INTRODUCTION
Hepatitis B is a worldwide public health problem. In Far-East Asia and tropical Africa, chronic carriers of hepatitis B virus (HBV) represent 10% or more of population[1]. Both chronic hepatitis B and hepatitis C are prevalent among the 12 million Asians and Pacific Islanders living in the USA[2]. It is still a potential risk for public health as a result of immigration from countries with a high prevalence of HBV infection although there is a low incidence rate among the indigenous population in North America and Scandinavian countries[3-5]. Chronic active hepatitis and liver cirrhosis are major causes of mortality[1,6]. Moreover, epidemiological studies have clearly shown the importance of HBV in hepatocellular carcinoma (HCC). In China, 500000 to one million new cases appear every year[1] and 73% of patients with chronic hepatitis and 78% and 71% of those with cirrhosis and HCC, respectively, are HBsAg positive[7]. Therefore, studies on transmission of HBV are of substantial importance in virology as well as in public health.
The transmission routes of HBV through blood transfusion[8,9]; body fluids including serum, saliva, vaginal secretions, breast milk, and semen[10-14]; intrauterine infection[15,16]; cell, tissue, and organ transplantation[17,18] and others including hemodialysis units and intravenous drug injection[19-23] have been documented. Hadchouel et al[24], using molecular hybridization, confirmed the presence of integrated HBV DNA sequences in spermatozoa from two of three patients with HBV infection. They assumed that the presence of integrated sequences in spermatozoa suggested the possibility of true vertical transmission of HBV via the germ line. Using interspecific in vitro fertilization between zona-free hamster ova and spermatozoa from nine patients with HBV infection, we prepared human sperm metaphases for fluorescence in situ hybridization (FISH) with the biotin-labeled full length HBV DNA probe. The specific fluorescent signal spots for HBV DNA were seen in sperm chromosomes of one patient with chronic persistent hepatitis. Our results suggest that there is evidence that HBV DNA can integrate into human sperm chromosomes and that human sperms with HBV DNA sequences can normally complete the fertilization with oocytes and zygotes can develop to the first cleavage. The above studies provided some evidence for the feasibility of HBV vertical transmission via spermatozoa. Zhong et al[25] analyzed HBV nucleotide sequences isolated from three sets of mother/child pairs and found that the nucleotide and deduced amino acid sequences were absolutely identical between the mother and children in two of three families. Their results suggested the vertical transmission linkage of HBV between the mother and children. An interesting question is that whether HBV vertically transmits from mother to offspring via oocytes. To answer this question, the presence and integration of HBV DNA in oocytes should be confirmed. This is the purpose of our present study.
MATERIALS AND METHODS
Preparation of mouse oocytes
Mature female mice were induced to superovulate by intramuscular injection of 0.2 IU/g body weight of pregnant mare serum gonadotropin (PMSG, Ningbo Hormone Products Co., Ltd, China) into the inguinal region on d 1 of estrous cycle followed by administration of 0.2 IU/g body weight of human chorionic gonadotropin (HCG, Ningbo Hormone Products Co., Ltd) after 48-58 h. Superovulated oocytes were collected from ampullar region of oviducts 16 h after HCG injection and freed from cumulus cells by gently pipetting in 0.1% hyaluronidase and then washed thrice in modified TYH medium (mTYH)[26]. The oocytes were divided into two groups. Group A was transferred into mTYH containing 0.2 μg/mL pBR322-HBV DNA plasmids and group B into HBV DNA-free mTYH. The oocytes of two groups were kept in a CO2 incubator (37 °C, 50 mL/L CO2 in air) for 2 h and then washed with mTYH eight times. The oocytes and washing solutions of the last three times were collected respectively.
Labeling of HBV DNA probe for Southern blot and dot hybridization
Recombinant plasmids, pBR322-HBV containing full length (3.2 kb) HBV genomic DNA, were taken to be amplified according to the routine PCR method. The PCR products, 591-bp HBV DNA fragments containing HBx coding region, were labeled with biotin using random primer biotin labeling of DNA (NEB® Phototope® Kit). Unincorporated nucleotides were separated by cold ethanol precipitation method.
Labeling of HBV DNA probe for FISH
Recombinant plasmids, pBR322-HBV containing full length (3.2 kb) HBV genomic DNA, were taken to be amplified according to the routine transformation method. The 3.2 kb HBV DNA probe was labeled with biotin-14-dATP by nick translation (BioNick DNA Labeling System, GIBCO BRL). Unincorporated nucleotides were separated by cold ethanol precipitation method.
DNA extraction
Cumulus-free oocytes from groups A and B were treated with 0.1% trypsin to remove the zona pellucida, then washed twice immediately in fresh mTYH. Genomic DNA was isolated from the zona-free oocytes through the digestion of lysis buffer, proteinase K and RNase I; the extraction of phenol-chloroform and the precipitation of ethanol.
Polymerase chain reaction (PCR)
The tested DNA samples were from group A oocytes in eight experiments. The positive and negative controls were from recombinant plasmids, pBR322-HBV and group B oocytes respectively.
Primers used for PCR amplification (Sangon Company, Shanghai, China) were as follows: P1 (up) (5’-gaactcctagcagc-ttgttt-3’); P2 (down) (5’-tgaacagtaggacatgaaca-3’). The expected size of the product was 591 bp and this fragment contained region X of HBV genome. A total of 50 μL reaction included 41.5 μL ddH2O, 5 μL 10×buffer, 1 μL dNTPs (10 mmol/L), 0.5 μL primer 1 (100 pmol/L), 0.5 μL primer 2 (100 pmol/L), 0.5 μL Taq DNA polymerase (5 IU/5 μL), 1 μL DNA template (1 μg/μL). The thermo-cycling conditions were at 94 °C for 5 min, and 35 cycles each at 94 °C for 1 min, at 50 °C for 1 min, at 72 °C for 1 min, and finally at 72 °C for 10 min in a Peltier thermal cycler (PTC 100). The amplification products were visualized by staining with ethidium bromide, after electrophoresis on 1% agarose gel.
Southern blot hybridization
Each 15 μL amplification product from PCR assays was separated by electrophoresis on 1% agarose gel and blotted onto a nylon membrane (Schleicher & Schuell BioScience, Inc., USA). Southern blot analysis was carried out using biotin DNA labeling and detection kits (New England Biolabs, Inc., USA) with the biotin-labeled HBV DNA sequence containing HBx coding region as a probe.
Dot hybridization
The tested samples were from the solutions of the last three washings of group A in each experiment. All of the 24 tested samples in eight experiments were collected for dot hybridization. The positive and negative controls were from recombinant plasmids, pBR322-HBV and the washing solutions of group B respectively.
Each 2 µL of the samples of washing solution was subjected to denaturing in boiling water bath for 5 min and dotted onto a nylon membrane (Schleicher & Schuell BioScience, Inc.). The blots were dried and baked for 2 h at 80 °C. Dot hybridization was carried out using the biotin-labeled HBV DNA sequence containing HBx coding region as a probe. After an overnight hybridization at 68 °C, the blots were treated successively with SSC, sodium dodecyl sulfate, blocking reagent, streptokinase, alkaline phosphatase, CDP-star assay buffer, followed by the procedures of exposure, development and fixation, and analysis.
Preparation of oocyte metaphases
Oocyte metaphases were prepared using a gradual fixation air-dry method[27]. In brief, oocytes were first exposed to a hypotonic solution containing 0.068 mol/L potassium chloride with 0.1% bovine serum albumin for about 30 min. The swollen oocytes were transferred to the bottom of a glass spot well pre-filled with fixative I (methanol:acetic acid:distilled water 5:1:3). When the color of oocytes changed from brown to white and subtransparent (after about 5-7 min), the oocytes were aspirated and then released gently onto the slide. Then fixative II (methanol:acetic acid 3:1) was dispensed carefully over the surface of the slide until it covered the oocytes. The slide was immersed in a Coplin jar containing fixative II for about 5 min or more. After that it was gently removed and placed in a Coplin jar containing fixative III (methanol:acetic acid:distilled water 3:3:1) for 1 min. The slide was slowly removed and dried gradually with a steam of moist air.
FISH of oocyte chromosomes
FISH and detection of signals were performed with the procedures described previously.
RESULTS
PCR of the DNA from oocytes
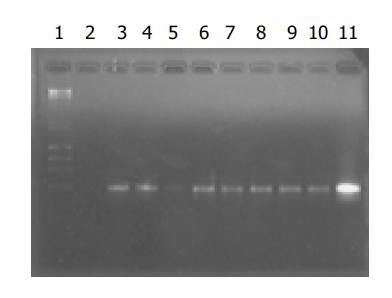
The positive bands for HBV DNA in eight samples from group A oocytes were observed. A bright positive band was seen in the positive control and no band in the negative control was seen (Figure 1).
Figure 1 Electrophoresis of amplification products of DNA from mouse oocytes.
Lane 1: markers; lane 2: negative control; lane 11: positive control; lanes 3-10: amplification products of DNA from group A oocytes in eight experiments.
Southern blot hybridization
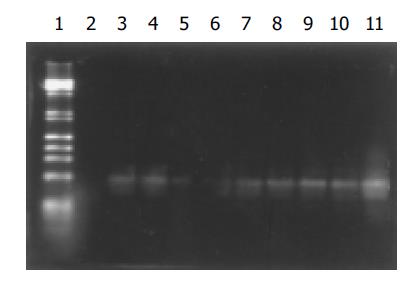
Consistent with electrophoresis of amplification products, Southern blot analysis revealed that the hybridization signals for HBV DNA in each of eight samples from group A oocytes were observed. A strong signal was seen in the positive control and no signal was found in the negative control (Figure 2).
Figure 2 Southern blot results of amplification products of DNA from mouse oocytes.
Lane 1: Markers; lane 2: negative control; lane 11: positive control; lanes 3-10: amplification products of DNA from group A oocytes in eight experiments.
Dot hybridization for washing solutions
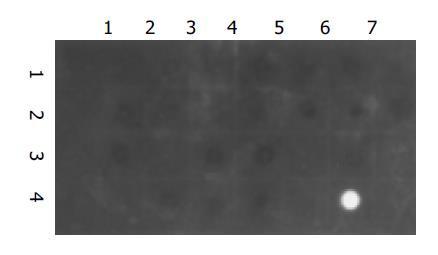
No signal for HBV DNA was observed in all of the 24 tested samples. A strong signal was seen in the positive control and no signal was observed in the negative control (Figure 3).
Figure 3 Dot hybridization for the washing solution.
Dots (1,1): negative control; Dots (4,6): positive control; Dots (1,2) to (1,7), (2,1) to (2,7), (3,1) to (3,7), (4,1) to (4,4): the washing solution samples from group A in eight experiments.
FISH of oocyte chromosomes
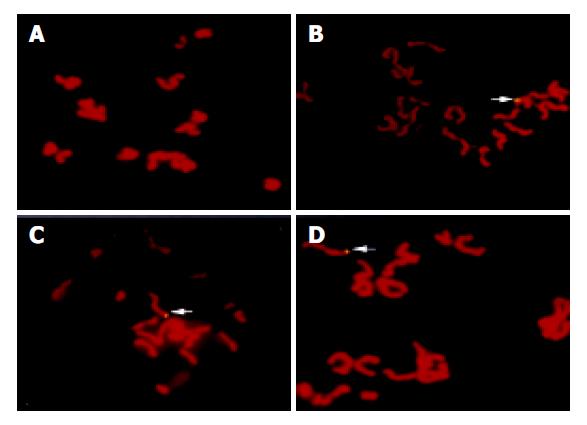
Using specific whole length HBV DNA as a probe to perform FISH with oocyte chromosomes, 36 of 1000 metaphases from group A oocytes presented positive signals for HBV DNA and no signal was found in 500 metaphases from group B oocytes (Figure 4).
Figure 4 FISH results.
A: No signal of HBV DNA integration in the metaphase from group B oocytes. B-D: Clear signals of HBV DNA integration in the metaphases from group A oocytes (arrow).
DISCUSSION
To confirm vertical transmission of HBV from mother to children via oocytes is an exciting program and of substantial importance. However, such studies have been hampered since neither experimental animals nor cell culture systems have been available. Thus it is very crucial to establish a model system for study on HBV transmission via oocytes. Zona pellucida is a glycoprotein coat surrounding the egg proper. It protects the fragile eggs and embryos from physical damage[28]. Can zona become a barrier to the entry of HBV DNA sequence into the oocytes? This is a key question for establishing such a model system, which we primarily have to answer.
In our study, the PCR products were visualized after electrophoresis on 1% agarose gel. A bright specific band for HBV DNA and no specific band were detected in the positive and negative controls respectively. The specific bands for HBV DNA were observed in all the samples from group A oocytes that were cocultured with HBV DNA in eight experiments (Figure 1). Consistent with PCR results, Southern blot analysis revealed that the hybridization signals for HBV DNA in all the amplification products from group A oocytes were observed. A strong signal was seen in the positive control and no signal was found in the negative control (Figure 2). It was impossible that the contamination of washing solutions gave rise to such positive results of PCR and Southern blot in the tested oocytes because they were washed eight times in each experiment and dot hybridization showed negative results in the last three of eight washing solutions in our study (Figure 3). Two reasons might contribute to the positive results in the tested oocytes. One was that some samples contained HBV-plasmid DNA that stuck firmly to the zona surface, the other was that some samples contained HBV DNA fragments that passed through the zona and oolemma and entered into the oocytes. The latter has been confirmed by our FISH experiments, the former was not excluded.
Although HBV replication is restricted to a more or less stringent host cell range, it is clear that viral integration is not restricted to any particular organ but occurs in many tissues including host somatic cells and spermatozoa[21,29-32]. However, so far no study on its integration into chromosomes of mammalian oocytes has been reported. We prepared metaphases of mouse oocytes cocultured with HBV DNA and performed FISH with a full length HBV DNA as a probe. Thirty-six of thousand metaphases presented positive signals and no FISH signal was detected in all the metaphases from 500 oocytes not cocultured with HBV DNA (Figure 4). This result indicated that the zona could not become a barrier to the entry of HBV DNA sequences into the oocytes and the oocytes cultured in vitro could passively take up exogenous DNA fragments. This result also indicated that HBV DNA fragments were able to enter into oocytes and integrate into their chromosomes. It is undoubted that HBV DNA sequences could be brought into embryos when the oocytes with the presence and integration of HBV DNA were fertilized with normal spermatozoa. So the in vitro culture system bringing HBV DNA into embryos via oocytes might be used as a model system for study on the exact vertical transmission of HBV via oocytes. Tsurimoto et al[33], demonstrated that HBV genome in an integrated state could act as a template for viral gene expression and replication. It would provide direct evidence for HBV transmission via oocytes if we could demonstrate the replication and expression of such genes of HBV in embryonic cells.
Hepatotropism is a prominent feature of HBV infection. It has been reported that cell lines of nonhepatic origin do not independently support HBV replication[34]. The reason why viral tropism was restricted to hepatocytes is largely unknown, although some investigations have indicated that HBV DNA methylation[35,36], liver-enriched transcription factors[37-39] and some receptors of hepatocytes[40,41] contribute to hepatotropism. Moreover, replication of HBV was found in transfected nonhepatic cells by ectopic expression of liver-enriched transcription factors[42]. DNA methylation is a very important manner for cells to regulate the expression of host genes and viral genes entering into cells. These genes would be almost fully methylated in sperms and probably the female germ line[43]. It was reported that DNA methylation patterns of the male and female pronuclei were erased in morula and early blastulae, when blastocysts formed, most of DNAs were demethylated. Following implantation, however, there was a surge of de novo methylation affecting the entire genome. During subsequent development, tissue-specific genes underwent programmed demethylation, which might cause their activation[44]. Preimplantation embryonic cells are non-specialized cells that are different from hepatic cells as well as specialized nonhepatic cells. It might be helpful for exploring the mechanism of hepatotropism and HBV vertical transmission to study on the contribution of methylation of HBV DNA, individual and multiple liver-enriched transcription factors and some membrane receptors in preimplantation embryonic cells with HBV sequences brought via oocytes in vitro culture system. This would be our further study in the future projects.