Published online Oct 6, 2021. doi: 10.12998/wjcc.v9.i28.8280
Peer-review started: January 26, 2021
First decision: June 4, 2021
Revised: June 15, 2021
Accepted: August 23, 2021
Article in press: August 23, 2021
Published online: October 6, 2021
Processing time: 244 Days and 19.1 Hours
The pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in more than 93 million cases and 2 million deaths in the world. SARS-CoV-2 respiratory tract infection and its main clinical manifestations such as cough and shortness of breath are well known to the scientific community. However, a growing number of studies have reported SARS-CoV-2-related gastrointestinal involvement based on clinical manifestations, such as diarrhea, nausea, vomiting, and abdominal pain as well as on the pathophysiological mechanisms associated with coronavirus disease 2019. Furthermore, current evidence suggests SARS-CoV-2 transmission via the fecal-oral route and aerosol dissemination. Moreover, studies have shown a high risk of contamination through hospital surfaces and personal fomites. Indeed, viable SARS-CoV-2 specimens can be obtained from aerosols, which raises the possibility of transmission through aerosolized viral particles from feces. Therefore, the infection by SARS-CoV-2 via fecal-oral route or aerosolized particles should be considered. In addition, a possible viral spread to sources of drinking water, sewage, and rivers as well as the possible risk of viral transmission in shared toilets become a major public health concern, especially in the least developed countries. Since authors have emphasized the presence of viral RNA and even viable SARS-CoV-2 in human feces, studies on the possible fecal-oral coronavirus disease 2019 transmission become essential to understand better the dynamics of its transmission and, then, to reinforce preventive measures against this infection, leading to a more satisfactory control of the incidence of the infection.
Core Tip: The pandemic caused by severe acute respiratory syndrome coronavirus 2 has become a global health problem. Transmission via the respiratory tract and the clinical manifestations related to that organ system are well known to the scientific community. In addition, current knowledge about the viral infection strongly suggests a possible transmission of the severe acute respiratory syndrome coronavirus 2 via the fecal-oral route. In addition, a possible spread of the virus to sources of drinking water, sewage, and rivers as well as the possibility of viral transmission in shared toilets become a major public health problem, especially in underdeveloped countries.
- Citation: Silva FAFD, de Brito BB, Santos MLC, Marques HS, da Silva Júnior RT, de Carvalho LS, de Sousa Cruz S, Rocha GR, Correa Santos GL, de Souza KC, Maciel RGA, Lopes DS, Silva NOE, Oliveira MV, de Melo FF. Transmission of severe acute respiratory syndrome coronavirus 2 via fecal-oral: Current knowledge. World J Clin Cases 2021; 9(28): 8280-8294
- URL: https://www.wjgnet.com/2307-8960/full/v9/i28/8280.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i28.8280
In December 2019, cases of pneumonia of unknown etiology were reported in Wuhan, China. The disease would be later named corona virus disease 2019 (COVID-19), and researchers found that it is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)[1]. According to the World Health Organization, from the beginning of the pandemic to the writing of this article, more than 93 million cases and 2 million deaths were reported globally[2]. The SARS-CoV-2 belongs to the β-genus of the Coronaviridae family. Its main target is the human respiratory system, and its transmission occurs mainly with personal contact through droplets originated from infected individuals. Droplet transmission involves exposure of an entry point such as mucosa or conjunctiva to potentially infectious particles[3,4]. This infection classically involves respiratory tract symptoms such as dry cough and shortness of breath as well as associated fever, myalgia, myalgia, and fatigue[5]. The patients can also present with gastrointestinal (GI) manifestations, including diarrhea, nausea, abdominal pain, and GI bleeding[6,7]. Interestingly, in January 2020, the first COVID-19 case in the Americas presented with diarrhea. Stools were collected from the patient, underwent analyses, and were later found to be positive for SARS-CoV-2 RNA through reverse transcriptase polymerase chain reaction (RT-PCR) test[8].
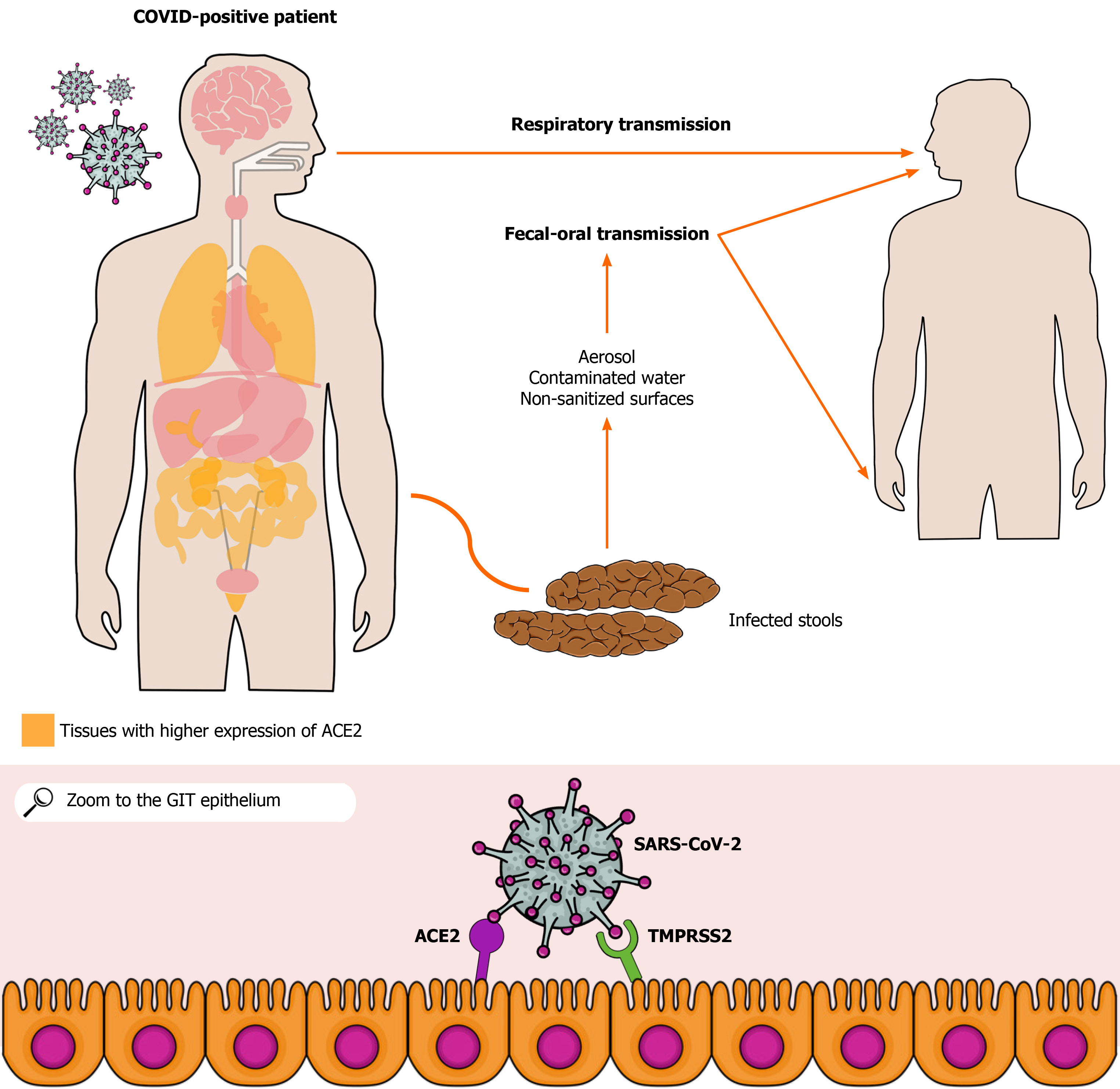
The angiotensin-converting enzyme type 2 (ACE2) receptor, which is expressed in alveolar[9], gastric, and rectal epithelial cells as well as in enterocytes from the small intestine[10,11], seems to play a crucial role in the infection pathogenesis[12,13]. Viral binding to ACE2 allows SARS-CoV-2 to enter host cells[14]. In addition, the viral presence in the feces of infected patients or asymptomatic people indicates a possible fecal-oral transmission route[11]. Studies have shown that the ACE2 receptor may contribute to this route, allowing the occurrence of GI infection. The colonization of the GIT via ACE2 may lead to viral spread through stools, which may or may not be preceded by the onset of diarrhea and occur before or after respiratory tract-related clinical manifestations[15,16]. RT-PCR tests of rectal and anal swabs collected from pediatric patients were positive within 10 d after hospital discharge following mild COVID-19, even after patients turned asymptomatic and RT-PCR from nasopharyngeal samples were negative[17]. In asymptomatic adult patients, there was viral detection in the stools for up to 42 d as well, whereas nasopharyngeal samples were negative, showing a possible GI involvement regardless of the occurrence of respiratory complaints[18]. In this sense, it has to be emphasized the possible significant implications of this potential transmission route and the possibility of its use for early identification of the SARS-CoV-2 infection as well.
The way SARS-CoV-2 infects GI tract (GIT) remains unclear. Of note, there is an evident genetic similarity between this virus and the SARS-CoV (70%), which infects the human body by attaching to the ACE2. Therefore, researchers hypothesized that this receptor could also play a role in SARS-CoV-2 manifestations[19]. Indeed, ACE2 was subsequently identified as the operative receptor of SARS-CoV-2, and, since it is distributed throughout almost all organs, including the small intestine, colon, rectum, and liver epithelia, this receptor may explain the viral presence in the GIT[20,21]. Besides ACE2, serine protease TMPRSS2 seems to be important in the SARS-CoV-2 infection[22]. That enzyme activates the S protein, which facilitates the entry of the virus into cells through the action of its two subunits, S1 and S2. The S1 unit is responsible for binding cell receptors, whereas S2 promotes the fusion between viral and cell membranes[23]. Therefore, the virus depends on both ACE2 and TMPRSS2 to invade host cells[24], as illustrated in Figure 1.
The pathophysiology in humans is not yet fully understood. Because clinical studies in humans are scarce, many experimental studies have been performed with mice. A study showed that a decrease in the levels of tryptophan absorption might be associated with the pathophysiology of the intestinal infection by SARS-CoV-2 in a murine model. This phenomenon can be explained by the need for ACE2 activation for the absorption of the aforementioned amino acid and the competitive binding of the receptor by the SARS-CoV-2. The mice that underwent the infection had an imbalance in their intestinal microbiota and a higher susceptibility to develop colitis, a condition that has been reversed with the administration of glycine tryptophan dipeptide during the study[25].
Another phenomenon that may contribute to the worsening of COVID-19 is the “cytokine storm”, an exacerbated expression of pro-inflammatory cytokines. Studies have observed higher levels of cytokines including interleukin (IL)1B, IL1RA, IL2, IL6, IL7, IL8, IL9, IL12p70, IL15, IL17A, interferon γ, and tumor necrosis factor α in patients with severe COVID-19 than individuals who experienced a milder disease[6]. Ye et al[26] highlighted the gut-lung axis as a potential cause of the intestinal manifestations in COVID-19. The effector CD4+ T cells play a role in GI immunity and chronic enteritis, using the C-C chemokine receptor type 9 to enter the GIT environment. Since C-C chemokine receptor type 9+ CD4+ T cells are highly expressed in the lung during infections with viral pathogens, this may explain intestinal damage and the onset of symptoms such as diarrhea. Moreover, the aggressions against the GIT mucosa can lead to damage to the intestinal barrier, which predisposes to infections by other external pathogens and the onset of GI symptoms[27].
Complementarily, the liver is also affected by the SARS-CoV-2 infection. The ACE2 is expressed in biliary epithelial cells, allowing viral access to the hepatobiliary system[28]. The most common repercussions associated with this organ are increased levels of alanine aminotransferase, aspartate aminotransferase, and aggravation of preexisting liver damage[29]. The intense systemic inflammatory response that can occur in COVID-19 patients, as previously discussed in this topic, may contribute to those repercussions[30,31]. Another important factor is the potential hepatotoxicity caused by drugs used in COVID-19 treatment. Medications such as lopinavir and ritonavir are associated with hepatic dysfunction among inpatients with SARS-CoV-2 infection[32].
A study analyzing 4243 patients from six countries observed the occurrence of GI symptoms such as loss of appetite, nausea/vomiting, diarrhea, and abdominal pain in 17.6% of the cases. The lack of appetite was the most common symptom (26.8%), followed by diarrhea (12.5%), nausea/vomiting (10.2%), and abdominal pain (9.2%)[33]. In another analysis including 6686 patients, the prevalence of digestive symptoms was 15%, and nausea/vomiting, diarrhea and loss of appetite were the most common manifestations, with incidences of 6%, 9%, and 21%, respectively. Of note, the symptoms are similar among adults and children. Moreover, patients with GI symptoms had their COVID-19 diagnosis delayed as compared to individuals without GI manifestations[34]. Concerning early GI manifestations of COVID-19, Redd et al[35] found that, among 318 patients, 61.8% reported at least one GI symptom at the time of hospital admission. Another study found that these manifestations were more common during hospitalization (49.5%) than at the moment of admission (11.6%)[36].
In a study performed with 1942 outpatients, 53.3% presented with at least one GI symptom, with loss of appetite affecting almost half of the patients (47%) and 24.2% of the individuals reporting diarrhea[37]. Pan et al[38] examined 204 patients with COVID-19 admitted to three hospitals in Hubei, China and noticed that 103 had digestive symptoms such as loss of appetite (n = 81), diarrhea (n = 35), vomiting (n = 4), and abdominal pain (n = 2) (Table 1). The diarrhea was the only GI symptom that occurred as a mild-to-severe manifestation in some patients, and, as the patients’ condition worsened, the digestive symptoms tended to become more intense. Moreover, abdominal pain can be considered as an indicator of severity in patients with COVID-19, being it important for decision-making during clinical management. Kumar et al[39] observed that patients experiencing severe disease have a 7-fold higher chance to present with abdominal pain than non-serious cases.
In China, in an analysis with 651 patients, 74 had at least one digestive symptom, including nausea (n = 10), vomiting (n = 11), and diarrhea (n = 53). Only three cases had all the aforementioned digestive symptoms, and four individuals had both nausea and vomiting. Among patients with GI manifestations, the more frequent complication was liver damage, which was observed in 17.57% of the cases. Moreover, one person developed shock and five had severe acute respiratory syndrome[40]. Sulaiman et al[41] confirmed that GI manifestations are common among individuals infected with SARS-CoV-2, and that they are reported mainly as initial symptoms, preceding respiratory symptoms. This study estimates that about half of COVID-19 patients may have GI symptoms along with fever and/or respiratory symptoms.
Regarding hepatic damage, high levels of important markers were registered, such as elevated serum bilirubin in 9% of 1471 patients and prolonged prothrombin time in 7% of 750 cases[42]. Similarly, 243 out of 1450 patients had an abnormally high level of aspartate aminotransferase, and 197 out of 1347 individuals had a high level of alanine aminotransferase[43].
Since March 2020, the detection of SARS-CoV-2 in intestinal biopsies and stool samples has been reported in several studies[28]. Therefore, some authors have highlighted the possibility of fecal transmission of the virus[44].
Currently, there are studies that prove and validate the detection of SARS-CoV-2 RNA in stool samples[45]. In addition, anal swabs (AS) are used for the diagnosis and monitoring of COVID-19, especially to evaluate the possibility of hospital discharge of patients[46]. A timeline that compares the viral load detected in AS and throat smears (TS) during the stages of the disease found that, although TS samples detect viral load earlier, AS detects the virus for longer periods[47].
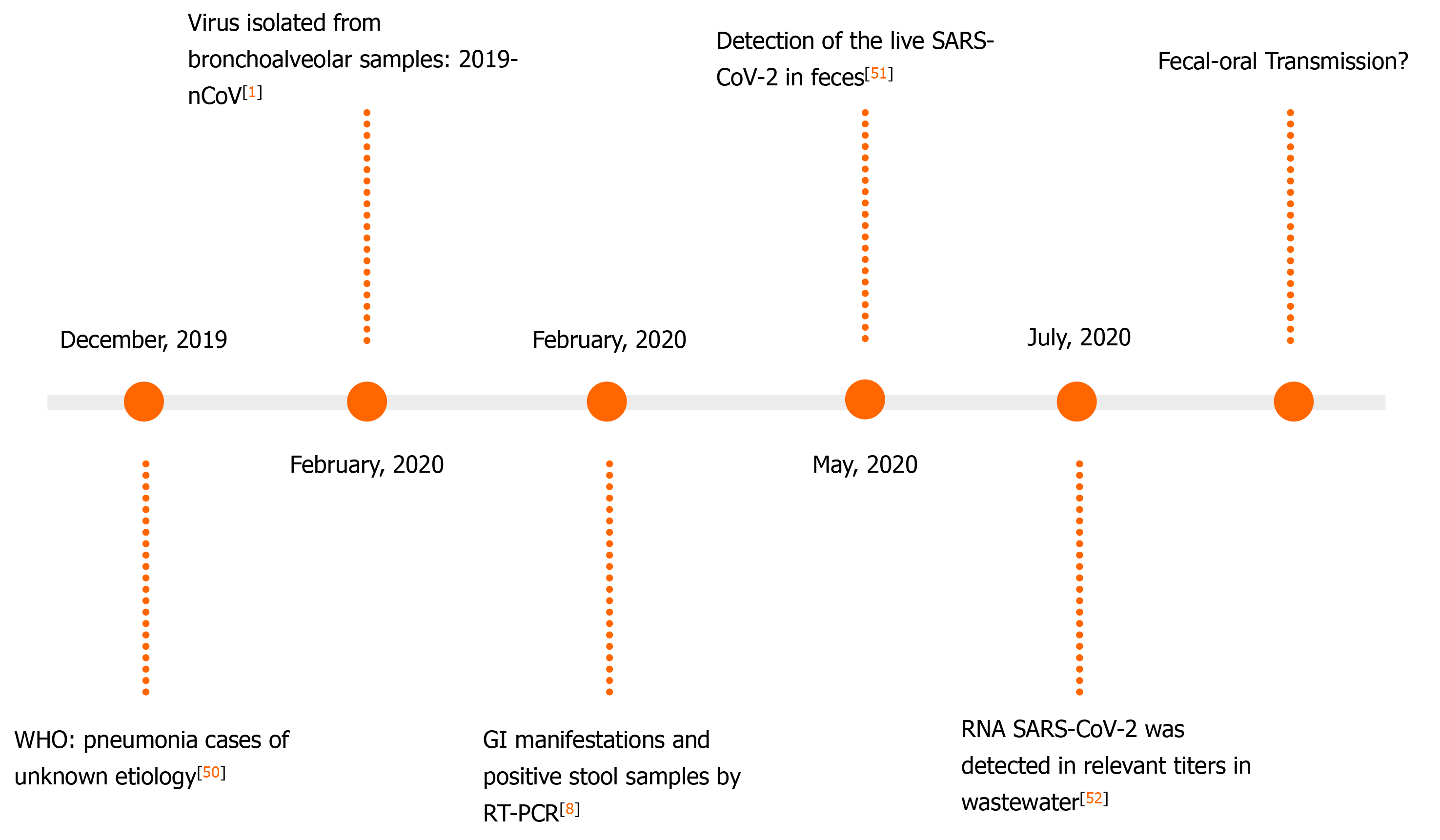
A Chinese study including 57 individuals has noticed the presence of SARS-CoV-2 viral RNA in extrapulmonary sites. They found that the RT-PCR detection of the viral RNA in stool samples or AS increased the likelihood of increased clinical severity[48]. A study from December 2020 indicates that the use of enteric samples may be effective for the monitoring of the natural course of COVID-19[49]. The findings regarding the detection of viral RNA in stool are shown in Figure 2. It demonstrates an evolution regarding the development of a new tool for the management of COVID-19 patients[1,8,50-52].
The detection of viral RNA in feces has already been observed in individuals infected with another type of coronavirus, the Middle East respiratory syndrome coronavirus[53]. A study that included 3028 patients observed a positivity of 85.8% for SARS-CoV-2 in fecal nucleic acid tests with COVID-19 patients. It was also reported that 71.2% of the patients were still positive for fecal nucleic acid after samples from the respiratory tract became negative for the virus[54]. A study that included children with COVID-19 detected SARS-CoV-2 fecal RNA in more than 91% of the cases, with viral detection in feces for up to 70 d[55]. Data from studies evaluating the duration of positivity of stools for SARS-CoV-2 among infected individuals are shown in Table 2[56-68]. These findings reinforce the hypothesis raised by several authors regarding possible transmission of SARS-CoV-2 via fecal-oral.
| SARS-CoV-2 RNA | ||||||||||
| n | Ref. | Country | n | Age | RT-PCR | Stool | PSDI | Respiratory | PRDI | Relevance information |
| 1 | Xie et al[56] | China | 4 | 4-9 yr | AS-PS | 4/4 | 8-33 d | 4/4 | 11-21 d | One patient with positive stool RNA 54 d after hospital admission |
| 2 | Ge et al[57] | China | 1 | 55 yr | FS-RS | 1/1 | 18 d | 1/1 | 7 d | Positive stool RNA 22 d after negative respiratory |
| 3 | Lo et al[58] | China | 10 | 27-64 yr | FS-PS | 10/10 | 5-19 d | 10/10 | 9-24 d | Some patients had positive stool even with negative respiratory |
| 4 | Zhang et al[17] | China | 3 | 6-9 yr | AS-PS | 3/3 | 16-20 d | 3/3 | 7-14 d | The anal swab was positive after 10 d of discharge |
| 5 | Liu et al[59] | China | 9 | NR | AS-PS | 8/9 | 28-66 d | 9/9 | 6–24 d | Positive stool RNA 46 d after discharge |
| 6 | Wang et al[60] | China | 5 | 35-56 yr | FS-PS | 5/5 | 11-30 d | 5/5 | 5-9 d | Even after a negative respiratory test, IgM was positive on 2 consecutive occasions |
| 7 | Wang X et al[61] | China | 3 | 24-42 yr | FS-PS | 2/3 | 30-36 d | 3/3 | 15-29 d | - |
| 8 | Xing et al[62] | China | 3 | ND | FS-PS | 3/3 | 8 d-4 wk | 3/3 | 2 wk | Positive stool RNA 8-20 d after negative respiratory |
| 9 | Chen et al[63] | China | 42 | 42-62 yr | AS-FS | 28/42 | 1-24 d | 42/42 | 1-19 d | Eighteen patients remained positive for viral RNA in the feces after the pharyngeal swabs turned negative |
| 10 | Wu et al[64] | China | 74 | ND | FS-PS | 33/74 | 27, 9 d | 74/74 | 16, 7 d | Fecal positive 47 d after the onset of the first symptoms |
| 11 | Li et al[65] | China | 13 | 33-73 yr | FS-RS | 5/13 | Until 38 d | 13/13 | 5-14 d | Positive stool RNA 14-15 d after negative respiratory |
| 12 | Du et al[66] | China | 10 | 9 mo-14 yr | FS-PS | 7/10 | Mean 34.43 d | 10/10 | Mean 9 d | Seven patients positive stool RNA 2 wk after discharge but negative respiratory and urine |
| 13 | Xiao et al[16] | China | 73 | 10 mo-78 yr | FS-PS | 39/73 | 1-12 d | ND | ND | Seventeen patients positive stool RNA after negative respiratory |
| 14 | Xu et al[67] | China | 10 | 2 mo-15 yr | AS-PS | 8/10 | 1-27 d | 10/10 | 1-21 | Eight patients positive stool RNA after negative respiratory |
| 15 | Fan et al[68] | China | 1 | 3 mo | AS-PS | 1/1 | 1-28 d | 1/1 | 1-14 | Positive stool RNA 14 d after negative respiratory |
Since the first detection of SARS-CoV-2-positive stool samples[6,16,48,66,68-72], the possibility of viral transmission through the fecal-oral route and fecal aerosols has been widely debated. Scientists have gathered their efforts in order to elucidate these issues and to determine the viability of viral particles found in air and stool samples.
Studies have emphasized the risk of COVID-19 contamination from hospital surfaces and personal fomites. In an evaluation of contaminated surfaces in hospital rooms, researchers found that most samples that were found to be positive for SARS-CoV-2 RNA were from bathrooms[73]. This could indicate the possibility of fecal viral loads being more likely to remain on surfaces than viral particles released from other biological secretions, reinforcing the possibility of fecal-oral transmissions. Another paper found that five out of 27 toilet flushes tested were positive for viral RNA and all the five patients that used these bathrooms had GI symptoms[74]. This could indicate a relationship between enteric manifestations and viral shedding through feces, but further studies are needed on the topic.
Complementarily, cytopathic effects have been observed in Vero cells infected by SARS-CoV-2 isolated from feces of COVID-19 patients[75]. Moreover, the latter analysis through electron microscopy found viral particles with similar morphology to the novel coronavirus in fecal samples[75-77]. These studies were essential to elucidate if the positive results from RT-PCR tests in fecal samples were due to actual virions or only represent RNA from the inactivated virus.
It is well established that SARS-CoV can be transmitted through fecal aerosols after the analysis of the Amoy Gardens incident in Hong Kong, in which multiple residents from an apartment complex were infected due to faulty pipelines that led to the spread of aerosols from the feces of infected patients[78]. The question raised with the new pandemic is whether or not SARS-CoV-2 is also able to remain viable through the process of aerosolization and to be transmitted.
Some works showed the presence of SARS-CoV-2 in air samples taken from patient’s rooms[74,79]. In two studies, researchers have observed cytopathic effects in Vero cells by viral particles collected in air samples. In one of these studies, the samples were observed through electron microscopy, which detected the presence of SARS-CoV-2 virions[80]. In the other investigation, a full genome sequencing was obtained from purified material from air samples and the viral genome was the same as the genome obtained from the infected patient’s nasopharyngeal swab[81]. This indicates that viable viral particles can be obtained from aerosols, and it raises the question of possible transmission from aerosolized virus shed through feces. Interestingly, in a study, researchers were not able to replicate viruses in Vero Cells from aerosol particles examined[82]. However, as Pan et al[83] have shown, the collection of viral particles using non-ideal air samplers could be the cause of viral inactivation of some specimens, which could impede the observation of cytopathic effects.
In two studies, researchers have tested the time of SARS-CoV-2 survival in aerosol suspensions. The first one found that the virus was able to survive for at least 3 h (the duration of the experiment)[84]. The second one searched for viral RNA through RT-quantitative PCR and found traces of the virus up to 16 h after the aerosolization of the particles, which were later visualized through an electron microscope and presented a shape consistent with the SARS-CoV-2 at the 10th minute of the experiment[85]. These studies show that SARS-CoV-2 is able to remain viable for long periods, and aerosols can be a dangerous form of infection.
Among studies evaluating the possibility of animal infection with virus from fecal samples, Jeong et al[86] intranasally inoculated viral particles that were isolated from the stool of confirmed COVID-19 patients in ferrets. Some animals showed an increase in viral load throughout the period of infection and tested positive for the virus a few days later. All ferrets had symptoms of the disease after inoculation. In another study, Lee et al[87] used Syrian Golden Hamsters to inoculate intranasally and orally SARS-CoV-2 in variable doses. They found that hamsters that underwent intranasal inoculation developed more severe respiratory symptoms and similar GI inflammation to those that received the virus orally, but these results were not statistically significant (P > 0.05). Both groups tested positive for the virus in fecal samples and saliva[87]. This finding could indicate that in the case of fecal-oral transmission, COVID-19 manifestations could be less severe than through aerosols, but viral shedding from the infected patients would still be an important factor to consider.
Research has also shown that SARS-CoV-2 persistence time depends on multiple factors such as pH, temperature, and humidity, with the longest viability time at 4 ºC temperature in a pH of 9[88]. Strategies with the use of this knowledge and standard disinfection procedures should be applied in order to reduce the risk of surface contamination, and special attention should be given to bathrooms in order to prevent transmission from their fomites. Ong et al[89] found contamination in 87% of surfaces in a patient’s room before cleaning as opposed to no contamination in rooms of patients after the cleaning, showing the importance of precaution during the disinfecting process. In the case of the first SARS coronavirus from 2003, studies have reported that virus viability is highly affected by the pH of the patients’ feces[90]. Analysis should be conducted to evaluate if this applies to SARS-CoV-2 as well.
Overall, although not totally proven, the possibility of infection by SARS-CoV-2 through the fecal-oral route or aerosolized particles should be considered. In any case, proper cleaning processes and prophylactic measures should be performed in order to avoid contact with the virus through fomites or aerosols until the possibility of both types of transmission are properly elucidated.
Studies that evidence the detection of SARS-CoV-2 in fecal samples raise attention to the risks of human exposure in the environment[91]. The indirect transmission through fomites has been already well documented, especially when individuals touch contaminated surfaces and then take the hands to the mouth, nose, or eyes without first cleaning them[92]. Besides, it is assumed that improper disposal of solid waste associated with viral stability on solid surfaces could lead to contamination of surface water from household or hospital waste[93]. Furthermore, a possible spread of the virus to drinking water sources is assumed when an infected person defecates in open environments. In addition, there is a possible risk of viral transmission in shared toilets[94]. Moreover, Li et al[95] showed that the flow of water during flushing the toilet can spread viral particles through aerosols.
Although the transmission of SARS-CoV-2 by sewage aerosol is not yet confirmed[93], some studies (Table 3)[96-107] show the detection of SARS-CoV-2 RNA in wastewater. Arslan et al[108] and Pandey et al[93] highlight that this can be beneficial from a wastewater-based epidemiology perspective because the viral presence in wastewater can be an early warning as well as a monitoring and surveillance tool for COVID-19. However, they highlight the risk of recurrent outbreaks due to the continued presence of the virus in those media.
| Ref. | Country | Wastewater type | Main findings |
| Medema et al[96] | The Netherlands | Sewage | Sewage samples of six cities and the airport were tested: (1) February: No SARS-CoV-2 RNA was detected; and (2) March: SARS-CoV-2 RNA detection increased concomitant with the increase in COVID-19 prevalence |
| Ahmed et al[97] | Australia | Wastewater in a catchment | Two positive detections within a 6-d period from the same wastewater treatment plant |
| Kumar et al[98] | India | Wastewater Treatment Plant | Increase in SARS-CoV-2 RNA samples was concomitant with the increase in the number of active COVID-19 patients in the city |
| Wurtzer et al[99] | France | Wastewater treatment plant (raw and treated wastewater samples) | Raw wastewater samples: all positive for SARS-CoV2; Treated wastewater samples: 6 out of 8 positive for SARS-CoV2 |
| Randazzo et al[100] | Spain | Wastewater treatments plants | (1) Influent samples: 35 of 42 were positive for SARS-CoV-2; (2) Effluent secondary treated samples: 2 of 18 were tested positive; and (3) Effluent tertiary treated samples: 0 of 12 tested positive |
| Kocamemi et al[101] | Turkey | Wastewater treatment plants (primary sludge and waste activated sludge) | SARS-CoV-2 RNA was detected quantitatively from all samples. |
| Guerrero-Latorre et al[102] | Ecuador | River | SARS-CoV-2 RNA was detected in the three locations |
| Zhang et al[103] | China | Septic tanks | SARS-CoV-2 RNA was detected in septic tanks after disinfection with sodium hypochlorite |
| Ahmed et al[104] | United States | Wastewater treatment plants | Two out of 15 wastewater samples tested positive |
| Haramoto et al[105] | Japan | River, wastewater treatment plants | One of 5 secondary-treated wastewater samples tested positive |
| La Rosa et al[106] | Italy | Sewage | Six out of 12 samples tested positive. |
| Hasan et al[107] | United Arab Emirates | Sewage, wastewater treatment plants, pumping stations | SARS-CoV-2 RNA was detected in 85% of untreated wastewater samples; SARS-CoV-2 RNA was not detected in wastewater treatment plants. |
Recent surveys have shown that around 2.3 billion people do not have access to a basic sanitation service, and 844 million people do not have a drinking water service worldwide. Another study reports that developing countries have inefficient wastewater treatment[108]. In addition, Pandey et al[93] emphasized the difficulty of inspection in the least developed countries owing to the absence of a proper sewage system. Besides, Arslan et al[109] suggested that various societies do not have the basic conditions to inactivate the coronavirus from the water. Therefore, the treatment of water[93], the use of disinfection methods with high doses of disinfectant products[110], and public guidelines on hygienic measures[111] are convenient in that context.
The current knowledge about the dynamics of the infection in the GI tract strongly suggests a possible transmission through the fecal-oral route. In addition, the spread of infected aerosolized feces in hospital environments, bathrooms, and surfaces draws attention to this issue, which has not been considerably taken into account by health agencies in discussions on infection prevention. The contamination of water by SARS-Cov-2 in sewers, wastewater treatment plants, and rivers, possibly from fecal samples, evidences a major public health problem, especially in developing countries. However, further studies are needed in order to elucidate completely the SARS-Cov-2 transmission via the fecal-oral route.
Manuscript source: Invited manuscript
Specialty type: Infectious diseases
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Navarro-Alvarez N, Poddighe D S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Li JH
| 1. | Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18987] [Cited by in RCA: 17622] [Article Influence: 3524.4] [Reference Citation Analysis (0)] |
| 2. | World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. Data as received by WHO from national authorities. [cited 18 January 2021]. Available from: https://covid19.who.int/. |
| 3. | Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5228] [Cited by in RCA: 5779] [Article Influence: 1155.8] [Reference Citation Analysis (2)] |
| 4. | Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2713] [Cited by in RCA: 2764] [Article Influence: 552.8] [Reference Citation Analysis (0)] |
| 5. | Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;324:782-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2691] [Cited by in RCA: 3174] [Article Influence: 634.8] [Reference Citation Analysis (0)] |
| 6. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30090] [Article Influence: 6018.0] [Reference Citation Analysis (3)] |
| 7. | Cha MH, Regueiro M, Sandhu DS. Gastrointestinal and hepatic manifestations of COVID-19: A comprehensive review. World J Gastroenterol. 2020;26:2323-2332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 124] [Cited by in RCA: 105] [Article Influence: 21.0] [Reference Citation Analysis (2)] |
| 8. | Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK; Washington State 2019-nCoV Case Investigation Team. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382:929-936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4155] [Cited by in RCA: 3820] [Article Influence: 764.0] [Reference Citation Analysis (1)] |
| 9. | Villapol S. Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome. Transl Res. 2020;226:57-69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 217] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 10. | Wong SH, Lui RN, Sung JJ. Covid-19 and the digestive system. J Gastroenterol Hepatol. 2020;35:744-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 512] [Cited by in RCA: 444] [Article Influence: 88.8] [Reference Citation Analysis (0)] |
| 11. | Zhang H, Li HB, Lyu JR, Lei XM, Li W, Wu G, Lyu J, Dai ZM. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int J Infect Dis. 2020;96:19-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 216] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 12. | Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. 2020;5:335-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 546] [Cited by in RCA: 554] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 13. | Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, Zhu J, Zhang Q, Wu J, Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020;92:595-601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 468] [Cited by in RCA: 443] [Article Influence: 88.6] [Reference Citation Analysis (0)] |
| 14. | Stopsack KH, Mucci LA, Antonarakis ES, Nelson PS, Kantoff PW. TMPRSS2 and COVID-19: Serendipity or Opportunity for Intervention? Cancer Discov. 2020;10:779-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 287] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 15. | Zhang J, Wang S, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol. 2020;92:680-682. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 278] [Cited by in RCA: 301] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 16. | Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158:1831-1833.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1985] [Cited by in RCA: 1993] [Article Influence: 398.6] [Reference Citation Analysis (1)] |
| 17. | Zhang T, Cui X, Zhao X, Wang J, Zheng J, Zheng G, Guo W, Cai C, He S, Xu Y. Detectable SARS-CoV-2 viral RNA in feces of three children during recovery period of COVID-19 pneumonia. J Med Virol. 2020;92:909-914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 164] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 18. | Jiang X, Luo M, Zou Z, Wang X, Chen C, Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J Med Virol. 2020;92:1807-1809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 19. | Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem. 2020;66:549-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 844] [Cited by in RCA: 891] [Article Influence: 178.2] [Reference Citation Analysis (0)] |
| 20. | Dong M, Zhang J, Ma X, Tan J, Chen L, Liu S, Xin Y, Zhuang L. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed Pharmacother. 2020;131:110678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 182] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 21. | Liang W, Feng Z, Rao S, Xiao C, Xue X, Lin Z, Zhang Q, Qi W. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 269] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 22. | Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, Talavera-López C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, Yoshida M, Barnes JL; HCA Lung Biological Network. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1985] [Cited by in RCA: 1953] [Article Influence: 390.6] [Reference Citation Analysis (0)] |
| 23. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-280.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14248] [Article Influence: 2849.6] [Reference Citation Analysis (0)] |
| 24. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14106] [Article Influence: 2821.2] [Reference Citation Analysis (1)] |
| 25. | Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, Wild B, Camargo SM, Singer D, Richter A, Kuba K, Fukamizu A, Schreiber S, Clevers H, Verrey F, Rosenstiel P, Penninger JM. ACE2 Links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477-481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 787] [Cited by in RCA: 984] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 26. | Ye Q, Wang B, Zhang T, Xu J, Shang S. The mechanism and treatment of gastrointestinal symptoms in patients with COVID-19. Am J Physiol Gastrointest Liver Physiol. 2020;319:G245-G252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (2)] |
| 27. | Gu J, Han B, Wang J. COVID-19: Gastrointestinal Manifestations and Potential Fecal-Oral Transmission. Gastroenterology. 2020;158:1518-1519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 954] [Cited by in RCA: 949] [Article Influence: 189.8] [Reference Citation Analysis (1)] |
| 28. | Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14113] [Cited by in RCA: 14759] [Article Influence: 2951.8] [Reference Citation Analysis (0)] |
| 29. | Jothimani D, Venugopal R, Abedin MF, Kaliamoorthy I, Rela M. COVID-19 and the liver. J Hepatol. 2020;73:1231-1240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 358] [Article Influence: 71.6] [Reference Citation Analysis (1)] |
| 30. | Sun J, Aghemo A, Forner A, Valenti L. COVID-19 and liver disease. Liver Int. 2020;40:1278-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 219] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 31. | Cravioto J, Matsubara M, Arrieta R. [Low birth weight and the functioning of the central nervous system in the first years of life]. Bol Med Hosp Infant Mex. 1988;45:718-728. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1348] [Cited by in RCA: 1295] [Article Influence: 259.0] [Reference Citation Analysis (4)] |
| 32. | Fan Z, Chen L, Li J, Cheng X, Yang J, Tian C, Zhang Y, Huang S, Liu Z, Cheng J. Clinical Features of COVID-19-Related Liver Functional Abnormality. Clin Gastroenterol Hepatol. 2020;18:1561-1566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 559] [Cited by in RCA: 555] [Article Influence: 111.0] [Reference Citation Analysis (0)] |
| 33. | Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, Ng YY, Chu MY, Chung TWH, Tam AR, Yip CCY, Leung KH, Fung AY, Zhang RR, Lin Y, Cheng HM, Zhang AJX, To KKW, Chan KH, Yuen KY, Leung WK. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology. 2020;159:81-95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1113] [Cited by in RCA: 1131] [Article Influence: 226.2] [Reference Citation Analysis (1)] |
| 34. | Mao R, Qiu Y, He JS, Tan JY, Li XH, Liang J, Shen J, Zhu LR, Chen Y, Iacucci M, Ng SC, Ghosh S, Chen MH. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:667-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 741] [Cited by in RCA: 751] [Article Influence: 150.2] [Reference Citation Analysis (0)] |
| 35. | Redd WD, Zhou JC, Hathorn KE, McCarty TR, Bazarbashi AN, Thompson CC, Shen L, Chan WW. Prevalence and Characteristics of Gastrointestinal Symptoms in Patients With Severe Acute Respiratory Syndrome Coronavirus 2 Infection in the United States: A Multicenter Cohort Study. Gastroenterology. 2020;159:765-767.e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 283] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 36. | Lin L, Jiang X, Zhang Z, Huang S, Fang Z, Gu Z, Gao L, Shi H, Mai L, Liu Y, Lin X, Lai R, Yan Z, Li X, Shan H. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997-1001. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 630] [Cited by in RCA: 657] [Article Influence: 131.4] [Reference Citation Analysis (0)] |
| 37. | Sierpiński R, Pinkas J, Jankowski M, Zgliczyński WS, Wierzba W, Gujski M, Szumowski Ł. Sex differences in the frequency of gastrointestinal symptoms and olfactory or taste disorders in 1942 nonhospitalized patients with coronavirus disease 2019 (COVID-19). Pol Arch Intern Med. 2020;130:501-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 38. | Pan L, Mu M, Yang P, Sun Y, Wang R, Yan J, Li P, Hu B, Wang J, Hu C, Jin Y, Niu X, Ping R, Du Y, Li T, Xu G, Hu Q, Tu L. Clinical Characteristics of COVID-19 Patients With Digestive Symptoms in Hubei, China: A Descriptive, Cross-Sectional, Multicenter Study. Am J Gastroenterol. 2020;115:766-773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1160] [Cited by in RCA: 1204] [Article Influence: 240.8] [Reference Citation Analysis (0)] |
| 39. | Suresh Kumar VC, Mukherjee S, Harne PS, Subedi A, Ganapathy MK, Patthipati VS, Sapkota B. Novelty in the gut: a systematic review and meta-analysis of the gastrointestinal manifestations of COVID-19. BMJ Open Gastroenterol. 2020;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 40. | Jin X, Lian JS, Hu JH, Gao J, Zheng L, Zhang YM, Hao SR, Jia HY, Cai H, Zhang XL, Yu GD, Xu KJ, Wang XY, Gu JQ, Zhang SY, Ye CY, Jin CL, Lu YF, Yu X, Yu XP, Huang JR, Xu KL, Ni Q, Yu CB, Zhu B, Li YT, Liu J, Zhao H, Zhang X, Yu L, Guo YZ, Su JW, Tao JJ, Lang GJ, Wu XX, Wu WR, Qv TT, Xiang DR, Yi P, Shi D, Chen Y, Ren Y, Qiu YQ, Li LJ, Sheng J, Yang Y. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002-1009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 944] [Cited by in RCA: 869] [Article Influence: 173.8] [Reference Citation Analysis (0)] |
| 41. | Sulaiman T, Algharawi AA, Idrees M, Alzaidy RH, Faris K, Cullingford G, Rasheed J. The prevalence of gastrointestinal symptoms among patients with COVID-19 and the effect on the severity of the disease. JGH Open. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 42. | Kumar A, Arora A, Sharma P, Anikhindi SA, Bansal N, Singla V, Khare S, Srivastava A. Gastrointestinal and hepatic manifestations of Corona Virus Disease-19 and their relationship to severe clinical course: A systematic review and meta-analysis. Indian J Gastroenterol. 2020;39:268-284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 43. | Parasa S, Desai M, Thoguluva Chandrasekar V, Patel HK, Kennedy KF, Roesch T, Spadaccini M, Colombo M, Gabbiadini R, Artifon ELA, Repici A, Sharma P. Prevalence of Gastrointestinal Symptoms and Fecal Viral Shedding in Patients With Coronavirus Disease 2019: A Systematic Review and Meta-analysis. JAMA Netw Open. 2020;3:e2011335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 301] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 44. | Green CA, Quraishi MN, Shabir S, Sharma N, Hansen R, Gaya DR, Hart AL, Loman NJ, Iqbal TH. Screening faecal microbiota transplant donors for SARS-CoV-2 by molecular testing of stool is the safest way forward. Lancet Gastroenterol Hepatol. 2020;5:531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 45. | Perchetti GA, Nalla AK, Huang ML, Zhu H, Wei Y, Stensland L, Loprieno MA, Jerome KR, Greninger AL. Validation of SARS-CoV-2 detection across multiple specimen types. J Clin Virol. 2020;128:104438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 46. | Sun M, Guo D, Zhang J, Teng HF, Xia J, Liu P, Ge QX, Wang MY. Anal swab as a potentially optimal specimen for SARS-CoV-2 detection to evaluate hospital discharge of COVID-19 patients. Future Microbiol. 2020;15:1101-1107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 47. | Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, Wang YY, Xiao GF, Yan B, Shi ZL, Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1192] [Cited by in RCA: 1219] [Article Influence: 243.8] [Reference Citation Analysis (0)] |
| 48. | Chen W, Lan Y, Yuan X, Deng X, Li Y, Cai X, Li L, He R, Tan Y, Gao M, Tang G, Zhao L, Wang J, Fan Q, Wen C, Tong Y, Tang Y, Hu F, Li F, Tang X. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. 2020;9:469-473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 280] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 49. | Li H, Ren L, Zhang L, Wang Y, Guo L, Wang C, Xiao Y, Rao J, Wang X, Liu Y, Huang C, Gu X, Fan G, Li H, Lu B, Cao B, Wang J. High anal swab viral load predisposes adverse clinical outcomes in severe COVID-19 patients. Emerg Microbes Infect. 2020;9:2707-2714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 50. | World Health Organization. WHO: Pneumonia of unknown cause, China. [cited 5 January 2020]. Available from: https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/. |
| 51. | Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323:1843-1844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1894] [Cited by in RCA: 2658] [Article Influence: 531.6] [Reference Citation Analysis (0)] |
| 52. | Wu F, Zhang J, Xiao A, Gu X, Lee WL, Armas F, Kauffman K, Hanage W, Matus M, Ghaeli N, Endo N, Duvallet C, Poyet M, Moniz K, Washburne AD, Erickson TB, Chai PR, Thompson J, Alm EJ. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. mSystems. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 534] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 53. | Corman VM, Albarrak AM, Omrani AS, Albarrak MM, Farah ME, Almasri M, Muth D, Sieberg A, Meyer B, Assiri AM, Binger T, Steinhagen K, Lattwein E, Al-Tawfiq J, Müller MA, Drosten C, Memish ZA. Viral Shedding and Antibody Response in 37 Patients With Middle East Respiratory Syndrome Coronavirus Infection. Clin Infect Dis. 2016;62:477-483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 270] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 54. | Wang JG, Cui HR, Tang HB, Deng XL. Gastrointestinal symptoms and fecal nucleic acid testing of children with 2019 coronavirus disease: a systematic review and meta-analysis. Sci Rep. 2020;10:17846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 55. | Hua CZ, Miao ZP, Zheng JS, Huang Q, Sun QF, Lu HP, Su FF, Wang WH, Huang LP, Chen DQ, Xu ZW, Ji LD, Zhang HP, Yang XW, Li MH, Mao YY, Ying MZ, Ye S, Shu Q, Chen EF, Liang JF, Wang W, Chen ZM, Li W, Fu JF. Epidemiological features and viral shedding in children with SARS-CoV-2 infection. J Med Virol. 2020;92:2804-2812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 56. | Xie J, Long X, Ren C, He R, Yan X, Li W, Luo Z, Li Q, Xu H, Liu E. Follow-up Study of Long-time Positive RT-PCR in Stool Specimens From Asymptomatic Children Infected With SARS-CoV-2. Pediatr Infect Dis J. 2020;39:e315-e317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 57. | Ge R, Chen Z, Liu X, Zhang Q, Zhu G, Xiao Q. Positive Stool Test Results Suggest that the Discharge Standard for COVID-19 Needs Improvement. Jpn J Infect Dis. 2021;74:76-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 58. | Lo IL, Lio CF, Cheong HH, Lei CI, Cheong TH, Zhong X, Tian Y, Sin NN. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci. 2020;16:1698-1707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 230] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 59. | Liu P, Cai J, Jia R, Xia S, Wang X, Cao L, Zeng M, Xu J. Dynamic surveillance of SARS-CoV-2 shedding and neutralizing antibody in children with COVID-19. Emerg Microbes Infect. 2020;9:1254-1258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 60. | Wang QX, Huang KC, Qi L, Zeng XH, Zheng SL. No infectious risk of COVID-19 patients with long-term fecal 2019-nCoV nucleic acid positive. Eur Rev Med Pharmacol Sci. 2020;24:5772-5777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 61. | Wang X, Zhou Y, Jiang N, Zhou Q, Ma WL. Persistence of intestinal SARS-CoV-2 infection in patients with COVID-19 Leads to re-admission after pneumonia resolved. Int J Infect Dis. 2020;95:433-435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 62. | Xing Y, Ni W, Wu Q, Li W, Li G, Wang W, Tong J, Song X, Wong GWK, Xing Q. Dynamics of faecal SARS-CoV-2 in infected children during the convalescent phase. J Infect. 2020;81:318-356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 63. | Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, Yang Y, Liu B, Wang W, Wei C, Yang J, Ye G, Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92:833-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 570] [Article Influence: 114.0] [Reference Citation Analysis (0)] |
| 64. | Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, Yin H, Xiao Q, Tang Y, Qu X, Kuang L, Fang X, Mishra N, Lu J, Shan H, Jiang G, Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434-435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1050] [Cited by in RCA: 1150] [Article Influence: 230.0] [Reference Citation Analysis (0)] |
| 65. | Li Y, Hu Y, Yu Y, Zhang X, Li B, Wu J, Li J, Wu Y, Xia X, Tang H, Xu J. Positive result of Sars-Cov-2 in faeces and sputum from discharged patients with COVID-19 in Yiwu, China. J Med Virol. 2020;92:1938-1947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 66. | Du W, Yu J, Liu X, Chen H, Lin L, Li Q. Persistence of SARS-CoV-2 virus RNA in feces: A case series of children. J Infect Public Health. 2020;13:926-931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 67. | Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, Guo Q, Sun X, Zhao D, Shen J, Zhang H, Liu H, Xia H, Tang J, Zhang K, Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502-505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 992] [Cited by in RCA: 1052] [Article Influence: 210.4] [Reference Citation Analysis (0)] |
| 68. | Fan Q, Pan Y, Wu Q, Liu S, Song X, Xie Z, Liu Y, Zhao L, Wang Z, Zhang Y, Wu Z, Guan L, Lv X. Anal swab findings in an infant with COVID-19. Pediatr Investig. 2020;4:48-50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 69. | Chen C, Gao G, Xu Y, Pu L, Wang Q, Wang L, Wang W, Song Y, Chen M, Yu F, Yang S, Tang Y, Zhao L, Wang H, Wang Y, Zeng H, Zhang F. SARS-CoV-2-Positive Sputum and Feces After Conversion of Pharyngeal Samples in Patients With COVID-19. Ann Intern Med. 2020;172:832-834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 168] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 70. | Ling Y, Xu SB, Lin YX, Tian D, Zhu ZQ, Dai FH, Wu F, Song ZG, Huang W, Chen J, Hu BJ, Wang S, Mao EQ, Zhu L, Zhang WH, Lu HZ. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl). 2020;133:1039-1043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 641] [Cited by in RCA: 577] [Article Influence: 115.4] [Reference Citation Analysis (0)] |
| 71. | Tang A, Tong ZD, Wang HL, Dai YX, Li KF, Liu JN, Wu WJ, Yuan C, Yu ML, Li P, Yan JB. Detection of Novel Coronavirus by RT-PCR in Stool Specimen from Asymptomatic Child, China. Emerg Infect Dis. 2020;26:1337-1339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 323] [Cited by in RCA: 368] [Article Influence: 73.6] [Reference Citation Analysis (0)] |
| 72. | Xing YH, Ni W, Wu Q, Li WJ, Li GJ, Wang WD, Tong JN, Song XF, Wing-Kin Wong G, Xing QS. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J Microbiol Immunol Infect. 2020;53:473-480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 240] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 73. | Ding Z, Qian H, Xu B, Huang Y, Miao T, Yen HL, Xiao S, Cui L, Wu X, Shao W, Song Y, Sha L, Zhou L, Xu Y, Zhu B, Li Y. Toilets dominate environmental detection of severe acute respiratory syndrome coronavirus 2 in a hospital. Sci Total Environ. 2021;753:141710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 95] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 74. | Chia PY, Coleman KK, Tan YK, Ong SWX, Gum M, Lau SK, Lim XF, Lim AS, Sutjipto S, Lee PH, Son TT, Young BE, Milton DK, Gray GC, Schuster S, Barkham T, De PP, Vasoo S, Chan M, Ang BSP, Tan BH, Leo YS, Ng OT, Wong MSY, Marimuthu K; Singapore 2019 Novel Coronavirus Outbreak Research Team. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat Commun. 2020;11:2800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 499] [Cited by in RCA: 598] [Article Influence: 119.6] [Reference Citation Analysis (0)] |
| 75. | Xiao F, Sun J, Xu Y, Li F, Huang X, Li H, Zhao J, Huang J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg Infect Dis. 2020;26:1920-1922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 346] [Cited by in RCA: 389] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 76. |
Zhang Y, Chen C, Zhu SL, Shu C, Wang DY, Song JD, Song Y, Zhen W, Feng ZJ, Wu GZ, Xu J, Xu WB, Isolation of 2019-nCoV from a Stool Specimen of a Laboratory-Confirmed Case of the Coronavirus Disease 2019 (COVID-19).
|
| 77. | Zhang Y, Chen C, Song Y, Zhu S, Wang D, Zhang H, Han G, Weng Y, Xu J, Yu P, Jiang W, Yang X, Lang Z, Yan D, Wang Y, Song J, Gao GF, Wu G, Xu W. Excretion of SARS-CoV-2 through faecal specimens. Emerg Microbes Infect. 2020;9:2501-2508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 78. | Yu IT, Li Y, Wong TW, Tam W, Chan AT, Lee JH, Leung DY, Ho T. Evidence of airborne transmission of the severe acute respiratory syndrome virus. N Engl J Med. 2004;350:1731-1739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 874] [Cited by in RCA: 756] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 79. | Guo ZD, Wang ZY, Zhang SF, Li X, Li L, Li C, Cui Y, Fu RB, Dong YZ, Chi XY, Zhang MY, Liu K, Cao C, Liu B, Zhang K, Gao YW, Lu B, Chen W. Aerosol and Surface Distribution of Severe Acute Respiratory Syndrome Coronavirus 2 in Hospital Wards, Wuhan, China, 2020. Emerg Infect Dis. 2020;26:1583-1591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 782] [Cited by in RCA: 683] [Article Influence: 136.6] [Reference Citation Analysis (0)] |
| 80. | Santarpia JL, Rivera DN, Herrera VL, Morwitzer MJ, Creager HM, Santarpia GW, Crown KK, Brett-Major DM, Schnaubelt ER, Broadhurst MJ, Lawler JV, Reid SP, Lowe JJ. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci Rep. 2020;10:12732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 375] [Cited by in RCA: 373] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 81. | Lednicky JA, Lauzardo M, Fan ZH, Jutla A, Tilly TB, Gangwar M, Usmani M, Shankar SN, Mohamed K, Eiguren-Fernandez A, Stephenson CJ, Alam MM, Elbadry MA, Loeb JC, Subramaniam K, Waltzek TB, Cherabuddi K, Morris JG Jr, Wu CY. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int J Infect Dis. 2020;100:476-482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 345] [Cited by in RCA: 425] [Article Influence: 85.0] [Reference Citation Analysis (0)] |
| 82. | Binder RA, Alarja NA, Robie ER, Kochek KE, Xiu L, Rocha-Melogno L, Abdelgadir A, Goli SV, Farrell AS, Coleman KK, Turner AL, Lautredou CC, Lednicky JA, Lee MJ, Polage CR, Simmons RA, Deshusses MA, Anderson BD, Gray GC. Environmental and Aerosolized Severe Acute Respiratory Syndrome Coronavirus 2 Among Hospitalized Coronavirus Disease 2019 Patients. J Infect Dis. 2020;222:1798-1806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 83. | Pan M, Lednicky JA, Wu CY. Collection, particle sizing and detection of airborne viruses. J Appl Microbiol. 2019;127:1596-1611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 154] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 84. | van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Engl J Med. 2020;382:1564-1567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5894] [Cited by in RCA: 5662] [Article Influence: 1132.4] [Reference Citation Analysis (0)] |
| 85. | Fears AC, Klimstra WB, Duprex P, Hartman A, Weaver SC, Plante KS, Mirchandani D, Plante JA, Aguilar PV, Fernández D, Nalca A, Totura A, Dyer D, Kearney B, Lackemeyer M, Bohannon JK, Johnson R, Garry RF, Reed DS, Roy CJ. Persistence of Severe Acute Respiratory Syndrome Coronavirus 2 in Aerosol Suspensions. Emerg Infect Dis. 2020;26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 217] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 86. | Jeong HW, Kim SM, Kim HS, Kim YI, Kim JH, Cho JY, Kim SH, Kang H, Kim SG, Park SJ, Kim EH, Choi YK. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin Microbiol Infect. 2020;26:1520-1524. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 159] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 87. | Lee AC, Zhang AJ, Chan JF, Li C, Fan Z, Liu F, Chen Y, Liang R, Sridhar S, Cai JP, Poon VK, Chan CC, To KK, Yuan S, Zhou J, Chu H, Yuen KY. Oral SARS-CoV-2 Inoculation Establishes Subclinical Respiratory Infection with Virus Shedding in Golden Syrian Hamsters. Cell Rep Med. 2020;1:100121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 88. | Chan KH, Sridhar S, Zhang RR, Chu H, Fung AY, Chan G, Chan JF, To KK, Hung IF, Cheng VC, Yuen KY. Factors affecting stability and infectivity of SARS-CoV-2. J Hosp Infect. 2020;106:226-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 172] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 89. | Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, Marimuthu K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA. 2020;323:1610-1612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1461] [Cited by in RCA: 1391] [Article Influence: 278.2] [Reference Citation Analysis (0)] |
| 90. | Lai MY, Cheng PK, Lim WW. Survival of severe acute respiratory syndrome coronavirus. Clin Infect Dis. 2005;41:e67-e71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 215] [Cited by in RCA: 224] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 91. | Olusola-Makinde OO, Reuben RC. Ticking bomb: Prolonged faecal shedding of novel coronavirus (2019-nCoV) and environmental implications. Environ Pollut. 2020;267:115485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 92. | Anelich LECM, Lues R, Farber JM, Parreira VR. SARS-CoV-2 and Risk to Food Safety. Front Nutr. 2020;7:580551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 93. | Pandey D, Verma S, Verma P, Mahanty B, Dutta K, Daverey A, Arunachalam K. SARS-CoV-2 in wastewater: Challenges for developing countries. Int J Hyg Environ Health. 2021;231:113634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 94. | Sunkari ED, Korboe HM, Abu M, Kizildeniz T. Sources and routes of SARS-CoV-2 transmission in water systems in Africa: Are there any sustainable remedies? Sci Total Environ. 2021;753:142298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 95. | Li YY, Wang JX, Chen X. Can a toilet promote virus transmission? Phys Fluids (1994). 2020;32:065107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 96. | Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ Sci Technol Lett. 2020;7:511-516. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1010] [Cited by in RCA: 1028] [Article Influence: 205.6] [Reference Citation Analysis (0)] |
| 97. | Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O'Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;728:138764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1259] [Cited by in RCA: 1257] [Article Influence: 251.4] [Reference Citation Analysis (0)] |
| 98. | Kumar M, Patel AK, Shah AV, Raval J, Rajpara N, Joshi M, Joshi CG. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci Total Environ. 2020;746:141326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 319] [Article Influence: 63.8] [Reference Citation Analysis (0)] |
| 99. | Wurtzer S, Marechal V, Mouchel JM, Maday Y, Teyssou R, Richard E, Almayrac JL, Moulin L. Time Course Quantitative Detection of SARS-CoV-2 in Parisian Wastewaters Correlates With COVID-19 Confirmed Cases. 2020 Preprint. Available from: medRxiv. [RCA] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 200] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 100. | Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 850] [Cited by in RCA: 816] [Article Influence: 163.2] [Reference Citation Analysis (0)] |
| 101. | Kocamemi BA, Kurt H, Sait A, Sarac F, Saatci AM, Pakdemirli B. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. 2020 Preprint. Available from: medRxiv. [DOI] [Full Text] |
| 102. | Guerrero-Latorre L, Ballesteros I, Villacrés-Granda I, Granda MG, Freire-Paspuel B, Ríos-Touma B. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci Total Environ. 2020;743:140832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 167] [Cited by in RCA: 172] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 103. | Zhang D, Ling H, Huang X, Li J, Li W, Yi C, Zhang T, Jiang Y, He Y, Deng S, Zhang X, Wang X, Liu Y, Li G, Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci Total Environ. 2020;741:140445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 204] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 104. | Sherchan SP, Shahin S, Ward LM, Tandukar S, Aw TG, Schmitz B, Ahmed W, Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci Total Environ. 2020;743:140621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 432] [Cited by in RCA: 376] [Article Influence: 75.2] [Reference Citation Analysis (0)] |
| 105. | Haramoto E, Malla B, Thakali O, Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci Total Environ. 2020;737:140405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 470] [Cited by in RCA: 424] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 106. | La Rosa G, Iaconelli M, Mancini P, Bonanno Ferraro G, Veneri C, Bonadonna L, Lucentini L, Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ. 2020;736:139652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 534] [Cited by in RCA: 535] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 107. | Hasan SW, Ibrahim Y, Daou M, Kannout H, Jan N, Lopes A, Alsafar H, Yousef AF. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: Surveillance of COVID-19 epidemic in the United Arab Emirates. Sci Total Environ. 2021;764:142929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 108. | Arslan M, Xu B, Gamal El-Din M. Transmission of SARS-CoV-2 via fecal-oral and aerosols-borne routes: Environmental dynamics and implications for wastewater management in underprivileged societies. Sci Total Environ. 2020;743:140709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 101] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 109. | World Health Organization and United Nations Children's Fund. Progress on drinking water, sanitation and hygiene: 2017 update and SDG baselines. [cited 15 December 2020]. Available from: https://apps.who.int/iris/handle/10665/258617. |
| 110. | Saawarn B, Hait S. Occurrence, fate and removal of SARS-CoV-2 in wastewater: Current knowledge and future perspectives. J Environ Chem Eng. 2021;9:104870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 111. | Mohan N, Deswal S. Corona Virus Disease (COVID-19) Fecal-oral transmission: Is it a potential risk for Indians? Indian J Gastroenterol. 2020;39:305-306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |