Published online Mar 6, 2021. doi: 10.12998/wjcc.v9.i7.1513
Peer-review started: October 13, 2020
First decision: November 26, 2020
Revised: December 8, 2020
Accepted: December 24, 2020
Article in press: December 24, 2020
Published online: March 6, 2021
Processing time: 138 Days and 14.5 Hours
An outbreak of a novel coronavirus was reported in Wuhan, China, in late 2019. It has spread rapidly through China and many other countries, causing a global pandemic. Since February 2020, over 28 countries/regions have reported confirmed cases. Individuals with the infection known as coronavirus disease-19 (COVID-19) have similar clinical features as severe acute respiratory syndrome first encountered 17 years ago, with fever, cough, and upper airway congestion, along with high production of proinflammatory cytokines (PICs), which form a cytokine storm. PICs induced by COVID-19 include interleukin (IL)-6, IL-17, and monocyte chemoattractant protein-1. The production of cytokines is regulated by activated nuclear factor-kB and involves downstream pathways such as Janus kinase/signal transducers and activators transcription. Protein expression is also regulated by post-translational modification of chromosomal markers. Lysine residues in the peptide tails stretching out from the core of histones bind the sequence upstream of the coding portion of genomic DNA. Covalent modification, particularly methylation, activates or represses gene transcription. PICs have been reported to be induced by histone modification and stimulate exudation of hyaluronic acid, which is implicated in the occurrence of COVID-19. These findings indicate the impact of the expression of PICs on the pathogenesis and therapeutic targeting of COVID-19.
Core Tip: An infection by a novel coronavirus which originated in Wuhan, China has spread rapidly, causing a global pandemic. Clinically the infection known as coronavirus disease-19 (COVID-19) is similar to severe acute respiratory syndrome, with fever, cough, and upper airway congestion, along with high production of proinflammatory cytokines (PICs), forming a cytokine storm. PICs induced include interleukin (IL)-6, IL-17, and monocyte chemoattractant protein-1. Their production is regulated by transcription factors, and post-translational modification of histones, which activates or represses gene transcription. PICs in turn stimulate exudation of hyaluronic acid, implicated in the pathogenesis of COVID-19. Their role as therapeutic targets is suggested.
- Citation: Zhang XN, Wu LJ, Kong X, Zheng BY, Zhang Z, He ZW. Regulation of the expression of proinflammatory cytokines induced by SARS-CoV-2. World J Clin Cases 2021; 9(7): 1513-1523
- URL: https://www.wjgnet.com/2307-8960/full/v9/i7/1513.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i7.1513
In December 2019, an infection manifesting as atypical pneumonia was discovered in Wuhan, China[1], and its rapid spread has been noted in China and many other countries[2-5]. So far, the 2019 novel coronavirus (2019-nCoV) has affected > 20 countries and has become a major global health problem. As of February 2020, approximately 51000 cases of the disease known as coronavirus disease 19 (COVID-19) have been documented globally, resulting in at least 1600 deaths since it first appeared in Central China's Hubei Province[4,6].
The pathogen, initially called 2019-nCoV is a beta coronavirus, and was later named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). It forms a clade among the subgenus Sarbecovirus[7]. Serious human infections have been caused by Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV, and SARS-CoV-2 is one of the seven members in the family of coronaviruses capable of infecting humans[8]. On confirming its origin from the bat coronavirus (BatCoV RaTG13), SARS-CoV-2 has been assigned to the subgenus Sarbecovirus, of the genus Betacoronavirus[9]. Evidence suggests that it is related to the bat and pangolin coronaviruses, as well as SARS-CoV, the SARS pathogen[8,9].
Clinically COVID-19 is characterized by fever, severe respiratory illness, and pneumonia similar to those produced by SARS-CoV, which led to an outbreak in Southern China in 2003[10,11]. The two biologically related coronaviruses contribute to a clinical picture attributed to the high production of proinflammatory cytokines (PICs), due to the interactions of the viral and host proteins when the virus enters the cell[12,13]. An acute respiratory illness and increased PICs have been described in COVID-19 patients; the cytokines include interleukin (IL)-2, IL-6, IL-10, granulocyte colony-stimulating factor (G-CSF), interferon (IFN)-g-induced protein-10 (IP-10), monocyte chemoattractant protein factor-1 (MCP-1), macrophage inflammatory protein 1 (MIP-1) and tumor necrosis factor-αα)[14-16].
Like most proteins, the expression of PICs is triggered by activation of transcription factors and post-translational modification (PTM) of the factors and histones, the basic units of the nucleosome where genomic DNA is housed. The expression of PICs is regulated by the association of transcription factors and chromatin markers in the promoter and enhancer sequences upstream of the coding portion of genes. The activation or repression of these elements is modulated by PTM. In this review, we discuss the impact of histone and nonhistone transcription factor PTM in the increased production of PICs.
Similar to the pathogenesis of SARS and MERS, higher levels of cytokines in plasma, including IL-2, IL-6, IL-7, IL-10, G-CSF, IFN-gα[15-19]. The production of PICs is induced by SARS-CoV-2, which forms a cytokine storm, leading to a fatal syndrome involving multiple organ dysfunction[20]. PICs induced during COVID-19 originate from inflammatory cells that constitute the innate immune defense, including monocytes, macrophages, neutrophils, and pulmonary and renal epithelial cells.
The major PICs induced by the novel coronavirus include IL-1β, IL-6 and MCP-1; they promote the pathogenesis of COVID-19 by triggering an inflammatory response[21]. Among these, elevated IL-6 levels are closely correlated with disease progression[18,19]. The production of these factors involves a complicated regulatory network of transcriptional activation by transcription factors, PTM of histones, and nonhistone transcription factors. IL-6 production depends on activated transcription factors, in particular, nuclear factor kappa-B (NF-kB) and the aggregated IL-6 receptor when ligated by IL-6 stimulates a number of downstream intracellular signaling pathways, the most common being Janus kinase (JAK)/signal transducers and activators transcription (STATs). Cytokine-coding gene promoter occupancy with chromatin markers has also been identified[22].
After being ligated by cytokines released by inflammatory cells, activated cognate receptors stimulate downstream signaling pathways, mediated by JAK[23]. IL-6, a PIC that has been shown to act as a critical part of the pathogenic process of COVID-19, activates the JAK/STAT signaling pathway to exert a variety of biological effects, including immune regulation, proliferation, and differentiation of lymphocytes and oxidative stress[23-25].
The JAK/STAT pathway that transduces growth signals transmitted by surface receptors has also been shown to contribute significantly to the pathogenesis of COVID-19. To date, four proteins of the JAK non-receptor family have been identified in mammalian cells; namely, JAK1, JAK2, JAK3 and tyrosine kinase 2 (TYK2). JAK1/2 and TYK2 are expressed in different cell types, while JAK3 appears to play an essential role in the function of lymphocytes. The human STAT family comprises seven members: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6[26].
The novel coronavirus-encoded spike (S) protein is expressed on the viral surface. Virus entry to host cells is mediated by S protein, which interacts with angiotensin-converting enzyme 2 (ACE2) that acts as the viral receptor.
ACE2 is an enzyme that cleaves angiotensin II into angiotensin 1-7, which possesses vasodilatory and anti-inflammatory effects. In patients with cardiac disease, upregulation of ACE2 may increase susceptibility to COVID-19[27].
ACE inhibitors or angiotensin receptor blockers may enhance immune cell infiltration, and they also increase local production of PICs such as TNF-α, IL-1, IL-6, and IFN-g[28].
S protein has been shown to activate NF-kB like some other viral proteins[29]. Latent membrane protein 1(LMP1), encoded by a human lymphotropic herpesvirus, Epstein–Barr virus, is a viral homolog of mammalian CD40 and activates NF-kB behaving like an aggregated TNF-α[30]. The molecular interaction involving activation of NF-kB by S protein remains to be characterized.
In patients with severe COVID-19, elevated IL-6 has been frequently detected in the serum and correlated with nonsurvival[31]. Consequently, clinical trials have been initiated to evaluate the therapeutic effects of IL-6 inhibitors in COVID-19 patients[29]. The JAK/STAT pathway has also been explored for use as a new therapeutic target against SARS-CoV-2 infection[31,32]. Available data provide a rationale for administering licensed JAK inhibitors for improving the clinical management of COVID-19 and meeting the urgent global demand to mitigate the infection.
Both constitutively active and noncanonical STAT functions are involved in the molecular processes of tumorigenesis and viral pathogenesis. STAT3 and STAT5 have been investigated due to their activities in controlling proliferation, survival, cellular adhesion, intercellular communication, and angiogenesis when triggered by a wide range of stimuli[33,34].
The activity of STAT3 is also modified by methylation at Lys 49 and Lys 180 by the enhancer of Zester homolog 2 (EZH2), a component with lysine methyltransferase activity in the polycomb repressive complex 2 (PRC2), which regulates gene transcription. EZH2 methylates STAT3 downstream of AKT signaling and increases the transcriptional activity of STAT3, which has been shown to be activated by modification with methylation[35]. We discuss the role of histone modification by the methylation involved in the control of gene expression in the next section.
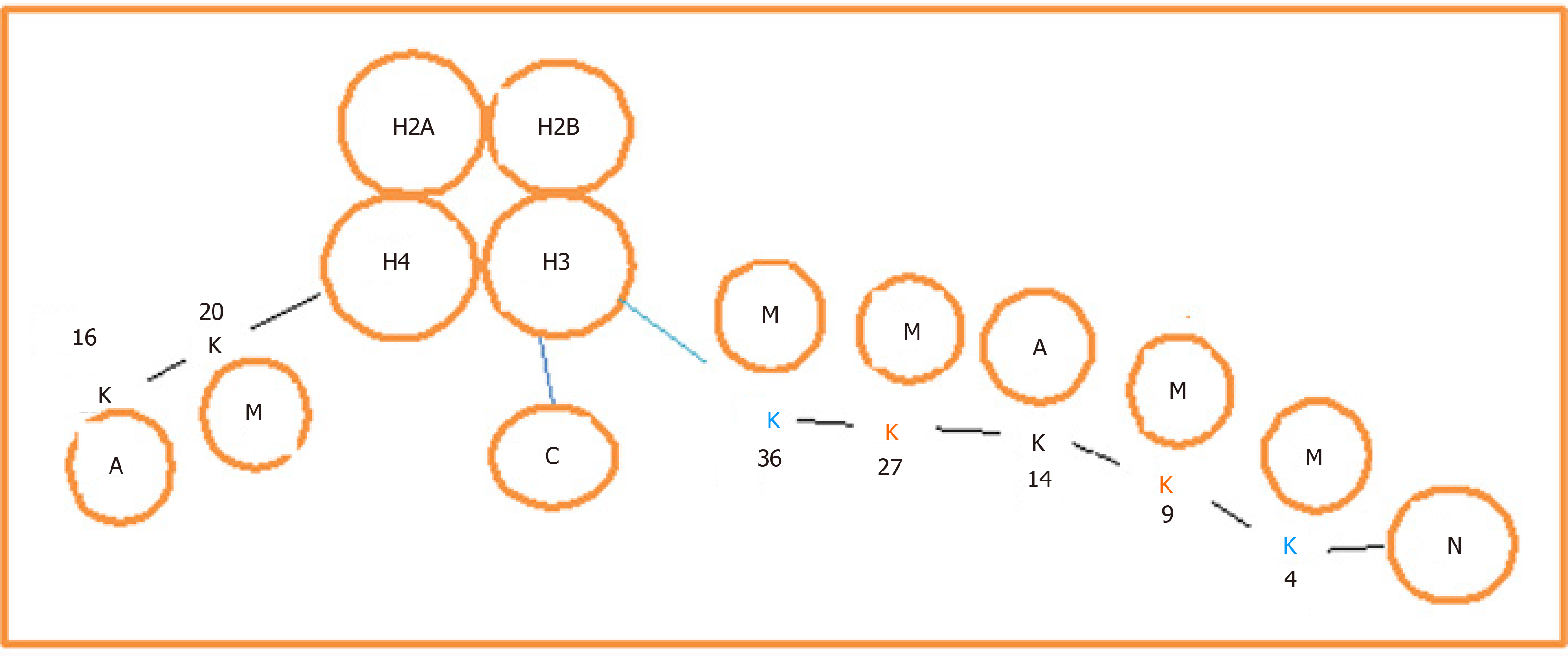
The activation and repression of gene transcription are regulated by chromatin modification. The basic units of the chromosome are the nucleosome, which is composed of a nucleus of histone octamer (H), heteromerized with four dimers of H1, H2A, H3 and H4, and the octamer is wrapped with strands of genomic DNA (Figure 1). Extended peptide tails contain a series of modifiable amino acid residues called chromatin marks. The main modification pattern for H3 is methylation, and for H4 it is acetylation, of lysine residues. They are important in the activation or repression of genes (Figure 1). The methylation of lysine residues in histones, mainly H3, is catalyzed by a group of enzymes called histone lysine methyltransferases (KMTs). The protein clusters are driven by the Suv39H1, G9a and SETDB1/SETDB2 families of enzymes known as lysine methyltransferase 1 (KMT1), and they function to catalyze mono-, bi- and trimethylation of lysine residues 4 and 9 (H3K4 and H3K9)[36,37].
Much remains to be elucidated with regard to the role of epigenetic modification of histones played by different modifiers in the regulation of inflammatory responses.
As mentioned above, methylation at various lysine residues on histone H3 leads to the activation or repression of gene transcription. While methylation at H3K9 or H3K27 represses gene expression, methylation at H3K4, H3K36 or H3K79 activates gene expression. The disruptor of telomeric silencing-1-like is a modifier that specifically catalyzes methylation of H3K79 to regulate proliferation and differentiation of tumor cells.
Chromosomal marks, for example, H3K4 and H3K9, with different modifications bind to the target sequence, in particular to regulatory elements such as promoters or enhancers, to modulate gene expression. The marks are recognized and incorporated into the target sequence in a process called reading[38]. Chromatin marks are modified by adding chemical groups with methylation, acetylation and SUMOylation, and ubiquitination, called writing. The modification can alter the ability of the marks to regulate gene expression. The modification is reversible, and the added groups are removed in a process called erasing. Chromatin marks such as H3K4 bind to the promoter upstream of the IL-6 coding sequence and methylated H3K4 is demethylated by a demethylase, lysine-specific demethylase 1A/lysine-specific demethylase 1[39].
The methyltransferase catalyzes adding different numbers of methyl groups to lysine, termed mono-, bi- and trimethylation. As mentioned above, six lysine residues on H3, i.e., K4, K9, K27, K36, K79 and K36, together with K20 from histone H4 and K26 from histone 1 have been recognized as the canonical sites of methylation.
Except for H3K79, all of these are found in the N-terminal tails of histone proteins. It has been identified that some proteins or the domain contained within the molecules serve as readers of methyllysine. Ankyrin, double chromodomain, tudor tandem domain, tudor and zinc finger, as well as the known motifs found in transcription regulators, malignant brain tumor, plant homeodomain and Pro-Trp-Trp-Pro.
The initiation of viral genome transcription, virion production, and spread of human immunodeficiency virus (HIV)-1 is triggered when host cell replication is stimulated by cytokines, drugs, or activation of T cell receptors.
The reactivation of provirus depends on the binding of NF-kB and/or nuclear factor of activated T cells with the specific target sequences on the viral long terminal repeat (LTR). The factors which initiate transcription of viral genome act by recruiting histone acetyltransferase p300, CBP-associated factor to the LTR of HIV; the modifying enzymes acetylate histones adjacent to the viral promoters[40,41].
The acetylation modifications on histones H3 and H4, like H3 lysine 4 acetylation (H3K4ac) and H3 lysine 27 acetylation (H3K27ac) create an open chromatin configuration, favoring binding of transcription factors and forming complexes of initiation and elongation. Trimethylation of H3 lysine 4, H3K4me3 and lysine H3 K27, H3K27me3 bind to gene promoters, activating and repressing gene transcription, respectively. Dimethylated H3K4 (H3K4me2) and di- and trimethylated H3K36 (H3K36 me2, me3) have been identified on chromatin sites being transcriptionally activated.
Modification of methylation is catalyzed by a panel of enzymes to add the chemical group to the lysine residue, and some of them are listed in Table 1.
| Category | Name of modifying enzymes | Other names | Targeting residue on histones | Effects |
| KMT3 | 3A | SETD2, SET2 | Trimethylates H3K36 | Transcription activation |
| 3B | NSD1 | Methylates H3K36 or H4K20 | Same as above | |
| 3C | SMYD2 | Monmethylates H3K4 or H3K36, monomethylates RB1 lys860 | Same as above | |
| 3D | SMYD1 | Mono-, di- and trimethylates histone H3 K4' | Transcriptional repression | |
| 3E | SMYD3 | Di- and trimethylates H3K4, also methylates H4K5 | Transcriptional activation | |
| KMT6 | KMT6 | EZH2 | Methylates H3K9 and H3K27 | Transcription repression |
| KMT7 | KMT7 | SET7/9 | Monomethylates histone H3K4 | Transcriptional activation |
Members of the KMT3 family, SET and MYND domain containing (SMYD)2 and SMYD3, function to catalyze methylation at the H3K4 or H3K36 marks, to activate gene transcription. SMYD2 has also been shown to regulate latency of HIV-1, and is associated with latent HIV-1 promoter in H4K20me1-enriched chromatin-harboring DNA, H4K20me1 enrichment is known as a mark lost in cells defected with SMYD2. A reader protein recognizing H4K20me1 Lethal 3 malignant brain tumor 1 (L3MBTL1), is recruited to the HIV latent promoter. The data suggest that an axis of SMYD2–H4K20me1–L3MBTL1 is implicated in the regulation of HIV-1 latency and may be targeted by small molecule inhibitors of SMYD2[42].
SMYD3, another member of the KMT3 family has been reported to be recruited to the inclusion bodies of Ebolavirus (EBOV) by interacting with the virally encoded nucleoprotein. A significant suppression of EBOV mRNA synthesis has been noted on deletion of SMYD3. A nonphosphorylated VP30 mimic, which is a transcription activator, can partially rescue viral mRNA production. Whether methyltransferase activity plays a role in this process remains to be determined, and nucleoprotein encoded by SARS-CoV-2 also interacts with SMYD3[43].
Methyltransferase SET7 generates H3K4me by monomethylation of H3K4. H3K4me represents a tag specific for epigenetic gene activation. It transcriptionally activates genes like collagenase or insulin on recruitment to the promoter sequence of the target genes. SET7 also methylates nonhistone proteins p53/TP53 and Transcription initiation factor TFIID subunit 10 (TAF10). When monomethylated on Lys/K-189, TAF10 has an enhanced affinity for RNA polymerase. Lys/K-372 monomethylation of p53/TP53 stabilizes p53/TP53 and increases p53/TP53-mediated transcriptional activation[44-46]. The relationship between methylation and demethylation and coronavirus infection has not been investigated. Human papillomavirus E6 has been reported to attenuate the transactivation function of p53 by decreasing the enzymatic activities of SET7[47]. SET7 was found to promote HIV transcription through monomethylated Tat protein[48]. Recent studies have demonstrated that the antiviral function of IFN-induced transmembrane protein 3 for vesicular stomatitis virus and influenza A virus could be attenuated by SET7[49]. Han et al[50] observed that SET7 expression in Huh7.5.1 cells was upregulated by hepatitis C virus (HCV) infection, and high levels of SET7 expression were also found in serum, peripheral blood mononuclear cells, and liver tissue of HCV patients compared with healthy individuals. Subsequent research showed that SET7 enhanced HCV replication in an enzyme-dependent manner. Furthermore, the data showed that SET7 decreased the expression of virus-induced IFN and IFN-related effectors.
As mentioned above, the production of proinflammatory cytokines is regulated by histone modification, and the modification also affects virus–host interaction.
The contribution of KMT histones to maintaining HIV latency remains to be elucidated[40,51]. EZH2/KMT6, which specifically methylates lysine residues H3K9 and H3K27, as repressive markers for gene transcription, is abundant on the LTR of the silenced proviruses of HIV, as demonstrated by chromatin immunoprecipitation in latently infected Jurkat T cells.
In proviral reactivation, it was rapidly displaced. The latent HIV-1 proviruses carried by methylated lysine residues on H3 are mainly trimethylated repressive genomic markers H3k9me3 and H3K27me3[52,53] or H3K9me2[54]. A histone KMT, SUV39H1, responsible for generation of H3K9me3, interacts with COUP-Transcription Factor-interacting protein 2 (CTIP-2) and Heterochromatin protein 1 homolog gamma (HP1γ) to maintain HIV-1 latency in microglial cells[52,53]. Removal of the two proteins contributes to HIV-1 activation. It has been proposed that HKMT G9a, which catalyzes the dimethylation of H3K9, may also contribute to the maintenance of HIV-1 latency[55].
The lungs of some patients with COVID-19 are filled with a clear liquid with jelly-like appearance. One of the components identified in this fluid is hyaluronic acid (HA), a polysaccharide seen in most connective tissues. In water, HA can trap its weight up to 1000 times[56]. It is present as a high-molecular-weight polymer in airway epithelial cells covering the apical surface. Depolymerization of HA initiates a cascade leading to the production of quinine and processing of growth factors. HA synthesis and degradation are mediated by integral membrane proteins HA synthase (HAS), including HAS1, HAS2 and HAS3, and hyaluronidases (HAases), respectively.
HYAL1, HYAL2 and PH20 are among six HAases that have been well characterized. HYAL1-like HAase was initially isolated in specimens of human urine and plasma[57,58]. HYAL1, alternatively termed Luca1 (lung cancer 1) is known as the main tumor-derived HAase expressed in bladder and prostate cancer tissues[59,60]. HYAL2 was originally identified in lysosomes, and it cleaves high-molecular-mass HA with release of a 20-kDa fragment[61]. HYAL3 is the third HAase, whose coding gene is mapped on the chromosomal region 3p21.3. Its transcripts have been detected in different tissues, and the sequence of the HYAL3-encoded product has only been predicted[62].
An enzyme that catalyzes the degradation of the jelly-like fluid HA, HYAL1, is not expressed in the bronchial alveolar lavage fluid (BALF) of COVID-19 patients and control individuals. This suggests that the amount of HA in the bronchoalveolar space of the lungs would be induced by the high PICs production during COVID-19 pathogenesis. In combination with the induced increase in vascular hyperpermeability due to the elevated PICs, a viscous hydrogel is generated to frustrate gas exchange[63]. The genesis of acute respiratory distress syndrome (ARDS) is associated with HA in BALF; the concentration of HA is significantly negatively correlated with the index of pulmonary oxygenation. HA is also responsible for pulmonary thrombosis, and ground-glass opacities; a characteristic finding of interstitial pneumonia on radiography[64-66].
In certain cell types, the effects of TNF-α –3 is present. Their expression and activity are augmented by IL-1 in synergy with TNF-α, HYALs are localized intracellularly, while HYAL2 is expressed in plasma-membrane-associated apical poles and in apical secretions in soluble form.
Also, increased HYAL expression and activity are noted in tracheal secretions and BALF derived from individuals with asthma in comparison with healthy controls with normal lungs. HYALs 1–3 expressed by airway epithelium coordinate to depolymerize HA during inflammation mediated by elevated TNF-α and IL-1[67].
Symptoms in patients with ARDS include rapid and exhaustive respiration and cyanosis. Severe cases of COVID-19 admitted to intensive care units normally require mechanical ventilation, and patients who are unable to breathe can be maintained with extracorporeal membrane oxygenation[68].
COVID-19 is characteristic on computed tomography images, with white spots known as ground glass opacities, with fluid in the lungs[69]. Autopsy confirms that the lungs are filled with clear liquid gelatin, similar to the lungs after drowning[70]. While further validation is needed, an association between HA and ARDS has been suggested[71], and the production and regulation of HA have been noted in SARS.
In the lungs of patients with COVID-19, the levels of PICs, IL-1 and TNF-α are elevated and they strongly induce HAS2 in epithelial-cell-adhesion-molecule-positive pulmonary alveolar epithelial cells and fibroblasts[72]. HA can absorb water up to 1000 times its molecular weight. Therefore, inhibition of HA production would greatly contribute to improvement of breathing in COVID-19 patients.
At present, specific therapy for COVID-19 has not been available, and the drugs administered mainly target viral infection and inflammation triggered by viral invasion, accompanied by supportive therapy[73]. The HIV protease inhibitors ritonavir and lopinavir have been used in anti-COVID-19 treatment[74]. High production of PICs such as IL-6 and helper T cytokine IL-17, is triggered, and the JAK/STAT signaling pathway is activated. The United States FDA and the European Medical Association have approved the use of JAK inhibitors, e.g. ruxolitinib[75] and baricitinib[76], for therapy of COVID-19[77].
More candidate JAK inhibitors are being tested in clinical trials[78]. A JAK1/2 inhibitor, ABT-494, has the potential to inhibit the regulation of endocytosis and may possess the potential to prevent entry to susceptible cells by SARS-CoV-2. In addition, its interaction with cytochrome enzymes is minimal and it has low serum binding. ABT-494 is active in combination with other drugs such as remdesivir in COVID-19 therapy[79].
IL-6 is a cytokine implicated in the cytokine release syndrome (CRS) seen in COVID-19. In its presence, immature T helper cells (Th0) differentiate into the Th17 subtype, and JAK2 is utilized by IL-6 to activate downstream signals[80].
As a characteristic disease of CRS, markedly high numbers of Th17 cells have been identified in COVID-19 patients, suggesting that CRS of Th17 type is involved in severe immune damage[73]. A highly selective JAK2 inhibitor, fedratinib, approved for myelofibrosis therapy, has been shown to inhibit expression of IL-17 in murine Th17 cells[81]. The data suggest that inhibition of JAK could be a therapeutic target against COVID-19 through an antagonistic effect on the activation of Th17-associated cytokines.
In summary, SARS-CoV-2 enters host cells via binding of viral surface proteins to ACE-2 as a receptor. Viral entry stimulates PIC production through a complicated network involving intracellular signaling pathways and histone PTMs. Therapeutic agents targeting the various stages of the virus–host interaction could be developed.
The authors wish to thank Professors Chiodi F, Karolinska Institutet, Sweden, and Ouyang SD, Guangdong Medical University, China for their interest in our work and their stimulating discussion.
Manuscript source: Invited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Farshadpour F S-Editor: Fan JR L-Editor: Webster JR P-Editor: Xing YX
| 1. | Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55:105924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3736] [Cited by in RCA: 3197] [Article Influence: 639.4] [Reference Citation Analysis (0)] |
| 2. | Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J Med Virol. 2020;92:401-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1670] [Cited by in RCA: 1769] [Article Influence: 353.8] [Reference Citation Analysis (0)] |
| 3. | Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14869] [Cited by in RCA: 12977] [Article Influence: 2595.4] [Reference Citation Analysis (1)] |
| 4. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30123] [Article Influence: 6024.6] [Reference Citation Analysis (3)] |
| 5. | Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470-473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4848] [Cited by in RCA: 4389] [Article Influence: 877.8] [Reference Citation Analysis (1)] |
| 6. | Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514-523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6483] [Cited by in RCA: 5423] [Article Influence: 1084.6] [Reference Citation Analysis (0)] |
| 7. | Cárdenas-Conejo Y, Liñan-Rico A, García-Rodríguez DA, Centeno-Leija S, Serrano-Posada H. An exclusive 42 amino acid signature in pp1ab protein provides insights into the evolutive history of the 2019 novel human-pathogenic coronavirus (SARS-CoV-2). J Med Virol. 2020;92:688-692. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 8. | Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18987] [Cited by in RCA: 17648] [Article Influence: 3529.6] [Reference Citation Analysis (0)] |
| 9. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14131] [Article Influence: 2826.2] [Reference Citation Analysis (1)] |
| 10. | Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8473] [Cited by in RCA: 7602] [Article Influence: 1520.4] [Reference Citation Analysis (0)] |
| 11. | Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6893] [Cited by in RCA: 7499] [Article Influence: 1499.8] [Reference Citation Analysis (0)] |
| 12. | He L, Ding Y, Zhang Q, Che X, He Y, Shen H, Wang H, Li Z, Zhao L, Geng J, Deng Y, Yang L, Li J, Cai J, Qiu L, Wen K, Xu X, Jiang S. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J Pathol. 2006;210:288-297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 323] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 13. | Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260-1263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5792] [Cited by in RCA: 6487] [Article Influence: 1297.4] [Reference Citation Analysis (0)] |
| 14. | Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ; HLH Across Speciality Collaboration; UK. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033-1034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6366] [Cited by in RCA: 6752] [Article Influence: 1350.4] [Reference Citation Analysis (0)] |
| 15. | Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front Microbiol. 2019;10:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 417] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 16. | Zhang H, Zhou P, Wei Y, Yue H, Wang Y, Hu M, Zhang S, Cao T, Yang C, Li M, Guo G, Chen X, Chen Y, Lei M, Liu H, Zhao J, Peng P, Wang CY, Du R. Histopathologic Changes and SARS-CoV-2 Immunostaining in the Lung of a Patient With COVID-19. Ann Intern Med. 2020;172:629-632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 308] [Cited by in RCA: 376] [Article Influence: 75.2] [Reference Citation Analysis (0)] |
| 17. | Poplutz MK, Wessels I, Rink L, Uciechowski P. Regulation of the Interleukin-6 gene expression during monocytic differentiation of HL-60 cells by chromatin remodeling and methylation. Immunobiology. 2014;219:619-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 18. | Qu G, Li X, Hu L, Jiang G. An Imperative Need for Research on the Role of Environmental Factors in Transmission of Novel Coronavirus (COVID-19). Environ Sci Technol. 2020;54:3730-3732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 197] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 19. | Lin L, Lu L, Cao W, Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect. 2020;9:727-732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 582] [Cited by in RCA: 559] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 20. | Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, Guo D, Hu W, Yang J, Tang Z, Wu H, Lin Y, Zhang M, Zhang Q, Shi M, Liu Y, Zhou Y, Lan K, Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 877] [Cited by in RCA: 856] [Article Influence: 171.2] [Reference Citation Analysis (0)] |
| 21. | Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O'Shea JJ. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat Rev Drug Discov. 2017;16:843-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 702] [Article Influence: 87.8] [Reference Citation Analysis (0)] |
| 22. | Kang S, Tanaka T, Narazaki M, Kishimoto T. Targeting Interleukin-6 Signaling in Clinic. Immunity. 2019;50:1007-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 569] [Article Influence: 94.8] [Reference Citation Analysis (0)] |
| 23. | Garbers C, Heink S, Korn T, Rose-John S. Interleukin-6: designing specific therapeutics for a complex cytokine. Nat Rev Drug Discov. 2018;17:395-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 465] [Article Influence: 66.4] [Reference Citation Analysis (0)] |
| 24. | Crowle AJ, Poche P. Inhibition by normal human serum of Mycobacterium avium multiplication in cultured human macrophages. Infect Immun. 1989;57:1332-1335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 590] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 25. | Tomasoni D, Italia L, Adamo M, Inciardi RM, Lombardi CM, Solomon SD, Metra M. COVID-19 and heart failure: from infection to inflammation and angiotensin II stimulation. Searching for evidence from a new disease. Eur J Heart Fail. 2020;22:957-966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 192] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 26. | Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846-848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2604] [Cited by in RCA: 3195] [Article Influence: 639.0] [Reference Citation Analysis (0)] |
| 27. | Seif F, Aazami H, Khoshmirsafa M, Kamali M, Mohsenzadegan M, Pornour M, Mansouri D. JAK Inhibition as a New Treatment Strategy for Patients with COVID-19. Int Arch Allergy Immunol. 2020;181:467-475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 138] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 28. | Xu X, Han M, Li T, Sun W, Wang D, Fu B, Zhou Y, Zheng X, Yang Y, Li X, Zhang X, Pan A, Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci U S A. 2020;117:10970-10975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1811] [Cited by in RCA: 1748] [Article Influence: 349.6] [Reference Citation Analysis (0)] |
| 29. | Dosch SF, Mahajan SD, Collins AR. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus Res. 2009;142:19-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 195] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 30. | Wang LW, Jiang S, Gewurz BE. Epstein-Barr Virus LMP1-Mediated Oncogenicity. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 111] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 31. | Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529-539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1895] [Cited by in RCA: 1791] [Article Influence: 223.9] [Reference Citation Analysis (0)] |
| 32. | Mazzoni A, Salvati L, Maggi L, Capone M, Vanni A, Spinicci M, Mencarini J, Caporale R, Peruzzi B, Antonelli A, Trotta M, Zammarchi L, Ciani L, Gori L, Lazzeri C, Matucci A, Vultaggio A, Rossi O, Almerigogna F, Parronchi P, Fontanari P, Lavorini F, Peris A, Rossolini GM, Bartoloni A, Romagnani S, Liotta F, Annunziato F, Cosmi L. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130:4694-4703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 380] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 33. | Ferbeyre G, Moriggl R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochim Biophys Acta. 2011;1815:104-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 34. | Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, Lee C, Joo KM, Rich JN, Nam DH, Lee J. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839-852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 643] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 35. | Dasgupta M, Dermawan JK, Willard B, Stark GR. STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc Natl Acad Sci USA. 2015;112:3985-3990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 36. | Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7446] [Cited by in RCA: 7672] [Article Influence: 426.2] [Reference Citation Analysis (0)] |
| 37. | Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 634] [Cited by in RCA: 583] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 38. | Musselman CA, Lalonde ME, Côté J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 626] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 39. | Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7019] [Cited by in RCA: 6891] [Article Influence: 287.1] [Reference Citation Analysis (0)] |
| 40. | Friedman J, Cho WK, Chu CK, Keedy KS, Archin NM, Margolis DM, Karn J. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J Virol. 2011;85:9078-9089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 234] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 41. | Lusic M, Marcello A, Cereseto A, Giacca M. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 2003;22:6550-6561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 201] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 42. | Boehm D, Jeng M, Camus G, Gramatica A, Schwarzer R, Johnson JR, Hull PA, Montano M, Sakane N, Pagans S, Godin R, Deeks SG, Krogan NJ, Greene WC, Ott M. SMYD2-Mediated Histone Methylation Contributes to HIV-1 Latency. Cell Host Microbe 2017; 21: 569-579. e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 43. | Chen J, He Z, Yuan Y, Huang F, Luo B, Zhang J, Pan T, Zhang H, Zhang J. Host factor SMYD3 is recruited by Ebola virus nucleoprotein to facilitate viral mRNA transcription. Emerg Microbes Infect. 2019;8:1347-1360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 44. | Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14:175-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 212] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 45. | Francis J, Chakrabarti SK, Garmey JC, Mirmira RG. Pdx-1 links histone H3-Lys-4 methylation to RNA polymerase II elongation during activation of insulin transcription. J Biol Chem. 2005;280:36244-36253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 75] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 46. | Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 497] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 47. | Hsu CH, Peng KL, Jhang HC, Lin CH, Wu SY, Chiang CM, Lee SC, Yu WC, Juan LJ. The HPV E6 oncoprotein targets histone methyltransferases for modulating specific gene transcription. Oncogene. 2012;31:2335-2349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 48. | Pagans S, Kauder SE, Kaehlcke K, Sakane N, Schroeder S, Dormeyer W, Trievel RC, Verdin E, Schnolzer M, Ott M. The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription. Cell Host Microbe. 2010;7:234-244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 49. | Shan Z, Han Q, Nie J, Cao X, Chen Z, Yin S, Gao Y, Lin F, Zhou X, Xu K, Fan H, Qian Z, Sun B, Zhong J, Li B, Tsun A. Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation. J Biol Chem. 2013;288:35093-35103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 50. | Han T, Wan Y, Wang J, Zhao P, Yuan Y, Wang L, She Y, Broering R, Lu M, Ye L, Zhu Y. Set7 facilitates hepatitis C virus replication via enzymatic activity-dependent attenuation of the IFN-related pathway. J Immunol. 2015;194:2757-2768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 51. | Tripathy MK, McManamy ME, Burch BD, Archin NM, Margolis DM. H3K27 Demethylation at the Proviral Promoter Sensitizes Latent HIV to the Effects of Vorinostat in Ex Vivo Cultures of Resting CD4+ T Cells. J Virol. 2015;89:8392-8405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 52. | Nguyen K, Das B, Dobrowolski C, Karn J. Multiple Histone Lysine Methyltransferases Are Required for the Establishment and Maintenance of HIV-1 Latency. mBio. 2017;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 53. | du Chéné I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, Bertrand E, Marcello A, Emiliani S, Kiernan R, Benkirane M. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007;26:424-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 260] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 54. | Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 2007;26:412-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 299] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 55. | Imai K, Togami H, Okamoto T. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J Biol Chem. 2010;285:16538-16545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 210] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 56. | Cowman MK, Matsuoka S. Experimental approaches to hyaluronan structure. Carbohydr Res. 2005;340:791-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 248] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 57. | Frost GI, Csóka AB, Wong T, Stern R. Purification, cloning, and expression of human plasma hyaluronidase. Biochem Biophys Res Commun. 1997;236:10-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 186] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 58. | Csóka AB, Frost GI, Wong T, Stern R. Purification and microsequencing of hyaluronidase isozymes from human urine. FEBS Lett. 1997;417:307-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 76] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 59. | Lokeshwar VB, Rubinowicz D, Schroeder GL, Forgacs E, Minna JD, Block NL, Nadji M, Lokeshwar BL. Stromal and epithelial expression of tumor markers hyaluronic acid and HYAL1 hyaluronidase in prostate cancer. J Biol Chem. 2001;276:11922-11932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 229] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 60. | Lokeshwar VB, Young MJ, Goudarzi G, Iida N, Yudin AI, Cherr GN, Selzer MG. Identification of bladder tumor-derived hyaluronidase: its similarity to HYAL1. Cancer Res. 1999;59:4464-4470. [PubMed] |
| 61. | Lepperdinger G, Müllegger J, Kreil G. Hyal2--less active, but more versatile? Matrix Biol. 2001;20:509-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 122] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 62. | Triggs-Raine B, Salo TJ, Zhang H, Wicklow BA, Natowicz MR. Mutations in HYAL1, a member of a tandemly distributed multigene family encoding disparate hyaluronidase activities, cause a newly described lysosomal disorder, mucopolysaccharidosis IX. Proc Natl Acad Sci U S A. 1999;96:6296-6300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 140] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 63. | Garvin MR, Alvarez C, Miller JI, Prates ET, Walker AM, Amos BK, Mast AE, Justice A, Aronow B, Jacobson D. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. Elife. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 256] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 64. | Bhagat R, Forteza RM, Calcote CB, Williams WT, Bigler SA, Dwyer TM. Pulmonary emboli from therapeutic sodium hyaluronate. Respir Care. 2012;57:1670-1673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 65. | Han SW, Park MJ, Lee SH. Hyaluronic acid-induced diffuse alveolar hemorrhage: unknown complication induced by a well-known injectable agent. Ann Transl Med. 2019;7:13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 66. | Jang JG, Hong KS, Choi EY. A case of nonthrombotic pulmonary embolism after facial injection of hyaluronic Acid in an illegal cosmetic procedure. Tuberc Respir Dis (Seoul). 2014;77:90-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 67. | Monzón ME, Manzanares D, Schmid N, Casalino-Matsuda SM, Forteza RM. Hyaluronidase expression and activity is regulated by pro-inflammatory cytokines in human airway epithelial cells. Am J Respir Cell Mol Biol. 2008;39:289-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 68. | Koga K, Miura I. A measurement of cerebral glucose uptake rate by 31P MRS. Biochem Biophys Res Commun. 1988;157:1258-1263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 276] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 69. | Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14113] [Cited by in RCA: 14767] [Article Influence: 2953.4] [Reference Citation Analysis (0)] |
| 70. | Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5228] [Cited by in RCA: 5786] [Article Influence: 1157.2] [Reference Citation Analysis (2)] |
| 71. | Hällgren R, Samuelsson T, Laurent TC, Modig J. Accumulation of hyaluronan (hyaluronic acid) in the lung in adult respiratory distress syndrome. Am Rev Respir Dis. 1989;139:682-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 120] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 72. | Bell TJ, Brand OJ, Morgan DJ, Salek-Ardakani S, Jagger C, Fujimori T, Cholewa L, Tilakaratna V, Östling J, Thomas M, Day AJ, Snelgrove RJ, Hussell T. Defective lung function following influenza virus is due to prolonged, reversible hyaluronan synthesis. Matrix Biol. 2019;80:14-28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 73. | Stebbing J, Phelan A, Griffin I, Tucker C, Oechsle O, Smith D, Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect Dis. 2020;20:400-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 726] [Cited by in RCA: 770] [Article Influence: 154.0] [Reference Citation Analysis (0)] |
| 74. | Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, Li X, Xia J, Chen N, Xiang J, Yu T, Bai T, Xie X, Zhang L, Li C, Yuan Y, Chen H, Li H, Huang H, Tu S, Gong F, Liu Y, Wei Y, Dong C, Zhou F, Gu X, Xu J, Liu Z, Zhang Y, Li H, Shang L, Wang K, Li K, Zhou X, Dong X, Qu Z, Lu S, Hu X, Ruan S, Luo S, Wu J, Peng L, Cheng F, Pan L, Zou J, Jia C, Wang J, Liu X, Wang S, Wu X, Ge Q, He J, Zhan H, Qiu F, Guo L, Huang C, Jaki T, Hayden FG, Horby PW, Zhang D, Wang C. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med. 2020;382:1787-1799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3386] [Cited by in RCA: 3627] [Article Influence: 725.4] [Reference Citation Analysis (0)] |
| 75. | Mascarenhas J, Hoffman R. Ruxolitinib: the first FDA approved therapy for the treatment of myelofibrosis. Clin Cancer Res. 2012;18:3008-3014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 205] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 76. | Mullard A. FDA approves Eli Lilly's baricitinib. Nat Rev Drug Discov. 2018;17:460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 77. | Luo W, Li YX, Jiang LJ, Chen Q, Wang T, Ye DW. Targeting JAK-STAT Signaling to Control Cytokine Release Syndrome in COVID-19. Trends Pharmacol Sci. 2020;41:531-543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 222] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 78. | Traynor K. FDA approves tofacitinib for rheumatoid arthritis. Am J Health Syst Pharm. 2012;69:2120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 79. | Kremer JM, Emery P, Camp HS, Friedman A, Wang L, Othman AA, Khan N, Pangan AL, Jungerwirth S, Keystone EC. A Phase IIb Study of ABT-494, a Selective JAK-1 Inhibitor, in Patients With Rheumatoid Arthritis and an Inadequate Response to Anti-Tumor Necrosis Factor Therapy. Arthritis Rheumatol. 2016;68:2867-2877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 153] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 80. | Tanaka Y, Takeuchi T, Tanaka S, Kawakami A, Iwasaki M, Song YW, Chen YH, Wei JC, Lee SH, Rokuda M, Izutsu H, Ushijima S, Kaneko Y, Akazawa R, Shiomi T, Yamada E. Efficacy and safety of peficitinib (ASP015K) in patients with rheumatoid arthritis and an inadequate response to conventional DMARDs: a randomised, double-blind, placebo-controlled phase III trial (RAJ3). Ann Rheum Dis. 2019;78:1320-1332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 95] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 81. | Wu D, Yang XO. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J Microbiol Immunol Infect. 2020;53:368-370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 523] [Cited by in RCA: 581] [Article Influence: 116.2] [Reference Citation Analysis (0)] |