PubMed] |
| 13. | Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2422] [Cited by in RCA: 2419] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 14. | Reddy P, Slack JL, Davis R, Cerretti DP, Kozlosky CJ, Blanton RA, Shows D, Peschon JJ, Black RA. Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. J Biol Chem. 2000;275:14608-14614. [PubMed] |
| 15. | Condon TP, Flournoy S, Sawyer GJ, Baker BF, Kishimoto TK, Bennett CF. ADAM17 but not ADAM10 mediates tumor necrosis factor-alpha and L-selectin shedding from leukocyte membranes. Antisense Nucleic Acid Drug Dev. 2001;11:107-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 53] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 16. | Akizu N, Estarás C, Guerrero L, Martí E, Martínez-Balbás MA. H3K27me3 regulates BMP activity in developing spinal cord. Development. 2010;137:2915-2925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Li C, Chu N, Wang B, Wang J, Luan J, Han L, Meng D, Wang Y, Suo P, Cheng L. Polymorphisms in the presumptive promoter region of the SLC2A9 gene are associated with gout in a Chinese male population. PLoS One. 2012;7:e24561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Han LY, Fan YC, Mu NN, Gao S, Li F, Ji XF, Dou CY, Wang K. Aberrant DNA methylation of G-protein-coupled bile acid receptor Gpbar1 (TGR5) is a potential biomarker for hepatitis B Virus associated hepatocellular carcinoma. Int J Med Sci. 2014;11:164-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 19. | Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427-1431. [PubMed] |
| 20. | Raimondo G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, Craxì A, Donato F, Ferrari C, Gaeta GB. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol. 2008;49:652-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 606] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 21. | Pollicino T, Saitta C, Raimondo G. Hepatocellular carcinoma: the point of view of the hepatitis B virus. Carcinogenesis. 2011;32:1122-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 22. | Rochtus A, Izzi B, Vangeel E, Louwette S, Wittevrongel C, Lambrechts D, Moreau Y, Winand R, Verpoorten C, Jansen K. DNA methylation analysis of Homeobox genes implicates HOXB7 hypomethylation as risk factor for neural tube defects. Epigenetics. 2015;10:92-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Yuan R, Zhi Q, Zhao H, Han Y, Gao L, Wang B, Kou Z, Guo Z, He S, Xue X. Upregulated expression of miR-106a by DNA hypomethylation plays an oncogenic role in hepatocellular carcinoma. Tumour Biol. 2015;36:3093-3100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 24. | Augello C, Gianelli U, Falcone R, Tabano S, Savi F, Bonaparte E, Ciboddo M, Paganini L, Parafioriti A, Ricca D. PDGFB hypomethylation is a favourable prognostic biomarker in primary myelofibrosis. Leuk Res. 2015;39:236-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 25. | Chan TA, Baylin SB. Epigenetic biomarkers. Curr Top Microbiol Immunol. 2012;355:189-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |