Published online Dec 6, 2021. doi: 10.12998/wjcc.v9.i34.10392
Peer-review started: February 1, 2021
First decision: March 29, 2021
Revised: March 10, 2021
Accepted: August 4, 2021
Article in press: August 4, 2021
Published online: December 6, 2021
Processing time: 301 Days and 17.4 Hours
The outbreak of coronavirus disease 2019 (COVID-19) is a significant challenge for clinicians, especially for immunocompromised cancer patients. By analyzing the impact of COVID-19 on the immune microenvironment of colorectal cancer (CRC) patients at the tissue level and single-cell level, we found that CRC patients are more easily infected by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), but promotion of infiltration and differentiation of monocytes makes them more likely to develop severe COVID-19. Because of the continuing activation of nuclear factor (NF)-κB and C-C chemokine receptor type 5 (CCR5) signaling pathways in monocytes, imbalance of macrophage polarization can aggravate the cytokine release syndrome. Therefore, regulating the infiltration and differentiation of monocytes is helpful for the treatment of COVID-19 in CRC patients.
Core Tip: Not only are colorectal cancer (CRC) patients susceptible to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) but their infiltrating monocytes are also affected by SARS-CoV-2. Promotion of infiltration and differentiation of monocytes after infection CRC patients are more likely to develop severe coronavirus disease 2019 (COVID-19). In severe COVID-19, because of activation of the nuclear factor (NF)-κB and C-C chemokine receptor type 5 (CCR5) signaling pathways, the imbalance of macrophage polarization can cause further aggravation of the cytokine release syndrome.
- Citation: Bai L, Yang W, Qian L, Cui JW. Regulating monocyte infiltration and differentiation: Providing new therapies for colorectal cancer patients with COVID-19. World J Clin Cases 2021; 9(34): 10392-10399
- URL: https://www.wjgnet.com/2307-8960/full/v9/i34/10392.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i34.10392
As a public health emergency of international concern, there are nearly 150 million coronavirus disease 2019 (COVID-19) cases worldwide. In particular, cancer patients are more vulnerable to virus infection because of their suppressed immune microenvironment. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) enters cells by recognizing angiotensin I converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2)[1], and other proteases, such as cathepsin B (CTSB) and cathepsin L (CTSL)[2], in the host cell. Clarifying the expression of those proteins unique to various human pathologies may be helpful in identifying susceptible populations.
Studies have shown that cancer patients not only have a 2.31 times higher risk of infection with SARS-CoV-2 than the general population[3] but also have a higher risk of developing severe COVID-19[3,4]. Compared with patients without cancer, those with cancer have a 3.56-fold increased risk of severe disease following COVID-19 infection[5]. However, the mechanism of the exacerbation of COVID-19 is not clear. Therefore, revealing the effects of COVID-19 infection on the human body may provide new ideas for preventing the deterioration of COVID-19 patients.
The leading cause of COVID-19 aggravation is multiorgan dysfunction caused by the cytokine release syndrome (CRS). By comparing the differences in proteomics and metabolomics of severe and nonsevere COVID-19 cases with healthy controls, it was found that the severe cases were associated with abnormal macrophage regulation[6,7]. The results of meta-analysis and bioinformatics analysis have shown that colorectal cancer (CRC) patients are more susceptible to SARS-CoV-2 than cancer patients with other tumors[8]. Studies have also shown that COVID-19 patients with CRC are more likely to have clinical characteristics with a poor COVID-19 prognosis than matched patients with COVID-19 but without cancer[9]. We used CRC as an example to elucidate the effect of SARS-CoV-2 on the cancer immune microenvironment, especially the impact on monocytes, with the goal of finding treatments to delay the progression of COVID-19.
After SARS-CoV-2 infection, viral envelope spike proteins bind to ACE2 and promote cellular recognition of the virus[2]. If transmembrane serine protease 2 (TMPRSS2) is present, it promotes the cleavage and activation of S proteins by host cells. SARS-CoV-2 usually enters cells through the endosomal pathway, but in the absence of TMPRSS2, it enters by cathepsin L (CTSL) and cathepsin B (CTSB) proteolysis and activation of S protein[10-12].
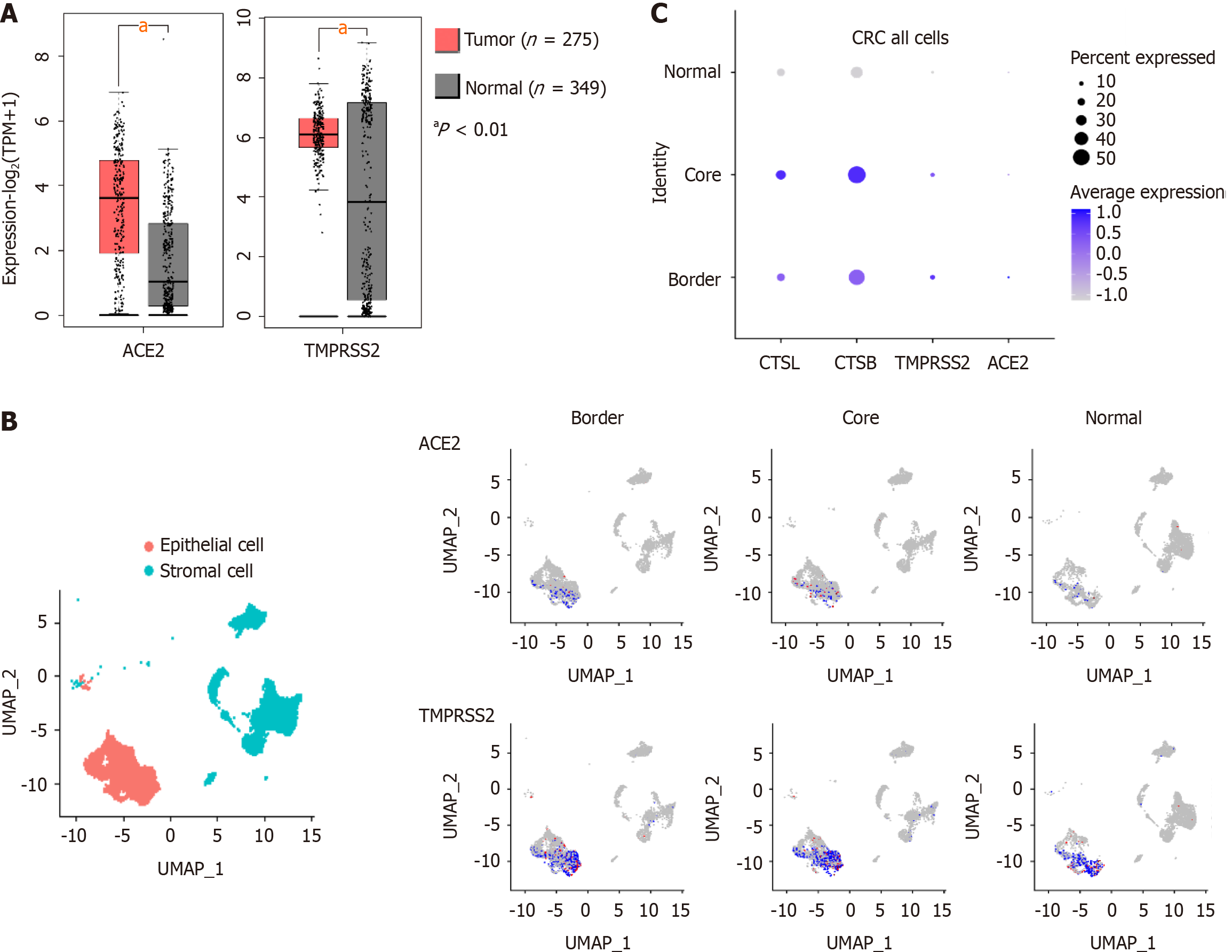
Data on mRNA expression in colorectal cancer tissue in the Cancer Genome Atlas (https://cancergenome.nih.gov/) and the Genotype-Tissue Expression project (https://www.gtexportal.org/home/) databases, we found that the expression of ACE2 and TMPRSS2 was higher in CRC than in healthy tissue (Figure 1A). In addition, single-cell sequencing data from 27,414 cells from six CRC patients in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (GSE144735), we found that ACE2, TMPRSS2, CTSL, and CTSB were primarily expressed in stromal and epithelial cells of CRC tissue (Figure 1B, C). Therefore, the susceptibility of CRC patients to COVID-19 may be associated with the high expression of SARS-CoV-2 recognition proteins.
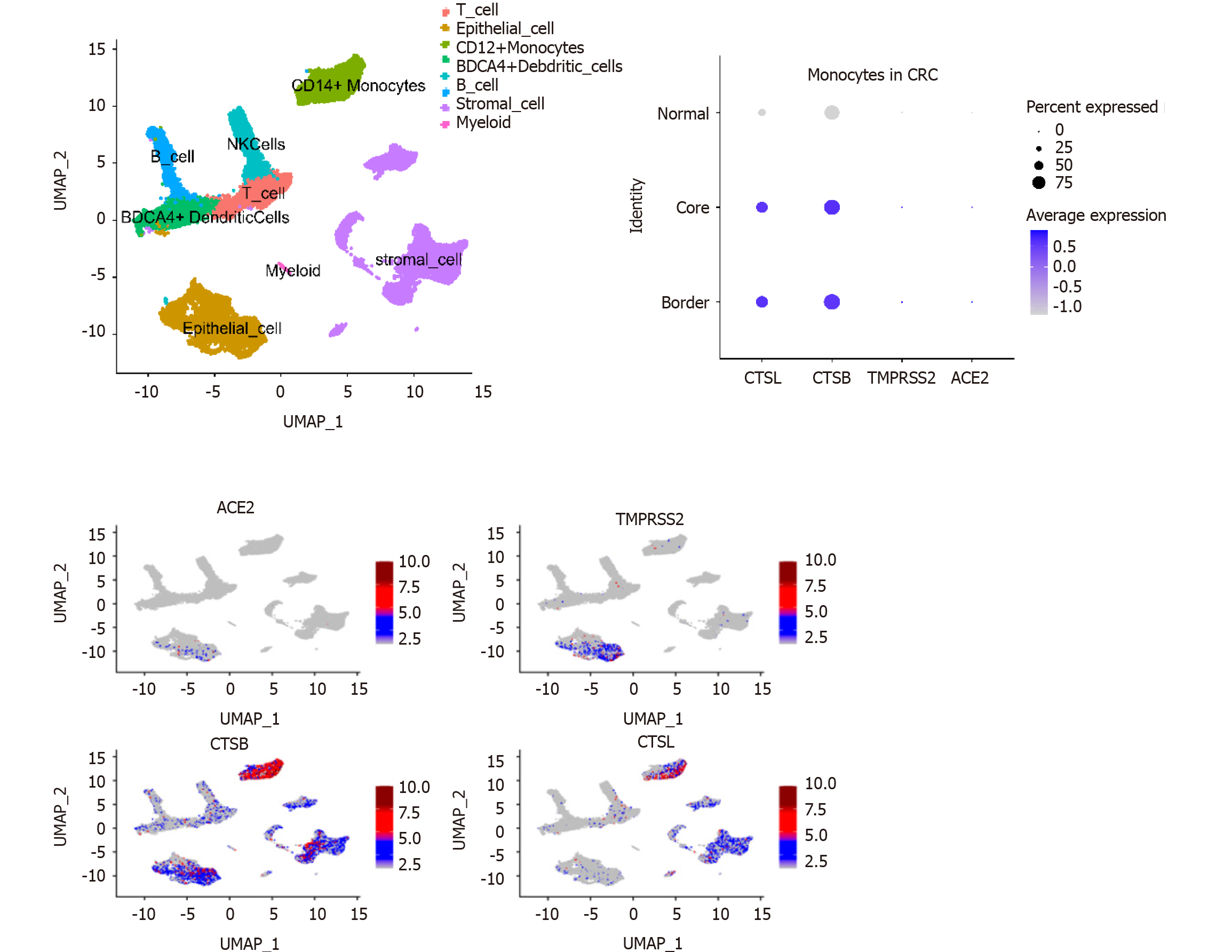
Analysis of GEO single-cell sequencing data (GSE144735) revealed that expression of the CTSL and CTSB SARS-CoV-2 recognition proteins was higher in monocytes compared with other immune cells in the tumor microenvironment (Figure 2). Consequently, SARS-CoV-2 infection may affect the function of monocytes. The analysis also showed that the expression of CTSL and CTSB mRNA was higher in infiltrated monocytes in cells from tumor than it was in cells from healthy tissue (Figure 2). Thus, when infected by COVID-19, monocytes in the tumor microenvironment may induce a stronger inflammatory effect.
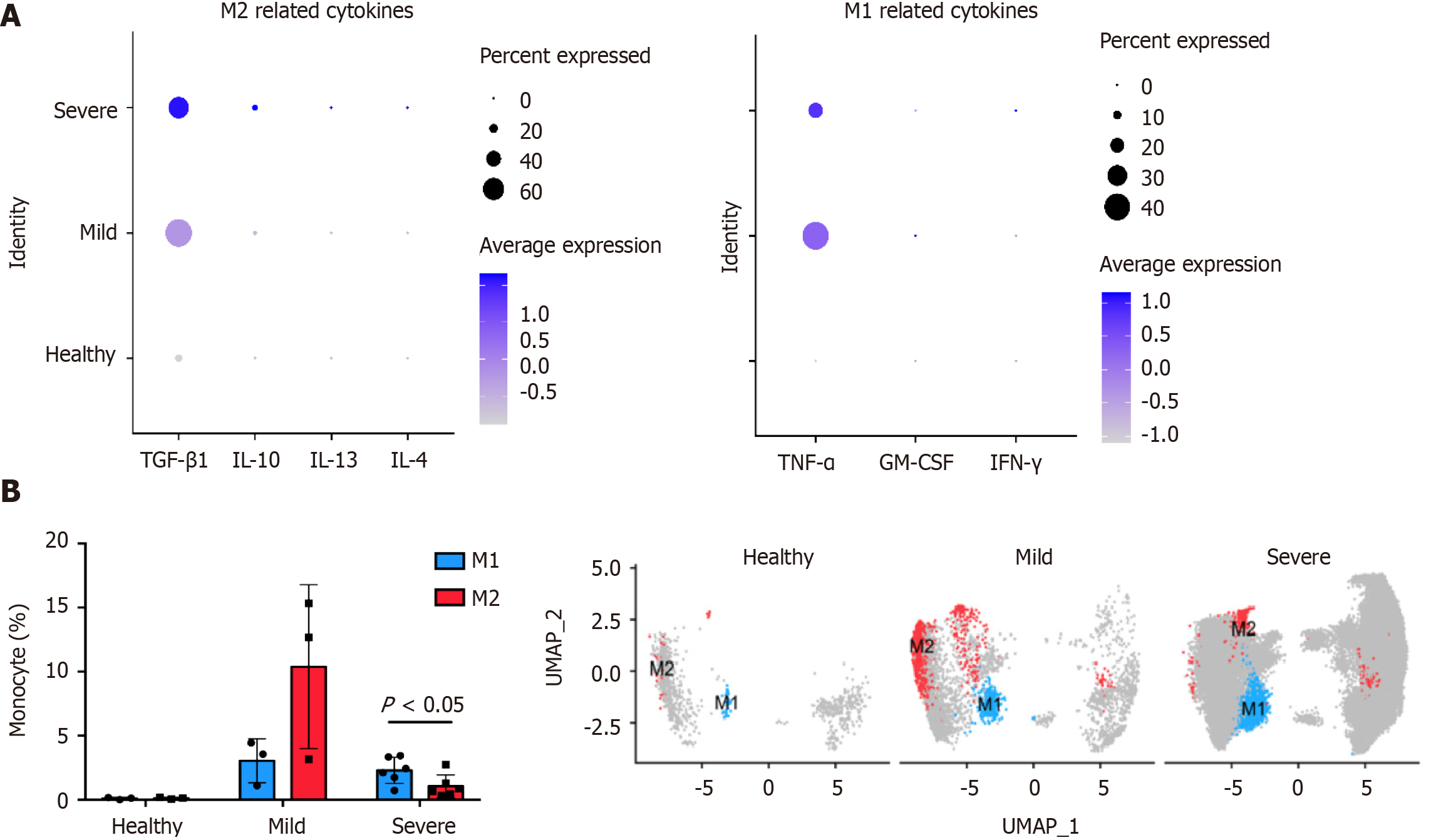
Monocytes constitute a pool of dendritic cells and macrophages, and differentiate into type 1 macrophages (M1) and type 2 macrophages (M2). M1 are proinflammatory macrophages and have an anti-tumor effect. M2 are anti-inflammatory macrophages that mediate tumor immune escape. The effects of monocyte cytokines produced following SARS-CoV-2 infection on macrophage polarization was investigated using GEO data (GSE145926). Cluster-based processing of cells from three healthy, three mild, and six severe COVID-19 patient using Harmony (https://www.harmony-alliance.eu/covid19/covid-19-news/open-call-for-data-partners-to-join-the-harmony-covid-19-data-platform) was used to remove batch effects and select 20 principal components. Differential expression analysis with Seurat (https://satijalab.org/seurat//archive/v3.2/de_vignette.html) identified CD14+ monocytes in the patient samples for subsequent analysis. The proportions of M1 and M2 in the samples were determined by counting the CD80- and CD86-positive M1 and MRC1- and CD163-positive M2 cells. Differences were compared with GraphPad Prism 7.0a for Mac OS X (GraphPad Inc., La Jolla, CA, United States) and the unpaired two-tailed Student’s t-test. Numeric data are reported as means ± SD. The gene set variation analysis (GSVA) for microarray and RNA-seq data (https://www.bioconductor.org/packages/release/bioc/html/GSVA.html) R statistics package was used to score gene set enrichment analysis data from 31,557 monocytes (c2.cp.v7.1.symbols.gmt: https://data.broadinstitute.org/gsea-msigdb/msigdb/release/7.1/).
Analysis of the cytokines expressed by monocytes in alveolar lavage fluid (GEO: GSE145926, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145926) showed that COVID-19 induced the differentiation of M1 and M2 by affecting the cytokines TGF-β1 and TNF-α (Figure 3A). Therefore, SARS-CoV-2 promotes macrophage polarization and enhances macrophage infiltration by stimulating cytokine production. By comparing the correlation between the proportion of macrophages and the degree of infection, it was found that patients with COVID-19 had higher macrophage infiltration than healthy people (Figure 3B). However, after developing into severe COVID-19, the M1/M2 ratio increased (Figure 3B), which may further aggravate the CRS.
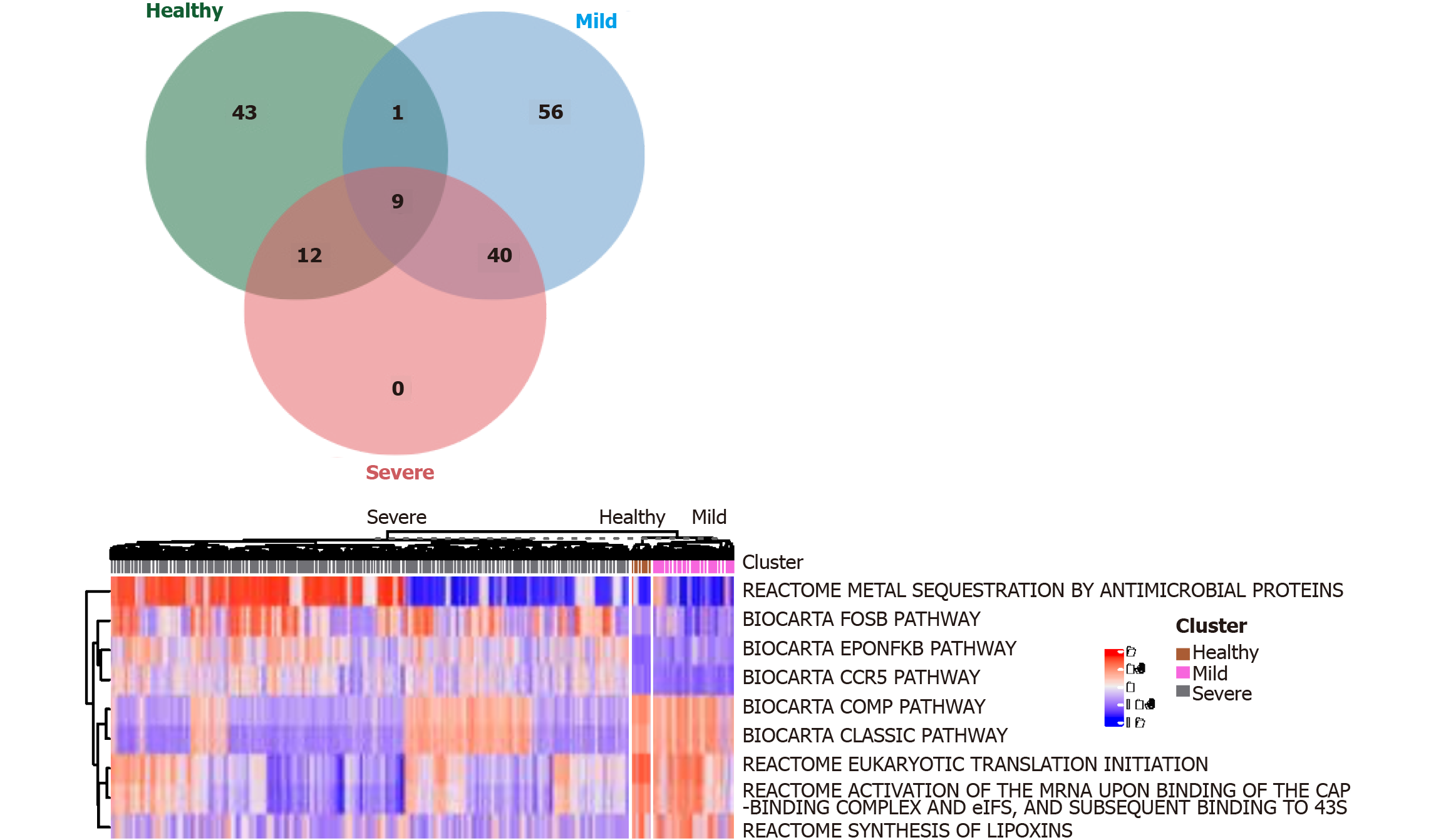
We use the Linear Models for Microarray (limma) R package (https://bioconductor.org/packages/release/bioc/html/limma.html) to distinguish signaling pathways (DEGenesets) in patients with severe and nonsevere COVID-19 infection and healthy controls and cutoffs of the average absolute between-group differences. We identified nine signaling pathways that were statistically different (≥ 0.35 false discovery rate-adjusted P < 0.001, Figure 4). The results suggest that COVID-19 infection promotes the activation of NF-κB and CCR5 signaling pathways in monocytes. Multiple metabolic pathways in monocytes are involved (Figure 4). Inhibiting the activation of the NF-κB or CCR5 signaling pathway is expected to maintain the balance of M1 and M2 monocytes and thus prevent progression to severe COVID-19 infection.
COVID-19 has many implications for the diagnosis and treatment of cancer. With the increasing understanding of the SARS-CoV-2 infection-related signaling pathway, a growing number of studies have described the association between tumors and the risk of CDVID-19 infection. Clinical studies and meta-analyses have also preliminarily confirmed the susceptibility of Chinese CRC patients to COVID-19[13,14], but reason needs further study. This study briefly described the expression of SARS-CoV-2 recognition proteins in the cells and tissues of COVID-19 patients, providing a reference for subsequent studies on the susceptibility of CRC patients to COVID-19.
Because SARS-CoV-2 recognition proteins are not only expressed in tumor cells but also immune cells, we analyzed the expression of essential proteins related to COVID-19 infection in various immune cells by the single-cell sequencing analysis of cells in CRC patients. We found that SARS-CoV-2 infection may affect the function of monocytes and that SARS-CoV-2 infection can promote the activation of NF-κB and CCR5 signaling pathways in monocytes. Suppressing the activation of NF-κB and CCR5 signaling pathways may reshape the balance of macrophage polarization, which can be helpful for the treatment of COVID-19.
Because of the relationship between monocyte-macrophage activation and the severity of COVID-19, the cytokines released by epithelial cells and fibroblasts following SARS-CoV-2 infection promote the recruitment of monocytes and the activation of macrophages, which promotes the CRS[15]. We also uncovered evidence that monocyte-macrophage infiltration was associated with COVID-19 severity and that M1/M2 ratio was higher in severe than in nonsevere COVID-19, which promotes disease progression. We believe that the high expression of SARS-CoV-2 recognition protein in monocytes affects monocytes function in direct or indirect ways.
Statins are inhibitors of cholesterol synthesis. They are also helpful in the treatment of COVID-19[16], although it is not clear how they do that. Statins can not only directly inhibit the growth and development of tumors, but also inhibit the release of pro-differentiation cytokines by M1 macrophages. They also cause a reduction of the M1/M2 ratio and inhibit macrophage infiltration by inhibiting TLR4/MYD88/NF-κB signaling[17,18]. Furthermore, lipids promote the upregulation of CCR5 in monocytes and enhance their proinflammatory phenotype[19]. Inhibitors of NF-κB and CCR5, such as statins, may offer a novel treatment for CRC patients with COVID-19 by regulating the changes in the M1/M2 ratio. Because lipopolysaccharides are a TLR4 agonist, active prevention and treatment of combined bacterial infections may be effective to prevent CRS occurrence. Leronlimab, a CCR5-specific antibody, has also been reported as a potential treatment of COVID-19. It inhibits macrophage polarization and CRS occurrence by blocking CCR5[20,21]. We believe that other drugs that block NF-κB or CCR5 may also be effective in treating COVID-19.
Provenance and peer review: Invited article; Externally peer reviewed.
Specialty type: Immunology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Trabelsi M S-Editor: Wu YXJ L-Editor: Filipodia P-Editor: Wang LYT
| 1. | Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, Talavera-López C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, Yoshida M, Barnes JL; HCA Lung Biological Network. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1985] [Cited by in RCA: 1952] [Article Influence: 390.4] [Reference Citation Analysis (0)] |
| 2. | Gkogkou E, Barnasas G, Vougas K, Trougakos IP. Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators. Redox Biol. 2020;36:101615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 3. | Yu J, Ouyang W, Chua MLK, Xie C. SARS-CoV-2 Transmission in Patients With Cancer at a Tertiary Care Hospital in Wuhan, China. JAMA Oncol. 2020;6:1108-1110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 626] [Cited by in RCA: 773] [Article Influence: 154.6] [Reference Citation Analysis (0)] |
| 4. | Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z, Zhang Z, You H, Wu M, Zheng Q, Xiong Y, Xiong H, Wang C, Chen C, Xiong F, Zhang Y, Peng Y, Ge S, Zhen B, Yu T, Wang L, Wang H, Liu Y, Chen Y, Mei J, Gao X, Li Z, Gan L, He C, Shi Y, Qi Y, Yang J, Tenen DG, Chai L, Mucci LA, Santillana M, Cai H. Patients with Cancer Appear More Vulnerable to SARS-CoV-2: A Multicenter Study during the COVID-19 Outbreak. Cancer Discov. 2020;10:783-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1142] [Cited by in RCA: 966] [Article Influence: 193.2] [Reference Citation Analysis (0)] |
| 5. | Liang W, Guan W, Chen R, Wang W, Li J, Xu K, Li C, Ai Q, Lu W, Liang H, Li S, He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3332] [Cited by in RCA: 3120] [Article Influence: 624.0] [Reference Citation Analysis (0)] |
| 6. | Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, Quan S, Zhang F, Sun R, Qian L, Ge W, Liu W, Liang S, Chen H, Zhang Y, Li J, Xu J, He Z, Chen B, Wang J, Yan H, Zheng Y, Wang D, Zhu J, Kong Z, Kang Z, Liang X, Ding X, Ruan G, Xiang N, Cai X, Gao H, Li L, Li S, Xiao Q, Lu T, Zhu Y, Liu H, Guo T. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020;182:59-72.e15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 728] [Cited by in RCA: 1085] [Article Influence: 217.0] [Reference Citation Analysis (0)] |
| 7. | Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1475] [Cited by in RCA: 1884] [Article Influence: 376.8] [Reference Citation Analysis (0)] |
| 8. | Wang B, Huang Y. Which type of cancer patients are more susceptible to the SARS-COX-2: Evidence from a meta-analysis and bioinformatics analysis. Crit Rev Oncol Hematol. 2020;153:103032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 9. | Liu C, Wang K, Zhang M, Hu X, Hu T, Liu Y, Hu Q, Wu S, Yue J. High expression of ACE2 and TMPRSS2 and clinical characteristics of COVID-19 in colorectal cancer patients. NPJ Precis Oncol. 2021;5:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 10. | Bittmann S, Weissenstein A, Villalon G, Moschuring-Alieva E, Luchter E. Simultaneous Treatment of COVID-19 With Serine Protease Inhibitor Camostat and/or Cathepsin L Inhibitor? J Clin Med Res. 2020;12:320-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-280.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14218] [Article Influence: 2843.6] [Reference Citation Analysis (0)] |
| 12. | Zhao MM, Yang WL, Yang FY, Zhang L, Huang WJ, Hou W, Fan CF, Jin RH, Feng YM, Wang YC, Yang JK. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Target Ther. 2021;6:134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 355] [Article Influence: 88.8] [Reference Citation Analysis (0)] |
| 13. | Antikchi MH, Neamatzadeh H, Ghelmani Y, Jafari-Nedooshan J, Dastgheib SA, Kargar S, Noorishadkam M, Bahrami R, Jarahzadeh MH. The Risk and Prevalence of COVID-19 Infection in Colorectal Cancer Patients: a Systematic Review and Meta-analysis. J Gastrointest Cancer. 2021;52:73-79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 14. | Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158:1831-1833.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1985] [Cited by in RCA: 1993] [Article Influence: 398.6] [Reference Citation Analysis (1)] |
| 15. | Gómez-Rial J, Rivero-Calle I, Salas A, Martinón-Torres F. Role of Monocytes/Macrophages in Covid-19 Pathogenesis: Implications for Therapy. Infect Drug Resist. 2020;13:2485-2493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 85] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 16. | Zhang XJ, Qin JJ, Cheng X, Shen L, Zhao YC, Yuan Y, Lei F, Chen MM, Yang H, Bai L, Song X, Lin L, Xia M, Zhou F, Zhou J, She ZG, Zhu L, Ma X, Xu Q, Ye P, Chen G, Liu L, Mao W, Yan Y, Xiao B, Lu Z, Peng G, Liu M, Yang J, Yang L, Zhang C, Lu H, Xia X, Wang D, Liao X, Wei X, Zhang BH, Zhang X, Zhao GN, Zhang P, Liu PP, Loomba R, Ji YX, Xia J, Wang Y, Cai J, Guo J, Li H. In-Hospital Use of Statins Is Associated with a Reduced Risk of Mortality among Individuals with COVID-19. Cell Metab. 2020;32:176-187.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 394] [Cited by in RCA: 363] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 17. | Methe H, Kim JO, Kofler S, Nabauer M, Weis M. Statins decrease Toll-like receptor 4 expression and downstream signaling in human CD14+ monocytes. Arterioscler Thromb Vasc Biol. 2005;25:1439-1445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 131] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 18. | Koushki K, Shahbaz SK, Mashayekhi K, Sadeghi M, Zayeri ZD, Taba MY, Banach M, Al-Rasadi K, Johnston TP, Sahebkar A. Anti-inflammatory Action of Statins in Cardiovascular Disease: the Role of Inflammasome and Toll-Like Receptor Pathways. Clin Rev Allergy Immunol. 2021;60:175-199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 225] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 19. | Renovato-Martins M, Matheus ME, de Andrade IR, Moraes JA, da Silva SV, Citelli Dos Reis M, de Souza AA, da Silva CC, Bouskela E, Barja-Fidalgo C. Microparticles derived from obese adipose tissue elicit a pro-inflammatory phenotype of CD16+, CCR5+ and TLR8+ monocytes. Biochim Biophys Acta Mol Basis Dis. 2017;1863:139-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 20. | Patterson BK, Seethamraju H, Dhody K, Corley MJ, Kazempour K, Lalezari J, Pang APS, Sugai C, Mahyari E, Francisco EB, Pise A, Rodrigues H, Wu HL, Webb GM, Park BS, Kelly S, Pourhassan N, Lelic A, Kdouh L, Herrera M, Hall E, Bimber BN, Plassmeyer M, Gupta R, Alpan O, O'Halloran JA, Mudd PA, Akalin E, Ndhlovu LC, Sacha JB. CCR5 inhibition in critical COVID-19 patients decreases inflammatory cytokines, increases CD8 T-cells, and decreases SARS-CoV2 RNA in plasma by day 14. Int J Infect Dis. 2021;103:25-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 101] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 21. | Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, Kazmierski J, Timmermann B, Twardziok S, Schneider S, Machleidt F, Müller-Redetzky H, Maier M, Krannich A, Schmidt S, Balzer F, Liebig J, Loske J, Suttorp N, Eils J, Ishaque N, Liebert UG, von Kalle C, Hocke A, Witzenrath M, Goffinet C, Drosten C, Laudi S, Lehmann I, Conrad C, Sander LE, Eils R. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38:970-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 596] [Cited by in RCA: 790] [Article Influence: 158.0] [Reference Citation Analysis (0)] |