Published online Dec 16, 2021. doi: 10.12998/wjcc.v9.i35.10884
Peer-review started: June 25, 2021
First decision: August 19, 2021
Revised: September 1, 2021
Accepted: October 27, 2021
Article in press: October 27, 2021
Published online: December 16, 2021
Processing time: 167 Days and 16.5 Hours
Pancreatic cancer is a highly heterogeneous disease, making prognosis prediction challenging. Altered energy metabolism to satisfy uncontrolled proliferation and metastasis has become one of the most important markers of tumors. However, the specific regulatory mechanism and its effect on prognosis have not been fully elucidated.
To construct a prognostic polygene signature of differentially expressed genes (DEGs) related to lipid metabolism.
First, 9 tissue samples from patients with pancreatic cancer were collected and divided into a cancer group and a para-cancer group. All patient samples were subjected to metabolomics analysis based on liquid tandem chromatography quadrupole time of flight mass spectrometry. Then, mRNA expression profiles and corresponding clinical data of pancreatic cancer were downloaded from a public database. Least absolute shrinkage and selection operator Cox regression analysis was used to construct a multigene model for The Cancer Genome Atlas.
Principal component analysis and orthogonal projections to latent structures-discriminant analysis (OPLS-DA) based on lipid metabolomics analysis showed a clear distribution in different regions. A Euclidean distance matrix was used to calculate the quantitative value of differential metabolites. The permutation test of the OPLS-DA model for tumor tissue and paracancerous tissue indicated that the established model was consistent with the actual condition based on sample data. A bar plot showed significantly higher levels of the lipid metabolites phosphatidy
This novel model comprising 4 lipid metabolism-related genes might assist clinicians in the prognostic evaluation of patients with pancreatic cancer.
Core Tip: Pancreatic malignant tumors are a highly heterogeneous disease and the seventh leading cause of cancer-related death. Lipid metabolomics analysis suggests differences in lipid metabolites in pancreatic cancer, and the occurrence and development of pancreatic cancer might be linked to lipid metabolism. A cohort from TCGA was used to construct a novel predictive model of a 4-lipid metabolism-related gene signature, which can be used to predict the prognosis of pancreatic cancer.
- Citation: Xu H, Sun J, Zhou L, Du QC, Zhu HY, Chen Y, Wang XY. Development of a lipid metabolism-related gene model to predict prognosis in patients with pancreatic cancer. World J Clin Cases 2021; 9(35): 10884-10898
- URL: https://www.wjgnet.com/2307-8960/full/v9/i35/10884.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i35.10884
Pancreatic cancer is one of the most common malignancies of the digestive system in China. Although the incidence of pancreatic cancer is not high, it is the seventh leading cause of cancer-related death[1]. Despite advances in surgical techniques and adjuvant therapy, mortality and morbidity due to the disease have not changed over the past 40 years. Furthermore, less than 5% of patients with pancreatic cancer have a survival rate of greater than 5 years; even among those who are able to undergo radical resection, the 5-year survival rate is only approximately 20%[2]. Ferlay et al[3] believed that pancreatic cancer would exceed breast cancer as the 3rd leading cause of cancer death in the future. One of the reasons for the low 5-year survival rate for pancreatic cancer is the lack of specific biomarkers for early diagnosis in clinical practice. Therefore, it is of particular importance to identify diagnostic hallmarks of significance for pancreatic cancer.
As the pancreas is an important organ for regulating lipid metabolism in the body, lipids and their metabolites might be used as indicators of health or disease. Lipidomics, proposed by Han in 2003, involves the study of all lipid molecules in the body and their role in the regulation of protein expression and gene expression[4]. Lipidomics has been widely used in the study of biomarkers for various tumors (such as ovarian cancer, prostate cancer, breast cancer)[5-7]. Recently, lipidomics has revealed that plasma concentrations of arachidonic acid, lysophosphatidylcholine, phosphatidylcholine (34:2) and phosphatidylethanolamine (26:0) are increased in patients with early pancreatic cancer[8]. Because lipid metabolism is involved in the proliferation of pancreatic cancer cells, detection of lipid contents in plasma might be helpful for the early diagnosis of pancreatic cancer. However, for pancreatic diseases, especially pancreatic cancer, lipidomics of pancreatic tissue samples has not been comprehensive.
Although our previous studies suggested that differences in serum lipid metabolites might serve as biomarkers for the early diagnosis of pancreatic cancer[9], tissue samples represent changes in the local environment of the tumor and are less affected by systemic factors. We aimed to investigate the characteristics of lipid metabolism profiles in tumor tissues and paracancerous tissues of pancreatic cancer patients using data from our center. Moreover, mRNA expression profiles and corresponding clinical data of pancreatic cancer patients were downloaded from public databases. A prognostic polygene profile of differentially expressed genes (DEGs) related to lipid metabolism was constructed using pancreatic adenocarcinoma (PAAD) data from The Cancer Genome Atlas (TCGA), and functional enrichment analysis was conducted to explore the underlying mechanisms.
All participants from our center signed an informed consent form, which was checked and verified by the Clinical Research Ethics Committee of Shanghai Fourth People’s Hospital Affiliated to Tongji University School of Medicine (No. 2019057-001).
In total, 9 patients with PAAD who had been treated and underwent surgery at the General Surgery, Shanghai Fourth People’s Hospital Affiliated to Tongji University School of Medicine, between October 2018 and March 2019 were enrolled in the study. All 9 patients were diagnosed with pancreatic cancer by computed tomography or magnetic resonance imaging before surgery and underwent laparoscopic pancreaticoduodenectomy; none of the 9 patients received neoadjuvant chemotherapy before surgery to exclude the influence of chemotherapy on the results. Postoperative pathology confirmed ductal adenocarcinoma of the pancreas. The mean age of the patients was 63, including 6 males and 3 females. Regarding clinical stage, 2 cases were T2N0Mx, 3 cases were T2N1Mx, 3 cases were T2N2Mx, and 1 case was T3N1Mx. Fresh frozen tumor tissues were selected as the experimental group, and adjacent tissues were selected as the control group. The exclusion criteria were diagnosis of benign tumors, chronic pancreatitis, other tumors, and radiotherapy and chemotherapy within 6 mo.
RNA-seq data and corresponding clinical information for 176 pancreatic cancer patients as of June 1, 2021, were downloaded from TCGA (https://portal.gdc.cancer.gov/); these data are publicly available. Therefore, this study did not involve relevant ethics or require approval of the local ethics committee. Our current study followed the access policy of TCGA and related guidelines. The “limma” package in R software (version R 4.1.0) was used to normalize the downloaded dataset, and the normalized and readable count values were obtained. A total of 197 genes related to lipid metabolism were retrieved from previous literature[10-12].
DEGs between tumor tissues and adjacent nontumor tissues in TCGA were screened by the “limma” package in R software, with a P value < 0.05 considered to be significant. Univariate Cox analysis of overall survival (OS) was performed to identify DEGs related to lipid metabolism with prognostic value. To minimize the risk of overfitting, a prognostic model was constructed using least absolute shrinkage and selection operator (LASSO) Cox regression analysis[13,14]. The “glmnet” package in R software was used to select variables and shrink them by the LASSO algorithm. The independent variable for regression was the standardized expression matrix of candidate prognosis-related DEGs. Dependent variables were OS and survival status of the pancreatic cancer patients in TCGA. In the LASSO regression model, the parameter λ value of the penalty was used to select the value with the minimum cross-validation error through tenfold cross-validation. The risk score value of patients was calculated according to the standardized expression level of each gene and the corresponding regression coefficient: risk score= esum (each gene’s expression × corresponding coefficient). Patients were divided into high-risk and low-risk groups according to the median risk score. Principal component analysis (PCA) was performed using the “prcomp” function of the “stats” package in R software according to the gene expression in the gene profile. Furthermore, the “Rtsne” package in R software was used to determine the distribution of each group by the t-distributed stochastic neighbor embedding (t-SNE) method. Survival analysis of each differentially expressed gene was performed with the “surv_cutpoint” function of the “survminer” package in R software to obtain the optimal cutoff value. Time-dependent receiver operating characteristic (ROC) curves were plotted using the “survivalROC” package in R software to assess the predictive power of the gene profile.
The “clusterProfiler” package in R software was used to perform Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional analyses for DEGs between the high-risk and low-risk groups based on a P value < 0.05 and false discovery rate (FDR) < 0.05. The corrected P value, or FDR value, was the expected ratio of false rejections (true hypothesis rejections) to the total number of rejected null hypotheses, which was also adjusted for the P value by the BH method. The infiltration scores of 16 types of immune cells and the activities of 13 immune-related pathways were calculated by single sample gene set enrichment analysis (ssGSEA) using the “gsva” package of R software [15].
All figures drawn from our central data were generated with SIMCA software (version 15.0.2, Sartorius Stedim Data Analytics AB, Umea, Sweden). SIMCA software was used to conduct data processing LOG conversion plus center formatting and then automatic modeling analysis[16]. Orthogonal projections to latent structures-discriminant analysis (OPLS-DA) can filter out orthogonal variables unrelated to categorical variables in metabolites and analyze nonorthogonal variables and orthogonal variables to obtain more reliable information about intergroup differences of metabolites and the degree of correlation between experimental groups. SIMCA software was employed to perform LOG conversion and UV formatting for the data. First, OPLS-DA modeling analysis was performed on the first principal component, and 7-fold cross-validation was used to verify the quality of the model. Then, the validity of the model was evaluated by R2Y (the model’s interpretability to the categorical variable Y) and Q2 (the model’s predictability) obtained after cross-validation. Finally, the permutation test was applied to randomly change the arrangement order of the classification variable Y several times to obtain different random Q2 values and further test the validity of the model. Statistical analysis of partial data was performed using SPSS software version 23.0 (IBM Corporation, 2015, Chicago, IL, United States). All continuous variables that followed a normal distribution were compared using Student’s t-test, and the results are shown as the mean ± standard deviation. A bilateral P value < 0.05 was considered statistically significant. The Pearson method was used to calculate the correlation coefficient of the quantitative value of differential metabolites.
All figures drawn with TCGA data were generated using R software (version R 4.1.0). Gene expression values of tumor tissues and adjacent nontumor tissues in TCGA were compared using Student's t-test. The chi-square test was utilized to compare data for classification variables. ssGSEA scores of immune cells or pathways in the high-risk group and the low-risk group were compared by the Mann-Whitney test with corrected P values. Kaplan-Meier analysis and the log-rank test were used to compare OS between the high-risk group and low-risk group, and potential predictors of OS were identified using Cox regression analysis. A P value < 0.05 was considered statistically significant, unless stated otherwise, and all P values were two-tailed.
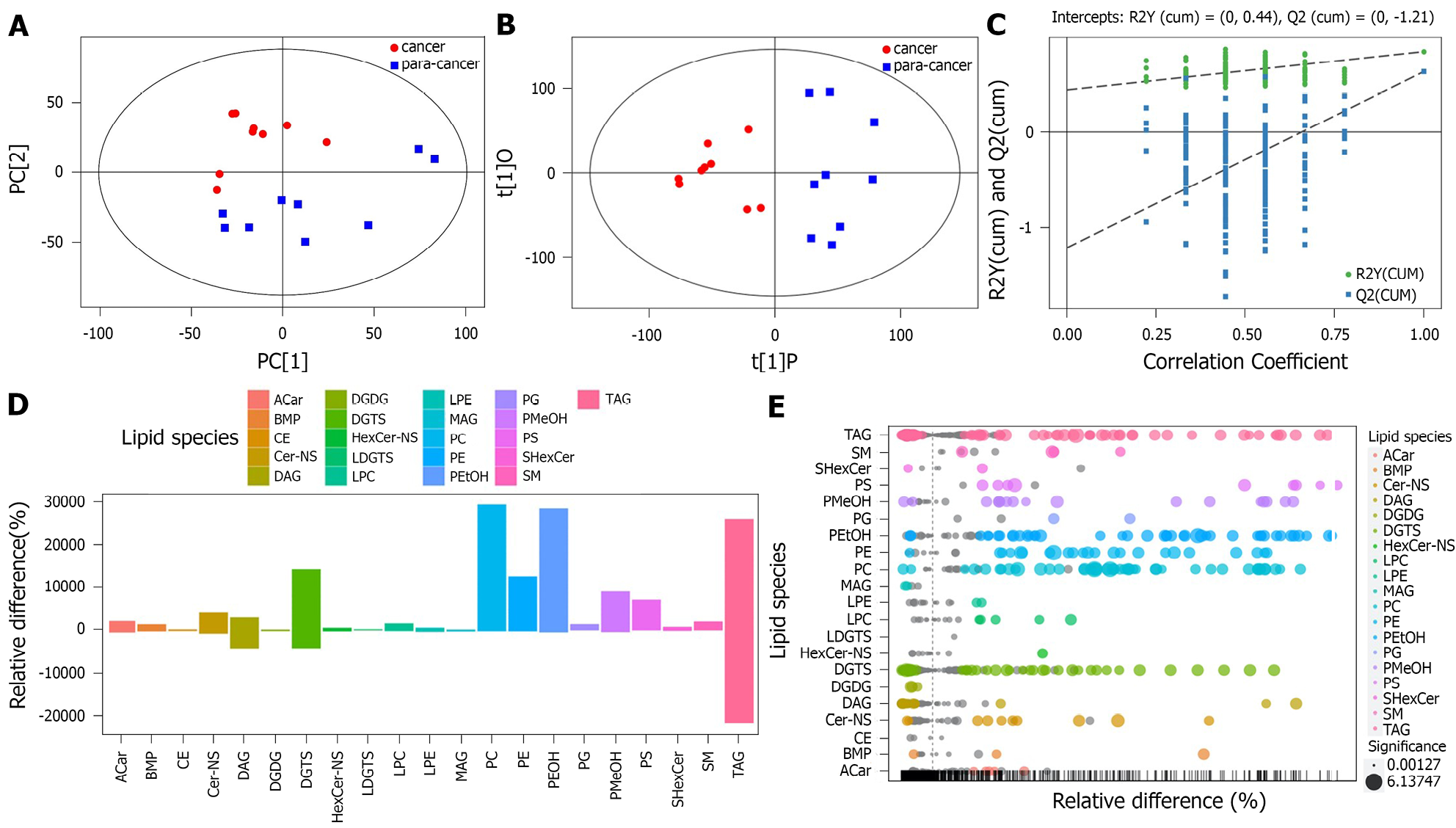
PCA based on lipidomics analysis clearly distinguished between pancreatic cancer and adjacent tissues (Figure 1A). The lipid metabolism of the two groups was distributed in different regions, indicating unique characteristics for the lipid metabolism of the two groups. The samples were all within the 95% confidence interval (Hotelling’s T-Squared Ellipse).
To investigate significant differences in lipid metabolites between the pancreatic cancer group and the paracancer group, the multivariate analysis model OPLS-DA was used. Data from the pancreatic group and the paracancer group were distributed in two opposite regions in the OPLS-DA model (Figure 1B). The results of the OPLS-DA score chart showed that the two groups of samples differed significantly. All samples were within the 95% confidence interval (Hotelling’s T-Squared Ellipse).
The permutation test established the corresponding OPLS-DA model to obtain R2 and Q2 values of the random model by randomly changing the ranking order of the categorical variable Y several times, with an important role in avoiding overfitting of the test model and evaluating its statistical significance. The Q2 values of the random model were all smaller than the Q2 values of the original model. The intercept of the Q2 regression line and vertical axis was less than zero; as the retention degree of displacement decreased gradually, the proportion of the Y variable of displacement increased, and Q2 of the random model decreased gradually. This indicated good robustness for the original model, and the original model could explain the difference between the two groups of samples (Figure 1C). The original Model R2Y was very close to 1, indicating that the established model conformed to the actual state of the sample data. The original Model Q2 was close to 1, indicating that if new samples were added to the model, a more approximate distribution might be obtained. Overall, the original model explained the difference between the two groups of samples well. The Q2 values of the random model were all smaller than those of the original model. The intercept of the Q2 regression line and vertical axis was less than zero. As the retention degree of displacement gradually decreased, the proportion of the Y variable of displacement increased, and Q2 of the random model decreased gradually, indicating that the original model had considerable robustness, with no overfitting observed.
The bar plot of the lipid group visualized the results of the cancer group and para-cancer group using the change degree of metabolite content and classification information (Figure 1D). As illustrated, the lipid metabolites phosphatidylcholine (PC), phosphatidyl ethanolamine (PE), phosphatidylethanol (PEtOH), phosphatidylmethanol (PMeOH), phosphatidylserines (PS) and diacylglyceryl trimethylhomoserine (DGTS) were significantly higher in tumor tissues than in paracancerous tissues.
A bubble plot was visualized by the degree of metabolite content change, difference significance and classification information of the cancer group and the paracancer group (Figure 1E). Each point in the bubble represents a metabolite, and the size of the point represents the P value of Student’s t-test (-log10 P value): the larger the dot, the smaller the P value. Gray points represent nonsignificant differences with a P value not less than 0.05, and colored points represent a P value less than 0.05 (different colors marked according to lipid classification). The relative change percentage of the content was 0, indicating the same content of the metabolite in the two groups. A negative percentage change in relative content indicated a higher content of the metabolite in the paracancer group. Thus, we concluded that the lipid metabolites PE, PEtOH, PMeOH, PS and DGTS were significantly higher in tumor tissues than in paracancer tissues.
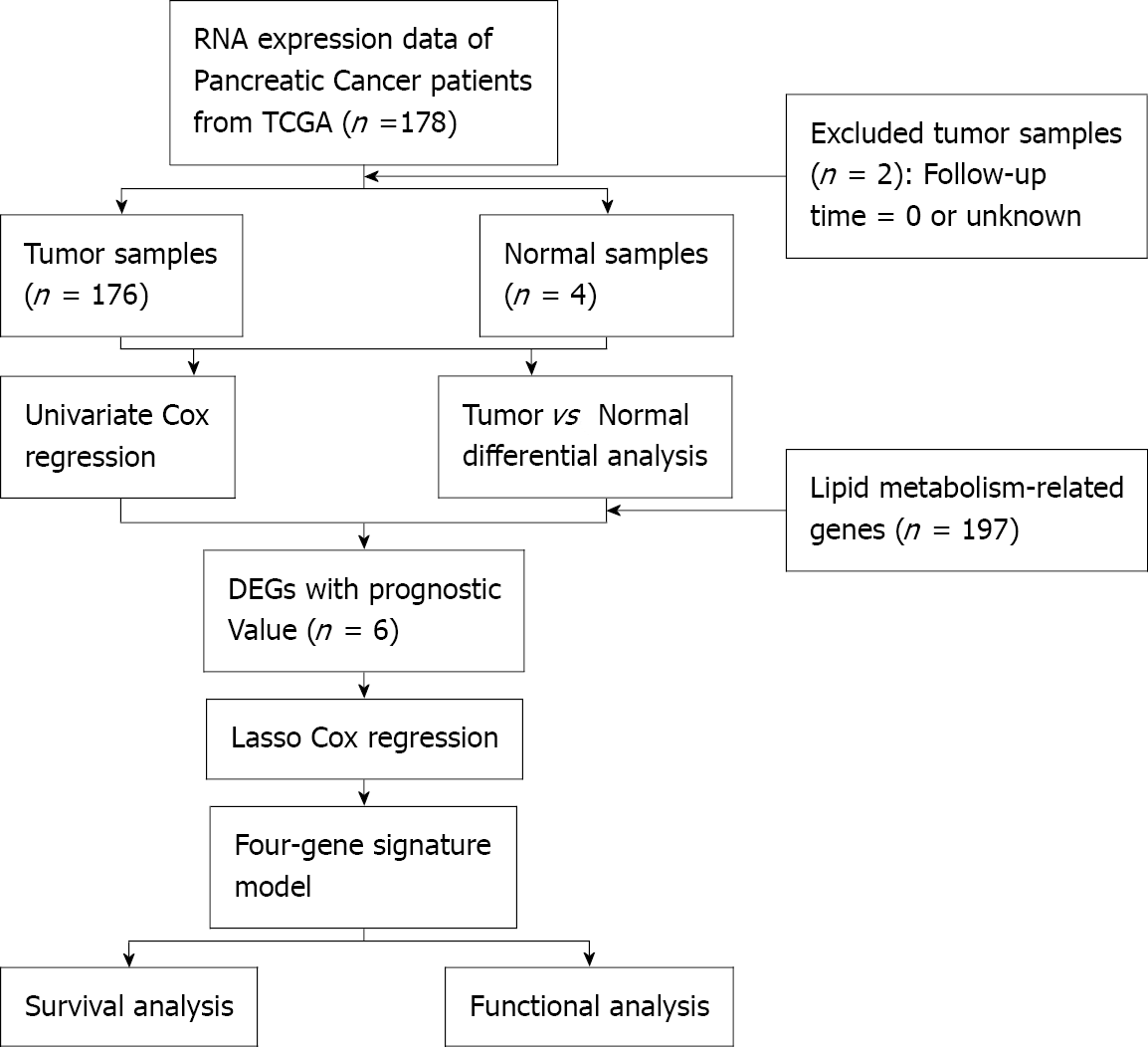
The research flow chart for TCGA is depicted in Figure 2. A total of 176 pancreatic cancer patients from the PAAD cohort of TCGA were enrolled. Baseline clinical data for these patients are shown in Table 1.
| Variables | TCGA data |
| Number of patients | 176 |
| Age (median, range) | 65 (35-88) |
| Sex (%) | |
| Female | 80 (45.5%) |
| Male | 96 (54.5%) |
| Grade (%) | |
| Grade 1 | 30 (17%) |
| Grade 2 | 94 (53.4%) |
| Grade 3 | 48 (27.3%) |
| Grade 4 | 2 (1.1%) |
| Unknown | 2 (1.1%) |
| Stage (%) | |
| I | 21 (11.9%) |
| II | 145 (82.4%) |
| III | 3 (1.7%) |
| IV | 4 (2.3%) |
| Unknown | 3 (1.7%) |
| Survival status | |
| OS days (median) | 464.5 |
| Censored (%) | 92 (52.3%) |
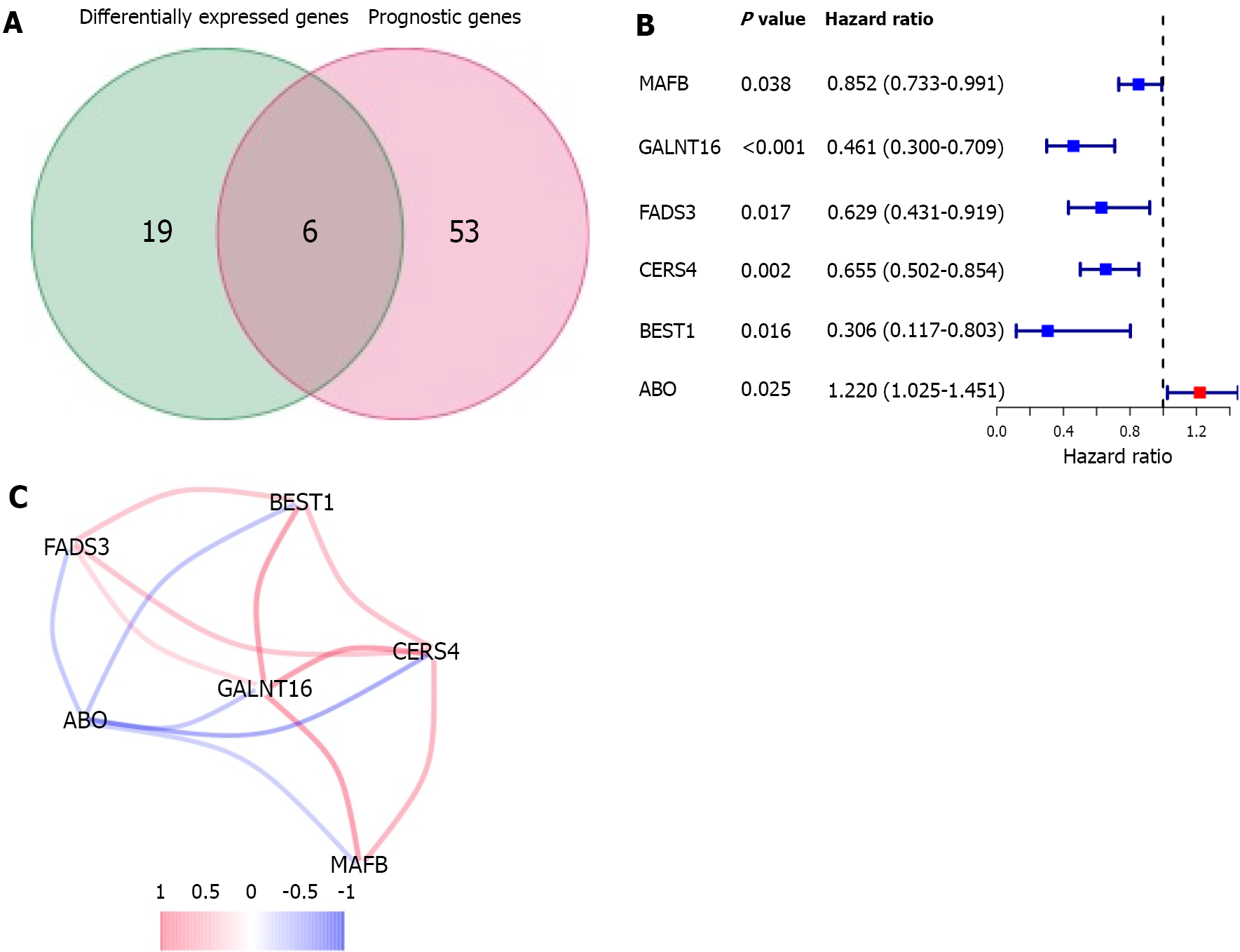
A total of 12.3% (25/197) of genes related to lipid metabolism were differentially expressed between tumor tissues and adjacent paracancerous tissues, and 6 of them were associated with OS in univariate Cox regression analysis (Figure 3A). A group of six prognostic DEGs related to lipid metabolism was revealed (Figure 3B), P value < 0.05). Among the DEGs, the ABO gene was highly expressed in tumor tissues, whereas MAFB, GALNT16, FADS3, CERS4 and BEST1 were expressed at low levels. The correlation between these genes is shown in Figure 3C. The network of interactions among these genes indicated GALNT16 to be the hub gene.
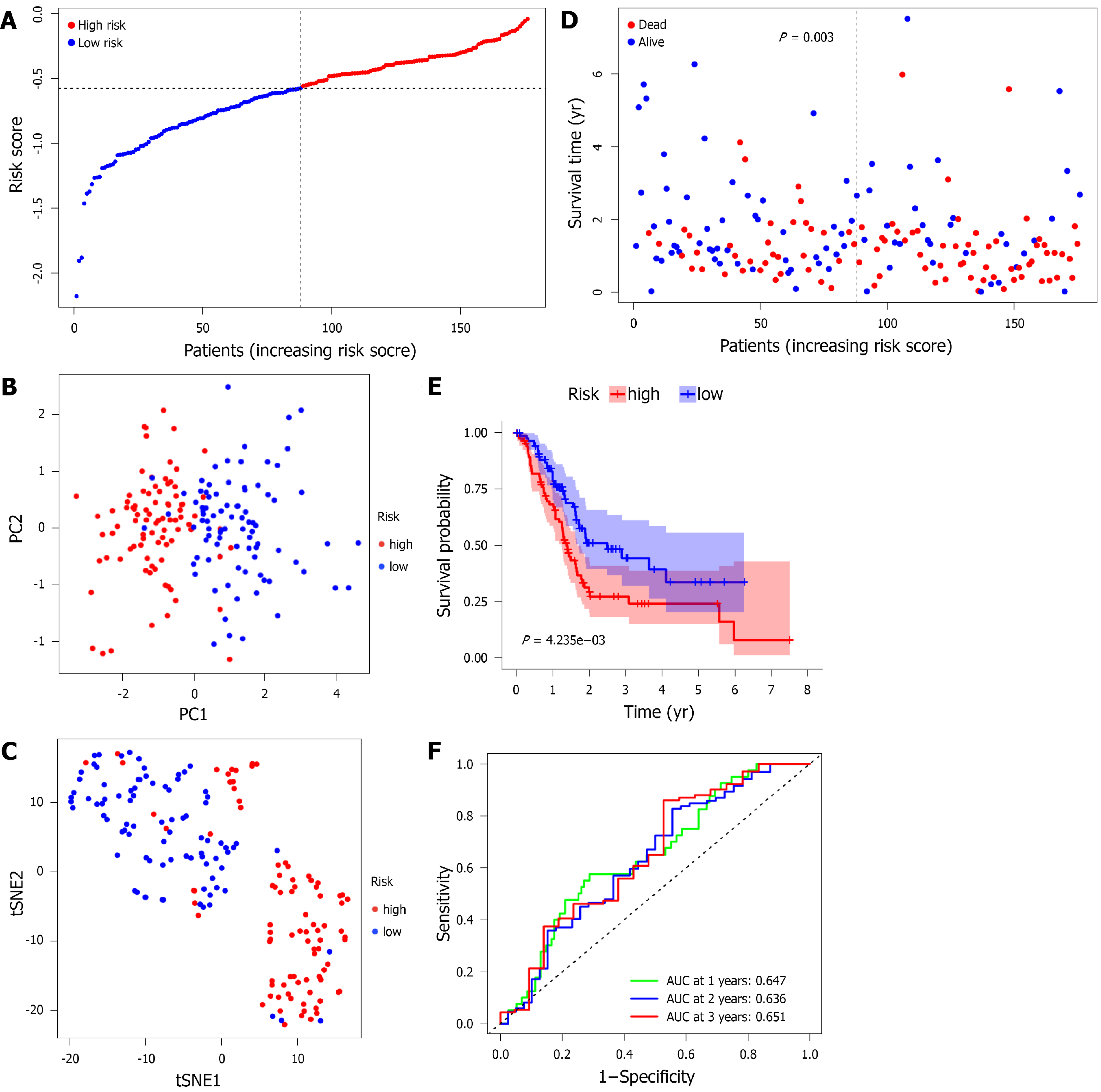
The above 6 lipid metabolism-related gene expression profiles were used to construct a prognostic model for pancreatic cancer patients by LASSO regression analysis. Four gene signature models were determined based on the optimal λ. Survival analysis showed that high expression of these genes was closely associated with poor prognosis in patients with pancreatic cancer according to the optimal cutoff expression value of each gene. The calculation formula of the risk score was as follows: e(0.034 × expression level of ABO - 0.551 × expression level of GALNT16 - 0.135 × expression level of FADS3 - 0.001 × expression level of CERS4). Patients were divided into a high-risk group (n = 88) and a low-risk group (n = 88) based on the median cutoff value (Figure 4A); baseline clinical characteristics for the groups are shown in Table 2. For TCGA pancreatic cancer data, PCA and t-SNE analyses showed that patients in different risk groups were distributed in two directions; the high- and low-risk groups could be distinguished according to these two analyses (Figure 4B and 4C). Figure 4D shows that patients in the high-risk group (63.6%) had a higher probability of death than those in the low-risk group (40.9%), a difference that was significant (P = 0.003). Kaplan-Meier curves showed that patients in the high-risk group had significantly worse OS than those in the low-risk group (Figure 4E, P = 0.004). The prediction performance of the OS risk score was evaluated by ROC curve analysis, and the area under the curve was 0.647 for 1 year, 0.636 for 2 years and 0.651 for 3 years (Figure 4F).
| Characteristics | TCGA-PAAD cohort | P value | |
| High risk | Low risk | ||
| Sex (%) | 0.354 | ||
| Female | 37 | 43 | |
| Male | 51 | 45 | |
| Age (%) | 0.450 | ||
| ≤ 65 yr | 44 | 49 | |
| > 65 yr | 44 | 39 | |
| Tumor grade (%) | 0.271 | ||
| G1+G2 | 59 | 65 | |
| G3+G4 | 27 | 23 | |
| Unknown | 2 | 0 | |
| TNM stage (%) | 0.779 | ||
| I + II | 84 | 82 | |
| III + IV | 3 | 4 | |
| Unknown | 1 | 2 | |
| Survival state | 0.003 | ||
| Alive | 32 | 52 | |
| Deceased | 56 | 36 | |
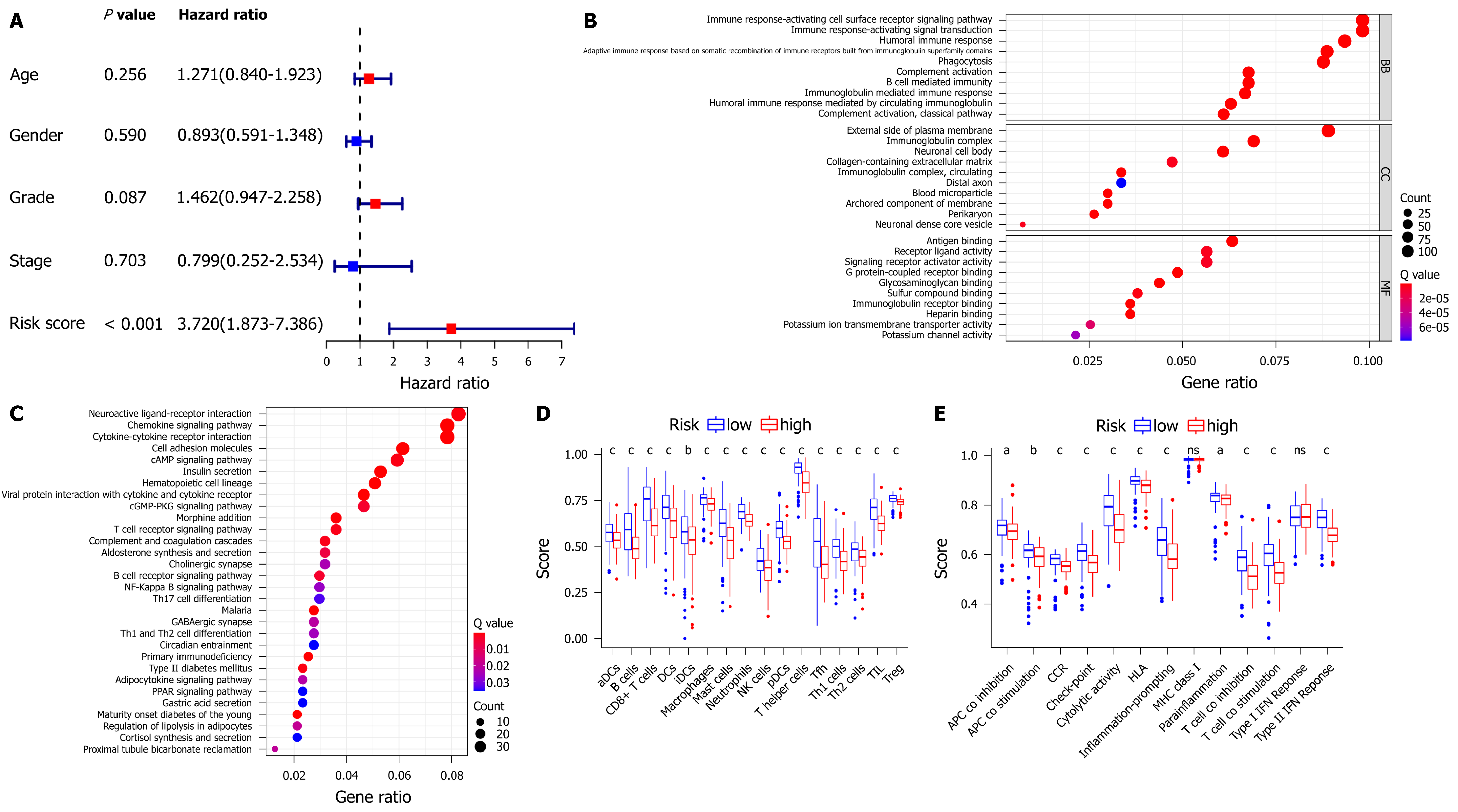
Univariate Cox regression analysis was performed for available variables to determine whether the risk score can serve as a prognostic predictor of OS. In univariate Cox regression analysis, TCGA data analysis for pancreatic cancer indicated a significant correlation between risk score and OS (HR = 3.720, 95% CI = 1.873-7.386, P < 0.001) (Figure 5A).
To further clarify the biological functions and pathways related to the risk score, GO function enrichment and KEGG pathway analyses were performed on DEGs between the high-risk group and the low-risk group. Based on GO functional enrichment analysis, DEGs were enriched in molecular functions related to transmembrane transport, including phagocytosis, receptor ligand activity, signaling receptor activator activity, and G protein-coupled receptor binding (Figure 5B, P value < 0.05). DEGs were also enriched in certain lipid metabolism-related signaling pathways, such as the adipocytokine signaling pathway, regulation of lipolysis in adipocytes and other lipid metabolism-related signaling pathways in TCGA (Figure 5C, P value < 0.05).
To further explore the correlation between the risk score and immune status, we used ssGSEA to quantify enrichment scores for different immune cell subsets and related functions or pathways. In the cohort from TCGA, differences in scores for 16 immune cells were found between the low-risk and high-risk groups, including aDCs, B cells, CD8+ T cells, DCs, iDCs, macrophages, mast cells, neutrophils, NK cells, pDCs, T helper cells, Tfhs, Th1 cells, Th2 cells, TILs and Tregs. Overall, significant differences in all immune cells were observed (Figure 5D, all adjusted P value < 0.05). Surprisingly, except for MHC class I related to antigen presentation and type I IFN response related to immune regulation, which had similar scores in the groups (Figure 5E, P value > 0.05), other related pathways showed relatively high scores in the low-risk group (Figure 5F, P value < 0.05).
The insidious onset, inconspicuous early symptoms, rapid progression and high fatality rate of pancreatic cancer results in considerable difficulties in the early detection, diagnosis and treatment of pancreatic cancer[17]. Although CA199 plays an important role in the diagnosis of pancreatic diseases, it is less sensitive for early pancreatic cancer and precancerous lesions. For example, in one study, CA199 did not serve as a biomarker for the screening of asymptomatic populations but only for the screening of symptomatic patients or clinical differential diagnosis of other diseases[18]. Therefore, finding specific biomarkers for pancreatic cancer has been a hot topic in pancreatic cancer research. In our study, we attempted to search for patient-specific molecular markers of pancreatic cancer by analyzing differences in lipid metabolites between tumor tissues and adjacent tissues. Preliminary analysis results suggest that the lipid metabolites PC, PE, PEtOH, PMeOH, PS and DGTS are significantly higher in tumor tissues than in paracancerous tissues.
Lipids are involved in regulating various biological processes, including energy conversion, material transport, information recognition and transmission, and abnormal lipid metabolism is closely related to certain diseases, such as diabetes mellitus, Alzheimer’s disease, and the occurrence and development of tumors[19-21]. Our results suggest that differences in lipid metabolites exist between tumor tissues and paracancerous tissues in pancreatic cancer patients. Nevertheless, how such differences in lipid metabolites lead to the occurrence and development of tumors is still unknown. Therefore, we systematically investigated the expression of 197 lipid metabolism-related genes in pancreatic cancer tissues and their relationship with OS. We developed a novel prognostic model integrating 4 lipid metabolism-related genes and performed functional analysis and enrichment of immune-related pathways.
The prognostic model proposed in this study consists of four lipid metabolism-related genes: GALNT16, FADS3, CERS4, and ABO. The main functions of these genes can be roughly divided into the following four categories: protein O-linked glycosylation via serine (GALNT16), involvement in the fatty acid biosynthetic process (FADS3), action upstream of or within the lipid metabolic process (CERS4) and involvement in lipid glycosylation (ABO)[22-26]. Tabassum et al[23] reported that GALNT16 is a novel lipid locus associated with cardiovascular diseases (CVDs). GALNT16 was enriched in proteins, lipid metabolism, insulin/IGF pathway-protein kinase B signaling cascade, prolactin signaling pathway, AMPK signaling pathway and other specific biological functions. However, how GALNT16 affects lipid metabolites remains unclear[27]. FADS3 is a novel mammalian membrane-bound fatty acid desaturase gene, and FADS3 single-nucleotide polymorphisms might be associated with changes in plasma levels of triglycerides, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol and sphingolipid metabolism[28]. High expression of FADS3 was associated with higher triglyceride levels. Karsai et al[29] reported a higher level of d18:2 sphingomyelin species (30%) in women than in men and a corresponding higher level of FADS3 activity. Ceramide synthases form a family of six different proteins, and ceramides are components of complex sphingo
Although the mechanisms of tumor susceptibility have been the focus of research recently, the potential regulatory mechanisms between tumor immunity and lipid metabolism remain unknown, especially in pancreatic cancer. We performed GO analysis of genes related to lipid metabolism based on differences in genes between risk groups and were pleasantly surprised to find that these DEGs are enriched in many immune-related biological processes and pathways. Therefore, we also provide evidence that lipid metabolism might be closely related to tumor immunity. An important finding in our study was significant differences in genes related to transmembrane transport in lipid metabolism between the low-risk group and the high-risk group. As alteration of energy metabolism to allow uncontrolled proliferation and metastasis is one of the most important markers of tumors, one possible hypothesis is that different signals released by lipid metabolism affect energy transfer by influencing transmembrane transport processes[33]. In addition, our study found no difference in MHC class I and type I IFN responses between the high-risk and low-risk groups. MHC class I is mainly related to the antigen presentation process in the immune response. Type I interferons, IFN-α and IFN-β, are mainly secreted by innate immune cells and are important effector molecules of antiviral immunity[34]. Type II interferon, IFN-γ, is primarily secreted by activated T cells[35]. Therefore, we believe that lipid metabolites, as opposed to nonantigens, play an important role in tumor development in pancreatic cancer patients.
The study had some limitations. First, we constructed a prognostic model using a public database; however, more prospective real-world data should be used to verify its clinical validity. Although we also attempted to use other public database data for verification, the sample size of these databases was small, and the available data were very limited. Second, our study developed a prognostic model by considering only one factor, lipid metabolism, and excluded many significant prognostic genes involved in other aspects of pancreatic cancer, which might constitute an inherent flaw. Third, 197 lipid metabolism-related genes mentioned in this paper were only summarized through a literature review, and some rare genes or functions involved in lipid metabolism are not included herein. Fourth, when we performed lipid metabolite analysis, our research only looked for differences in lipid metabolites between cancer tissues and adjacent para-cancer tissues and did not analyze differences between cancer tissues and healthy control pancreatic tissues. Given the high risk of pancreatic surgery, it was difficult to obtain healthy controls for pancreatic tissue. However, our previous studies showed differences in lipid metabolites between pancreatic cancer patients and healthy controls in blood samples[36]. In addition, the association between the risk score and immune activity was not further validated in the corresponding experiments.
In conclusion, our study developed a novel prognostic model composed of 4 lipid metabolism-related genes, which was analytically confirmed to be associated with OS in patients with pancreatic cancer and to be an independent risk factor for predicting the prognosis of pancreatic cancer.
Finding specific prognostic markers is important for pancreatic cancer. Understanding the relationship between lipid metabolism-related genes and pancreatic cancer is helpful to improve its prognosis.
To construct a novel model to predict the prognosis of pancreatic cancer.
To investigate the characteristics of lipid metabolites in pancreatic cancer and construct a prognostic polygene signature of differentially expressed genes related to lipid metabolism.
Lipid metabolomics analysis was conducted to explore differences in lipid metabolites between pancreatic cancer tissues and paracancerous tissues. A predictive model of lipid metabolism genes associated with pancreatic cancer was established using a cohort from The Cancer Genome Atlas.
Lipid metabolomics analysis showed that the lipid metabolites phosphatidylcholine, phosphatidyl ethanolamine, phosphatidylethanol, phosphatidylmethanol, phosphatidylserines and diacylglyceryl trimethylhomoserine were significantly higher in cancer tissues. A 4-gene signature model, including GALNT16, FADS3, CERS4 and ABO, was developed to predict the prognosis of pancreatic cancer.
Differentially expressed genes related to lipid metabolism reflected abnormal lipid metabolism in pancreatic cancer. A novel predictive model of a 4-lipid metabolism-related gene signature contributed to the prediction of pancreatic cancer prognosis.
New gene markers and models are needed to predict prognosis because of the high heterogeneity of pancreatic cancer.
The authors would like to thank Prof. Xiang-Dong Wang and Shanghai Zhongshan Hospital for guidance on the design of the study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Yoshizawa T S-Editor: Chang KL L-Editor: A P-Editor: Chang KL
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55758] [Article Influence: 7965.4] [Reference Citation Analysis (132)] |
| 2. | Conroy T, Bachet JB, Ayav A, Huguet F, Lambert A, Caramella C, Maréchal R, Van Laethem JL, Ducreux M. Current standards and new innovative approaches for treatment of pancreatic cancer. Eur J Cancer. 2016;57:10-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 135] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 3. | Ferlay J, Partensky C, Bray F. More deaths from pancreatic cancer than breast cancer in the EU by 2017. Acta Oncol. 2016;55:1158-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 311] [Article Influence: 34.6] [Reference Citation Analysis (1)] |
| 4. | Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44:1071-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 668] [Cited by in RCA: 631] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 5. | Liu Y, Chen Y, Momin A, Shaner R, Wang E, Bowen NJ, Matyunina LV, Walker LD, McDonald JF, Sullards MC, Merrill AH Jr. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol Cancer. 2010;9:186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 99] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 6. | Eberlin LS, Dill AL, Costa AB, Ifa DR, Cheng L, Masterson T, Koch M, Ratliff TL, Cooks RG. Cholesterol sulfate imaging in human prostate cancer tissue by desorption electrospray ionization mass spectrometry. Anal Chem. 2010;82:3430-3434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 7. | Min HK, Lim S, Chung BC, Moon MH. Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal Bioanal Chem. 2011;399:823-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 8. | Urayama S, Zou W, Brooks K, Tolstikov V. Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rapid Commun Mass Spectrom. 2010;24:613-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 109] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 9. | Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, Zhou C, Reungwetwattana T, Cheng Y, Chewaskulyong B, Shah R, Cobo M, Lee KH, Cheema P, Tiseo M, John T, Lin MC, Imamura F, Kurata T, Todd A, Hodge R, Saggese M, Rukazenkov Y, Soria JC; FLAURA Investigators. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med. 2020;382:41-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1120] [Cited by in RCA: 1901] [Article Influence: 380.2] [Reference Citation Analysis (0)] |
| 10. | Ding HR, Wang JL, Ren HZ, Shi XL. Lipometabolism and Glycometabolism in Liver Diseases. Biomed Res Int. 2018;2018:1287127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 11. | Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610-2623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 839] [Cited by in RCA: 1009] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 12. | Park JK, Coffey NJ, Limoges A, Le A. The Heterogeneity of Lipid Metabolism in Cancer. Adv Exp Med Biol. 2018;1063:33-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 13. | Simon N, Friedman J, Hastie T, Tibshirani R. Regularization Paths for Cox's Proportional Hazards Model via Coordinate Descent. J Stat Softw. 2011;39:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1263] [Cited by in RCA: 1352] [Article Influence: 96.6] [Reference Citation Analysis (0)] |
| 14. | Ternès N, Rotolo F, Michiels S. Empirical extensions of the lasso penalty to reduce the false discovery rate in high-dimensional Cox regression models. Stat Med. 2016;35:2561-2573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 15. | Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2009] [Cited by in RCA: 2768] [Article Influence: 276.8] [Reference Citation Analysis (0)] |
| 16. | Wiklund S, Johansson E, Sjöström L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80:115-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 906] [Cited by in RCA: 898] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 17. | Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, Neale RE, Tempero M, Tuveson DA, Hruban RH, Neoptolemos JP. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1442] [Cited by in RCA: 1333] [Article Influence: 148.1] [Reference Citation Analysis (2)] |
| 18. | Jelski W, Mroczko B. Biochemical diagnostics of pancreatic cancer - Present and future. Clin Chim Acta. 2019;498:47-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Meyer zu Heringdorf D, Jakobs KH. Lysophospholipid receptors: signalling, pharmacology and regulation by lysophospholipid metabolism. Biochim Biophys Acta. 2007;1768:923-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 295] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 20. | Brinkmann V. Sphingosine 1-phosphate receptors in health and disease: mechanistic insights from gene deletion studies and reverse pharmacology. Pharmacol Ther. 2007;115:84-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 446] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 21. | Iessi E, Marconi M, Manganelli V, Sorice M, Malorni W, Garofalo T, Matarrese P. On the role of sphingolipids in cell survival and death. Int Rev Cell Mol Biol. 2020;351:149-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 22. | Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449-462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 477] [Cited by in RCA: 648] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 23. | Tabassum R, Rämö JT, Ripatti P, Koskela JT, Kurki M, Karjalainen J, Palta P, Hassan S, Nunez-Fontarnau J, Kiiskinen TTJ, Söderlund S, Matikainen N, Gerl MJ, Surma MA, Klose C, Stitziel NO, Laivuori H, Havulinna AS, Service SK, Salomaa V, Pirinen M; FinnGen Project, Jauhiainen M, Daly MJ, Freimer NB, Palotie A, Taskinen MR, Simons K, Ripatti S. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat Commun. 2019;10:4329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 122] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 24. | Riebeling C, Allegood JC, Wang E, Merrill AH Jr, Futerman AH. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J Biol Chem. 2003;278:43452-43459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 240] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 25. | Marquardt A, Stöhr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 220] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 26. | Raman J, Guan Y, Perrine CL, Gerken TA, Tabak LA. UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferases: completion of the family tree. Glycobiology. 2012;22:768-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 27. | Gao Y, Jiang J, Yang S, Hou Y, Liu GE, Zhang S, Zhang Q, Sun D. CNV discovery for milk composition traits in dairy cattle using whole genome resequencing. BMC Genomics. 2017;18:265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 28. | Rioux V, Legrand P. Fatty Acid Desaturase 3 (FADS3) Is a Specific ∆13-Desaturase of Ruminant trans-Vaccenic Acid. Lifestyle Genom. 2019;12:18-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Karsai G, Lone M, Kutalik Z, Brenna JT, Li H, Pan D, von Eckardstein A, Hornemann T. FADS3 is a Δ14Z sphingoid base desaturase that contributes to gender differences in the human plasma sphingolipidome. J Biol Chem. 2020;295:1889-1897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 30. | Ebel P, Imgrund S, Vom Dorp K, Hofmann K, Maier H, Drake H, Degen J, Dörmann P, Eckhardt M, Franz T, Willecke K. Ceramide synthase 4 deficiency in mice causes lipid alterations in sebum and results in alopecia. Biochem J. 2014;461:147-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 31. | Hartmann D, Lucks J, Fuchs S, Schiffmann S, Schreiber Y, Ferreirós N, Merkens J, Marschalek R, Geisslinger G, Grösch S. Long chain ceramides and very long chain ceramides have opposite effects on human breast and colon cancer cell growth. Int J Biochem Cell Biol. 2012;44:620-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 32. | Ureme S, Anioke IC, Emenuga V, Eluke BC. Lipid Profiles in Different ABO Blood Groups in Owerri Metropolis, South East Nigeria: A Preliminary Study. Asian Journal of Research in Biochemistry 2018; 10:1-7. [DOI] [Full Text] |
| 33. | Zhu S, Dong Z, Ke X, Hou J, Zhao E, Zhang K, Wang F, Yang L, Xiang Z, Cui H. The roles of sirtuins family in cell metabolism during tumor development. Semin Cancer Biol. 2019;57:59-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 34. | Wu D, Sanin DE, Everts B, Chen Q, Qiu J, Buck MD, Patterson A, Smith AM, Chang CH, Liu Z, Artyomov MN, Pearce EL, Cella M, Pearce EJ. Type 1 Interferons Induce Changes in Core Metabolism that Are Critical for Immune Function. Immunity. 2016;44:1325-1336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 251] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 35. | Kosmidis C, Sapalidis K, Koletsa T, Kosmidou M, Efthimiadis C, Anthimidis G, Varsamis N, Michalopoulos N, Koulouris C, Atmatzidis S, Liavas L, Strati TM, Koimtzis G, Tsakalidis A, Mantalovas S, Zarampouka K, Florou M, Giannakidis DE, Georgakoudi E, Baka S, Zarogoulidis P, Man YG, Kesisoglou I. Interferon-γ and Colorectal Cancer: an up-to date. J Cancer. 2018;9:232-238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | Zhou D, Mu D, Cheng M, Dou Y, Zhang X, Feng Z, Qiu G, Yu H, Chen Y, Xu H, Sun J, Zhou L. Differences in lipidomics may be potential biomarkers for early diagnosis of pancreatic cancer. Acta Cir Bras. 2020;35:e202000508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |