Published online Aug 26, 2021. doi: 10.12998/wjcc.v9.i24.7099
Peer-review started: December 18, 2020
First decision: March 27, 2021
Revised: May 14, 2021
Accepted: July 12, 2021
Article in press: July 12, 2021
Published online: August 26, 2021
Processing time: 240 Days and 21.7 Hours
Colorectal liver metastases (CLM) occur in 15%-30% of patients with colorectal cancer (CRC). Advancements in next generation sequencing (NGS) can provide more precise prognoses for cancer patients and help guide clinical treatment. However, the genetic variants that predict high sensitivity to neoadjuvant chemotherapy remain unclear, especially in patients with CLM. The aim of this study was to identify the relevant genetic variants in a single CLM patient and to summarize the current evidence on mutations and single nucleotide polymor
A 76-year-old male patient, who was diagnosed as stage IV colon cancer with liver metastases, was found to have APC/TP53/KRAS mutations. He showed a good therapeutic response to 12 courses of oxaliplatin regimens combined with Bevacizumab. Genetic analysis of the patient identified 5 genes with 7 detected SNPs that may be related to a better response to chemotherapy drugs. In addition, a critical literature review was performed based on a standardized appraisal form after selecting the articles. Ultimately, 21 eligible studies were appraised to assess the association between gene mutations and good prognosis. Mutations in KRAS, TP53, SMAD4, and APC were identified as being associated with a poor response to chemotherapy drugs, whereas mutations of CREBBP and POLD1 were associated with longer overall survival.
NGS can identify precise predictors of response to neoadjuvant chemotherapy, leading to improved outcomes for CRC patients.
Core Tip: Colorectal cancer has high incidence and mortality rates, with liver metastases as the main cause of death. Although chemotherapy is an effective treatment, some patients with specific gene mutations are not sensitive to chemotherapy. With advancements in next generation sequencing, we detected multiple somatic mutations in one patient with colorectal cancer who was sensitive to chemotherapy. Based on the genetic mutation of this patient, we conducted a literature review, which identified KRAS, TP53, SMAD4, and APC as being associated with a poor response to chemotherapy, whereas mutations of CREBBP and POLD1 were associated with longer overall survival.
- Citation: Zhao L, Wang Q, Zhao SD, Zhou J, Jiang KW, Ye YJ, Wang S, Shen ZL. Genetic mutations associated with sensitivity to neoadjuvant chemotherapy in metastatic colon cancer: A case report and review of literature. World J Clin Cases 2021; 9(24): 7099-7109
- URL: https://www.wjgnet.com/2307-8960/full/v9/i24/7099.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i24.7099
Colorectal cancer (CRC) has the second highest incidence rate and the third highest mortality rate globally of all forms of malignant tumor[1]. In CLM patients, colorectal liver metastases (CLM) are the main cause of death. CLM occurs in approximately 15%-30% of patients with CRC[2]. Liver resection has been proven to be an effective treatment for CLM, and it has a survival benefit over chemotherapy alone in terms of the 5-year overall survival (OS) rate (approximately 50%)[3,4].
Currently, European Society for Medical Oncology[5] and National Comprehensive Cancer Network[6] guidelines clearly state that the multiple disciplinary team (MDT) model should be adopted in the diagnosis and treatment of CLM. Notably, the Chinese Society of Clinical Oncology guidelines (version 2019)[7] recommend “some high-level medical units should try their best to treat CRC patients under the management of MDT, especially metastatic colorectal cancer (metastatic colorectal cancer, mCRC)”. Subsequently, according to clinical tolerance to intensive treatments, CLM patients are divided into two categories based upon their general physical condition. For patients in poor health, the recommendations are low toxicity chemotherapy or the best supportive treatment, with re-assessment if necessary to improve quality of life and prolong survival as much as possible. For patients in good health who can tolerate intense treatment, the recommendation is to develop an active comprehensive treatment plan, or different treatment goals and individualized treatments, in order to minimize tumor size or maintain as much residual liver volume as safely possible[7].
Many studies have shown that chemotherapy combined with targeted therapies can significantly increase the response rate to treatment and improve survival time[8,9]. Heinemann et al[8] concluded that both FOLFIRI plus cetuximab and FOLFIRI plus bevacizumab patient groups did not differ significantly in the proportion of patients who achieved an objective response, even though FOLFIRI plus cetuximab is the preferred first-line regimen for patients with KRAS exon 2 wild-type metastatic colorectal cancer. However, for patients with a RAS gene family (KRAS, NRAS, and HRAS) mutation metastatic colorectal cancer, a lack of response to anti-EGRF therapy has been reported[10-12], and Bevacizumab is used as an alternative targeted therapy. Subsequently, the association between mutations in RAS and BRAF and a worse prognosis after CLM resection was also confirmed[13].
With advancements in next generation sequencing (NGS), it is now possible to detect multiple somatic mutations in clinical practice[13]. Such information can help achieve a better understanding of the effectiveness of and sensitivity to chemotherapy in cancer patients and even the emergence of drug resistance at the molecular level. The present article reviews the possible association of somatic gene mutations with the effectiveness and sensitivity of a chemotherapy regimen [oxaliplatin (XELOX) combined with Bevacizumab] through a case study of a patient with APC/TP53/KRAS mutations and microsatellite stable (MSS) status who was diagnosed with metastatic colorectal cancer (liver and lung). This patient showed a better objective response after 12 cycles of this chemotherapy regimen. The aim of this study is to identify possible predictors for the effectiveness of and sensitivity to this chemotherapy regimen through a panel of multi-gene testing [an 825-gene panel for solid tumors and 45 single nucleotide polymorphisms (SNPs) related to chemotherapy]. We then summarize the current best evidence of genes related to CLM through a systematic review of the literature to guide future research on predictors of the effectiveness of and sensitivity to XELOX and Bevacizumab treatment and to facilitate better clinical decision-making. This work was carried out according to the Cooperative for American Relief Everywhere (CARE) criteria[14], which is proven to improve the quality of reporting[15,16].
In January 2019, a 76-year-old male patient presented with the chief complaint of slight changes in bowel habits without any pain or other discomfort for 1 mo. He was admitted and treated at the Department of Gastroenterological Surgery, Peking University People’s Hospital, Beijing, China.
There were no obvious symptoms of pain and discomfort.
The patient’s medical history included liver cysts and gallstones for more than 20 years without specific treatment. He also had a history of hypertension for 3 years and took medication regularly. He denied a history of diabetes or other comorbidities.
Parents are deceased, but his siblings are alive. He denied having any family history of genetic disease.
Physical examination found no obvious abnormalities.
The tumor markers were reexamined in our hospital, and the carcinoma embryonic antigen was found to be elevated (388.1 ng/mL), while the remaining tumor markers were not abnormal.
Based on chest and abdominal computed tomography (CT) and magnetic resonance imaging (MRI), he was diagnosed with descending colon cancer with multiple metastases located in the lung and liver.
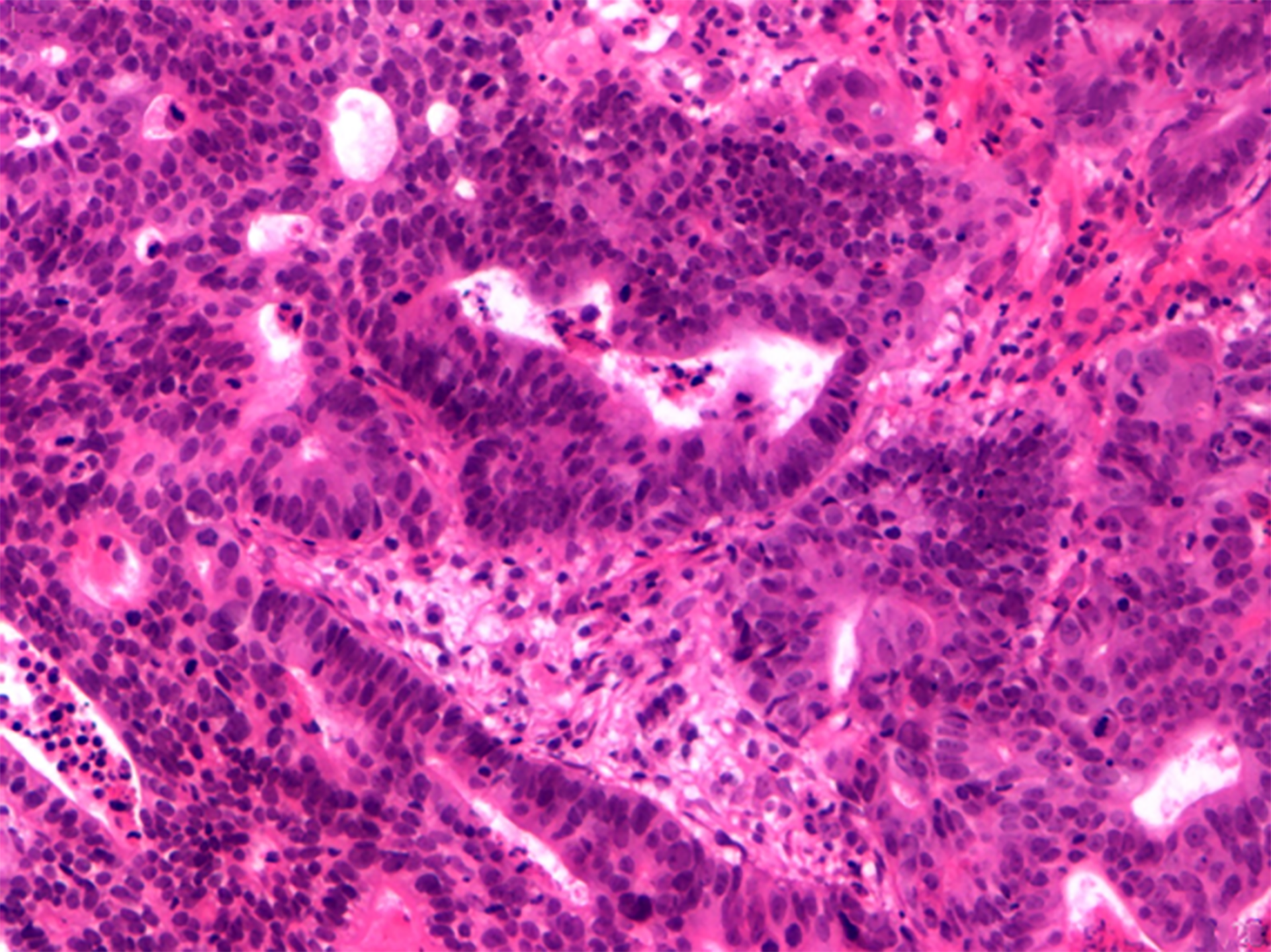
After discussion among the MDT, an endo-biopsy was determined to be necessary to confirm malignancy before neoadjuvant chemotherapy (Figure 1). The team also decided to determine the patient’s microsatellite instable status, genetic mutations and SNPs, and tumor mutation burden (TMB) in the primary tumor tissue to guide selection of the best chemotherapeutic agent.
Colorectal cancer with liver metastasis.
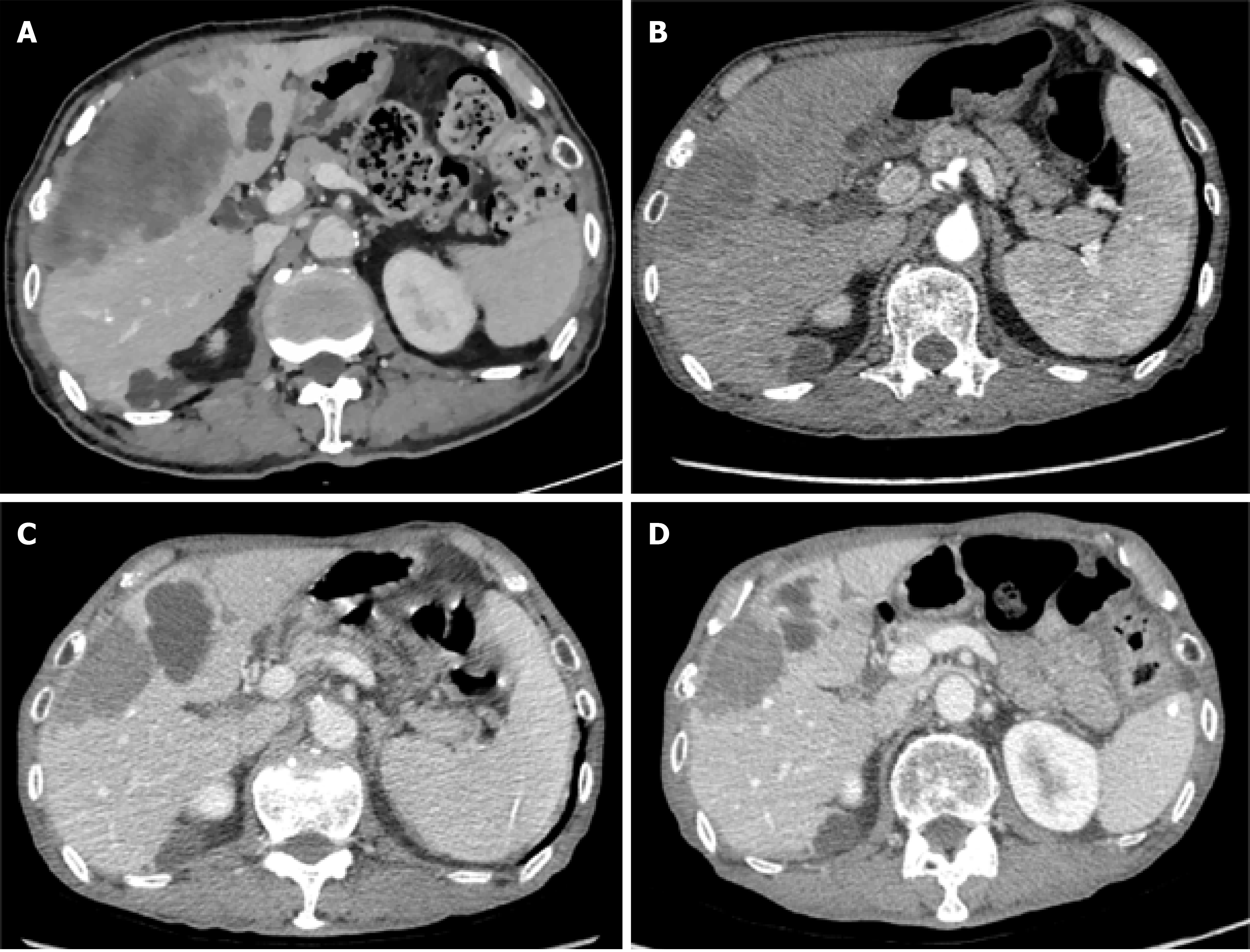
The patient was ultimately identified as having an APC/TP53/KRAS mutation, MSS status, and TMB with 4.27/Mb. The XELOX regimen (XELOX 150 mg iv dl q3w, Capecitabine 1500 mg bid po d2-d15 q3w) combined with Bevacizumab (300 mg iv dl q3w) was selected for chemotherapy. After three courses of systemic combination chemotherapy, chest and abdominal CT and MRI revealed a good therapeutic response. The above-mentioned chemotherapy regimen was continued for 9 more courses (12 courses in total). The patient continued to respond well to the chemotherapy regimen (Figure 2).
Despite his complete response to chemotherapy, the patient ultimately passed away due to intestinal obstruction and serious cachexia on June 15, 2020, having declined to continue the chemotherapy regimen during the coronavirus disease 2019 pandemic.
In light of the outcomes for this patient, we ask the following questions: (1) What is the association between these specific somatic mutations and prognosis? (2) What SNP(s) can predict treatment response to the XELOX regimen combined with Bevacizumab in patients with multiple metastases of colorectal cancer?
Search strategy: A systematic review of the literature was conducted using the PubMed, EMBASE, and Cochrane databases through 8 September 2020. The search for specific somatic mutations was conducted using the following keywords and algorithm: “(colorectal or rectal or colon or colonic) and (liver or hepatic) and (metastasis or metastases) and (gene or mutation or KRAS or TP53 or APC or IKZF1 or POLD1 or PRKCB or SMAD4 or CREBBP).”
Literature screening and evaluation: For somatic gene mutations and their functions, we included studies that focused on the association between a specific gene mutation and overall survival (OS) or recurrence-free survival (RFS) in resectable and unresectable patients with CLM, no matter whether chemotherapy was clearly described. Better OS and RFS were defined as the effectiveness of chemotherapy in this study. We included systematic reviews, meta-analyses, and cohort studies. We excluded studies that included the above-mentioned mutations but failed to report OS as a measure of prognosis, or studies in which patients received another form of intervention. Of the included studies, due to the lack of direct evidence of “genetic mutations of sensitivity to platinum-based neoadjuvant chemotherapy”, we regarded a genetic mutation of sensitivity to neoadjuvant chemotherapy to be when a published record showed a good prognosis regardless of whether the patient received adjuvant chemotherapy.
For SNPs predicting treatment response to XELOX regimens combined with Bevacizumab, the PharmGKB database (http://www.pharmgkb.org) was used to reference SNPs and chemotherapeutic drugs. This database focuses on genetic pharmacology and genomic pharmacology. Genetic evidence is graded using the following criteria: Level IA: Recognized by Guidelines from the Clinical Pharmacogenetics Implementation Consortium or the Genetic Pharmacology, or recognized by the International Genetic Pharmacology Research Network or other major health systems; Level IB: The relevance to drugs is supported by multiple studies that found consistent results with statistically significant differences; Level IIA: The relevance to drugs is supported by multiple studies, and the gene is known as an important drug metabolism gene with a clear function; Level IIB: The relevance to drugs is supported by multiple studies, but some studies have a small sample size or found no statistically significant differences; and Level III: The relevance to drugs is supported by a single study, but a consensus has not yet been reached.
Based on the criteria, 21 studies were identified. We appraised these 21 eligible studies using the critical appraisal questions developed by the Centre of Evidence-Based Medicine, University of Oxford.
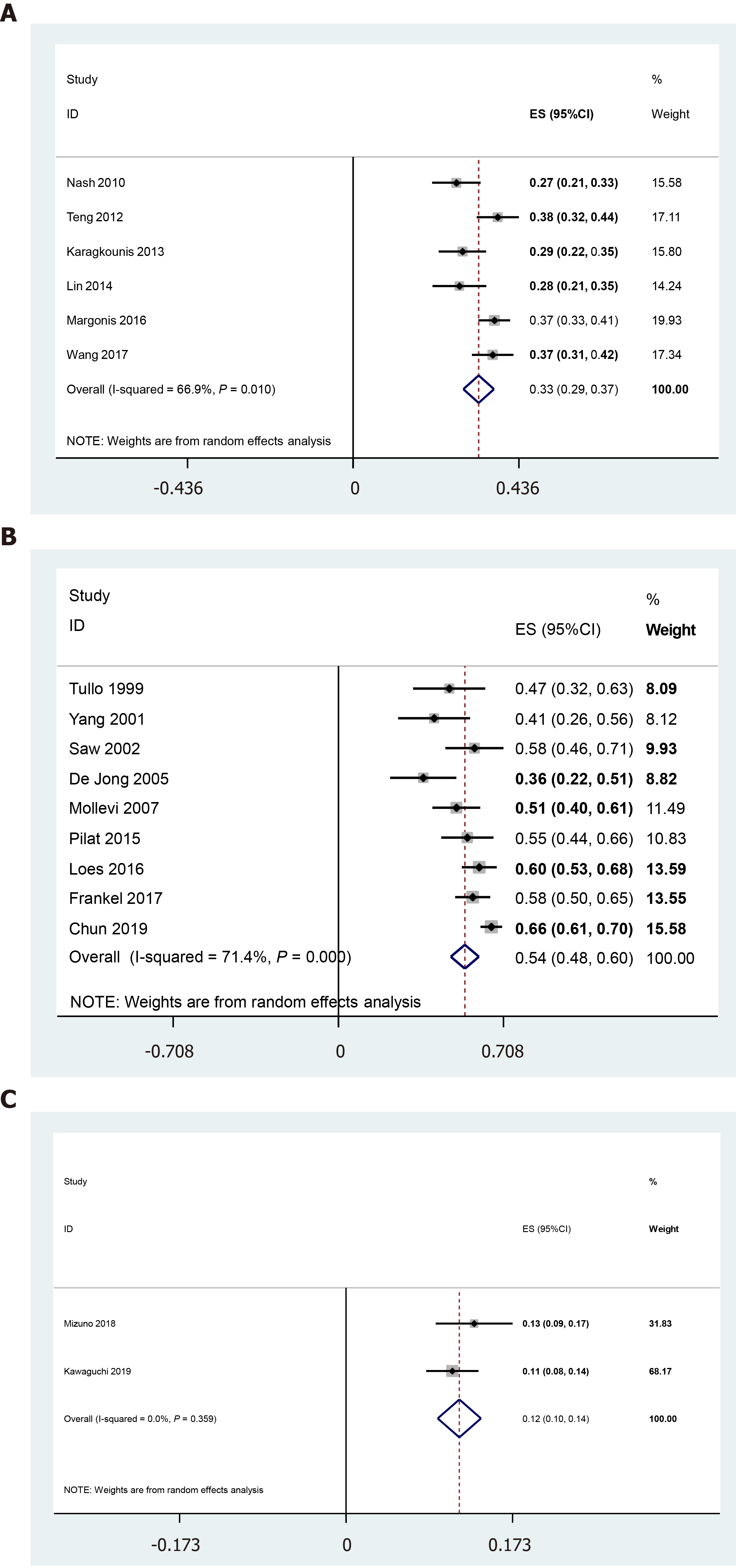
According to the literature review, KRAS, TP53, and SMAD4 mutations may be harmful to prognosis, with pooled mutation rates of [33% (29%, 37%), n = 1648], [54% (48%, 60%), n = 1080], and [12% (10%, 14%), n = 785], respectively (Figure 3 and Table 1). The frequency of APC mutation was 11% (45/396), and it showed a negative association with better prognosis. For the other mutations, there is a lack of cohort studies focused on gene mutations and prognosis in CLM patients. Table 2 shows the current evidence of CREBBP, IKZF1, POLD1, and PRKCB mutations and gene functions. CREBBP and POLD1 mutations may be related to longer OS and RFS.
| Genes mutation | Ref. | Total No. of patients | Frequency | Prognosis | ||
| n | % | OS | RFS | |||
| KRAS1 | Nash et al[19], 2010 | 188 | 51 | 27 | Worse | |
| Teng et al[24], 2012 | 292 | 111 | 38 | None | ||
| Karagkounis et al[20], 2013 | 202 | 58 | 29 | Worse | Worse | |
| Lin et al[21], 2014 | 154 | 43 | 28 | None | ||
| Margonis et al[22], 2016 | 512 | 190 | 37 | None | ||
| Wang et al[23], 2017 | 300 | 110 | 37 | Worse | ||
| TP53 | Tullo et al[25], 1999 | 40 | 19 | 48 | Worse | |
| Yang et al[27], 2001 | 39 | 16 | 41 | Better | Better | |
| Saw et al[26], 2002 | 60 | 35 | 58 | None | ||
| De Jong et al[30], 2005 | 44 | 16 | 36 | None | None | |
| Molleví et al[28], 2007 | 91 | 46 | 51 | Worse | ||
| Pilat et al[29], 2015 | 76 | 42 | 55 | Worse | ||
| Løes et al[33], 2016 | 164 | 99 | 60 | None | None | |
| Frankel et al[31], 2017 | 165 | 95 | 58 | None | ||
| Chun et al[32], 2019 | 401 | 263 | 66 | None | ||
| SMAD4 | Mizuno et al[35], 2018 | 278 | 37 | 13 | Worse | |
| Kawaguchi et al[36], 2019 | 507 | 56 | 11 | Worse | Worse | |
| APC2 | Yamashita et al[34], 2020 | 396 | 45 | 11 | Worse | Worse |
| Genes mutation | Ref. | Summary | Conclusion |
| CREBBP | Lin et al[37], 2020 | TMB-high (> 11 mutations/Mb), male, mutation of RNF43, CREBBP, NOTCH3, PTCH1, CIC, DNMT1 and SPEN were all related to longer OS | CREBBP mutation may be related to higher immunogenicity such as TMB, high expression of mRNA related to immune response, highly infiltrating immune-active cells such as CD8+T cells, active immune-active pathways, and DNA damage repair pathways with an increased number of mutations |
| Douglas et al[38],2020 | When looking at the complete responder group, mutations were noted in endoscopic biopsy specimens from at least two patients in genes including ARID1A, JAK1, CREBBP, and MTOR (three patients each), that were not seen to be mutated in PR specimens | The authors identified multiple genetic variations in tumor DNA from rectal cancer patients who are poor responders to neoadjuvant chemoradiation, compared to complete responders | |
| POLD1 | Hühns et al[40], 2019 | The authors performed POLE and POLD1 exonuclease domain Sanger sequencing of 271 unselected colorectal carcinomas and finally identified two microsatellite-stable tumors with somatic POLE p.P286R variants, both with ultrahigh TMBs as demonstrated by whole exome sequencing. In two tumors, a somatic POLE p.V411L and a POLD1 p.E279K, respectively, were found only focally, and TMBs were low. It is commonly assumed that compromise of one allele is sufficient, but this has not been specifically addressed | Taken together, including at least the more common DNA polymerase mutations in NGS panels allows for straightforward identification of hypermutator-type colorectal carcinomas which often may be "immunoreactive". This is important at least in young patients or when a metastasizing stage of disease has been reached and immune-checkpoint therapy enters deliberation |
| IKZF1 | None | No eligible studies | No evidence |
| PRKCB | None | No eligible studies | No evidence |
Regarding chemotherapy-related SNPs, 5 genes with 7 detected loci were identified as potentially being related to better response to chemotherapy drugs according to the PharmGKB database; however, the grade of this evidence is relatively low (Table 3).
| Genes | Detected loci | SNP results | Evidence | Summary | |
| Drugs | Grades | ||||
| DPYD | rs3918290 | CC (wild-type) | 5Fu-based | Level III | For patients with tumor, CC genotype may have a higher drug remission rate than CT genotype. However, there are also studies with inconsistent conclusions |
| rs1801159 | TT (wild-type) | Level III | For a patient with tumor, TT genotype may have a higher drug remission rate than CT or CC genotype | ||
| GSTP1 | rs1695 | AG (heterozygous mutants) | 5Fu + XELOX | Level IIA | For a patient with tumor, AG genotype may have a higher drug remission rate than AA genotype, but comparing with GG genotype, AG genotype may reduce drug remission rate and relate to a poor OS |
| ERCC2 | rs13181 | TT (wild-type) | 5Fu + Leucovorin + XELOX | Level III | For a patient with CRC, TT genotype may have a lower risk of recurrence and a longer PFS than GG genotype |
| rs1052555 | GG (wild-type) | Platinum compounds | Level III | For a patient with NSCLC, GG genotype may have a higher drug remission rate than AA or AG genotype | |
| ABCB1 | rs1045642 | GG (homozygous mutants) | Level III | For a patient with CRC, GG genotype may have a higher drug remission rate than AA or AG genotype | |
| VEGFA | rs25648 | CC (wild-type) | Level III | For a patient with gastric cancer, CC genotype may have a higher drug remission rate than CT or TT genotype | |
Advancements in NGS have enabled more precise prognosis of patients with CLM and have expanded the information available in clinical practice. It is reported that 10% of patients with CLM have mutations in TP53, APC, RAS, PIK3CA, or SMAD4. Notably, TP53, RAS, and SMAD4 are associated with worse survival in patients undergoing CLM resection[17,18].
In this study, we reported a stage IV colon cancer patient with APC/TP53/KRAS mutations who showed a good therapeutic response after 12 courses of XELOX regimen combined with Bevacizumab. Genetic testing identified 8 relevant genes in this patient (Tables 1 and 2). In this case report, we detected 5 genes with 7 SNPs that might result in a better response to chemotherapy drugs (Table 3). However, the genetic mutations and SNPs which could be used to predict high sensitivity to neoadjuvant chemotherapy in patients with metastatic colorectal cancer remain unclear.
Through reviewing the literature, we found that KRAS[19-24], TP53[25-33], and APC[34] were potentially related to a poor prognosis after adjuvant chemotherapy or radical surgery, and so is the mutational status of SMAD4[35,36], which were out of eight mutations detected in the patient. However, mutations like IKZF1 and PRKCB have not been studied comprehensively, especially in association with sensitivity to neoadjuvant chemotherapy. CREBBP seems to be mutated in patients who achieve complete remission. Lin et al[37] found that a high tumor mutation burden (TMB-high, mutation count > 11 mutation/Mb), male sex, RNF43-mutant status, and CREBBP-mutant status were associated with longer OS, while age ≤ 65 years, APC-mutant status, and TP53-mutant status were associated with shorter OS. Moreover, Douglas et al[38] identified 195 variants in 83 genes in tissue specimens implicated in colorectal cancer bio-pathways. Relative to partial responders, complete responders showed mutations in 10 genes, namely ARID1A, PMS2, JAK1, CREBBP, MTOR, RB1, PRKAR1A, FBXW7, ATM C11orf65, and KMT2D. Partial responders showed mutations in four genes: KDM6A, ABL1, DAXX-ZBTB22, and KRAS. In terms of advantages like longer OS or greater sensitivity to chemotherapy, these genes might be associated with higher immunogenicity, such as TMB, high expression of messenger ribonucleic acid related to immune responses, highly infiltrating immune-active cells like CD8+ T cells, active immune-active pathways, and deoxyribonucleic acid (DNA) damage repair pathways with an increased number of mutations[37]. Yamauchi et al[39] investigated changes in the amount and constitution of circulating tumor DNA (ctDNA) in serial peripheral blood samples collected from patients during anti-vascular endothelial growth factor (VEGF) chemotherapy, as understanding molecular changes in tumors in response to chemotherapy is crucial for optimization of the treatment strategy for metastatic colorectal cancer. They found that mutations in CREBBP and FBXW7 genes were newly detected in ctDNA at a low frequency of around 1% in the post-progression period. Moreover, they suggested that changes in ctDNA levels may be a useful predictive biomarker for survival. Furthermore, mutations in CREBBP and FBXW7 genes newly detected in ctDNA in the late treatment period might reveal the rise of a minor tumor clone that could show resistance to anti-VEGF therapy.
The remaining mutation identified in this study, POLD1, seems to be of unknown clinical significance because of a lack of published studies. Hühns et al[40] performed POLE and POLD1 exonuclease domain Sanger sequencing of 271 CRCs and identified two microsatellite-stable tumors with ultrahigh TMBs related to young patients (< 50-years-old, non-syndromic) and prominent T-cell infiltration. They also identified a somatic POLE p.A465T in a Lynch-associated tumor. Somatic POLE p.V411L and POLD1 p.E279K were only found focally and with low TMBs, resulting in the assumption that the compromise of one allele might be sufficient to increase sensitivity to neoadjuvant chemotherapy.
In the results for chemotherapy-related SNPs, 5 genes with 7 loci were identified in the PharmGKB database that might predict a better response. However, the grade of the evidence is relatively low, and a consensus has not yet been reached for SNPs identified in a single study. Multiple studies or a genome-wide association study (GWAS) are needed to confirm that an SNP is relevant to drug responses, and that there is a statistically significant difference in outcomes for patients with specific SNPs.
In the present case, a weak association was observed between mutations in CREBBP and POLD1 and high sensitivity to neoadjuvant chemotherapy. Future studies should identify a subgroup of patients who are excellent candidates for chemotherapy, including immune checkpoint inhibitor therapy. In addition to validating this weak evidence, a priority for future studies might be to confirm these findings in a larger sample. A GWAS of chemotherapy-related SNPs could examine whether favorable prognosis is independent of adjuvant chemotherapy.
In terms of the limitations, as a case report, this study focused on genetic mutations in only one patient, and the small sample size is an obvious limitation. However, as shown by evidence from the literature review, although the overall grade of evidence for chemotherapy-related SNPs is relatively low, an evidence-based case report and literature review might be useful for identifying predictors of sensitivity to chemotherapy in patients with metastatic colorectal cancer.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D, D
Grade E (Poor): 0
P-Reviewer: Casella C, Cerwenka H S-Editor: Zhang L L-Editor: Filipodia P-Editor: Xing YX
| 1. | Van Cutsem E, Nordlinger B, Cervantes A; ESMO Guidelines Working Group. Advanced colorectal cancer: ESMO Clinical Practice Guidelines for treatment. Ann Oncol. 2010;21 Suppl 5:v93-v97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 353] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 2. | Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 1009] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 3. | Choti MA, Sitzmann JV, Tiburi MF, Sumetchotimetha W, Rangsin R, Schulick RD, Lillemoe KD, Yeo CJ, Cameron JL. Trends in long-term survival following liver resection for hepatic colorectal metastases. Ann Surg. 2002;235:759-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1105] [Cited by in RCA: 1118] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 4. | Abdalla EK, Vauthey JN, Ellis LM, Ellis V, Pollock R, Broglio KR, Hess K, Curley SA. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann Surg. 2004;239:818-25; 825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1364] [Cited by in RCA: 1287] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 5. | Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson A, Bodoky G, Ciardiello F, D'Hoore A, Diaz-Rubio E, Douillard JY, Ducreux M, Falcone A, Grothey A, Gruenberger T, Haustermans K, Heinemann V, Hoff P, Köhne CH, Labianca R, Laurent-Puig P, Ma B, Maughan T, Muro K, Normanno N, Österlund P, Oyen WJ, Papamichael D, Pentheroudakis G, Pfeiffer P, Price TJ, Punt C, Ricke J, Roth A, Salazar R, Scheithauer W, Schmoll HJ, Tabernero J, Taïeb J, Tejpar S, Wasan H, Yoshino T, Zaanan A, Arnold D. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386-1422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2286] [Cited by in RCA: 2424] [Article Influence: 269.3] [Reference Citation Analysis (31)] |
| 6. | National Comprehensive Cancer Network Guidelines. NCCN Clinical Practice Guidelines in Oncology-Colon Cancer. (Version 2, 2019 Guidelines). [cited 20 May 2021]. Available from: https://www.nccn.org/patientresources/patient-resources. |
| 7. | Guidelines of Chinese Society of Clinical Oncology (CSCO): Colorectal Cancer (Version 1, 2019 Guidelines). Beijing: People's Medical Publishing House, 2019; 1-139.. |
| 8. | Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, Kullmann F, Stauch M, Scheithauer W, Hielscher J, Scholz M, Müller S, Link H, Niederle N, Rost A, Höffkes HG, Moehler M, Lindig RU, Modest DP, Rossius L, Kirchner T, Jung A, Stintzing S. FOLFIRI plus cetuximab vs FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1117] [Cited by in RCA: 1329] [Article Influence: 120.8] [Reference Citation Analysis (0)] |
| 9. | Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408-1417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2901] [Cited by in RCA: 3122] [Article Influence: 195.1] [Reference Citation Analysis (1)] |
| 10. | Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 736] [Cited by in RCA: 726] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 11. | Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, Canon JL, Van Laethem JL, Maurel J, Richardson G, Wolf M, Amado RG. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1444] [Cited by in RCA: 1471] [Article Influence: 81.7] [Reference Citation Analysis (0)] |
| 12. | Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2504] [Cited by in RCA: 2403] [Article Influence: 141.4] [Reference Citation Analysis (0)] |
| 13. | Kawaguchi Y, Lillemoe HA, Vauthey JN. Gene mutation and surgical technique: Suggestion or more? Surg Oncol. 2020;33:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 14. | Gagnier JJ, Kienle G, Altman DG, Moher D, Sox H, Riley D; CARE Group. The CARE guidelines: consensus-based clinical case report guideline development. J Clin Epidemiol. 2014;67:46-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 407] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 15. | Yang KL, Lu CC, Sun Y, Cai YT, Wang B, Shang Y, Tian JH. How about the reporting quality of case reports in nursing field? World J Clin Cases. 2019;7:3505-3516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | An GH, Tang XT, Chen YL, Zhao Y. Reporting characteristics of case reports of acupuncture therapy with CARE guidelines. Chin J Integr Med. 2018;24:56-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Kawaguchi Y, Velasco JD, Arvide EM, Wei SH, Vauthey JN. Interactions of multiple gene alterations in colorectal liver metastases. Chin Clin Oncol. 2019;8:50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 18. | Lang H, Baumgart J, Heinrich S, Tripke V, Passalaqua M, Maderer A, Galle PR, Roth W, Kloth M, Moehler M. Extended Molecular Profiling Improves Stratification and Prediction of Survival After Resection of Colorectal Liver Metastases. Ann Surg. 2019;270:799-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 19. | Nash GM, Gimbel M, Shia J, Nathanson DR, Ndubuisi MI, Zeng ZS, Kemeny N, Paty PB. KRAS mutation correlates with accelerated metastatic progression in patients with colorectal liver metastases. Ann Surg Oncol. 2010;17:572-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 140] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 20. | Karagkounis G, Torbenson MS, Daniel HD, Azad NS, Diaz LA Jr, Donehower RC, Hirose K, Ahuja N, Pawlik TM, Choti MA. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer. 2013;119:4137-4144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 21. | Lin Q, Ye Q, Zhu D, Wei Y, Ren L, Ye L, Feng Q, Xu P, Zheng P, Lv M, Fan J, Xu J. Determinants of long-term outcome in patients undergoing simultaneous resection of synchronous colorectal liver metastases. PLoS One. 2014;9:e105747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 22. | Margonis GA, Kim Y, Sasaki K, Samaha M, Amini N, Pawlik TM. Codon 13 KRAS mutation predicts patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Cancer. 2016;122:2698-2707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 23. | Wang K, Liu W, Yan XL, Li J, Xing BC. Long-term postoperative survival prediction in patients with colorectal liver metastasis. Oncotarget. 2017;8:79927-79934. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 24. | Teng HW, Huang YC, Lin JK, Chen WS, Lin TC, Jiang JK, Yen CC, Li AF, Wang HW, Chang SC, Lan YT, Lin CC, Wang HS, Yang SH. BRAF mutation is a prognostic biomarker for colorectal liver metastasectomy. J Surg Oncol. 2012;106:123-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 25. | Tullo A, D'Erchia AM, Honda K, Mitry RR, Kelly MD, Habib NA, Saccone C, Sbisà E. Characterization of p53 mutations in colorectal liver metastases and correlation with clinical parameters. Clin Cancer Res. 1999;5:3523-3528. [PubMed] |
| 26. | Saw RP, Koorey D, Painter D, Gallagher PJ, Solomon MJ. p53, DCC and thymidylate synthase as predictors of survival after resection of hepatic metastases from colorectal cancer. Br J Surg. 2002;89:1409-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 27. | Yang Y, Forslund A, Remotti H, Lönnroth C, Andersson M, Brevinge H, Svanberg E, Lindnér P, Hafström L, Naredi P, Lundholm K. P53 mutations in primary tumors and subsequent liver metastases are related to survival in patients with colorectal carcinoma who undergo liver resection. Cancer. 2001;91:727-736. [PubMed] |
| 28. | Molleví DG, Serrano T, Ginestà MM, Valls J, Torras J, Navarro M, Ramos E, Germà JR, Jaurrieta E, Moreno V, Figueras J, Capellà G, Villanueva A. Mutations in TP53 are a prognostic factor in colorectal hepatic metastases undergoing surgical resection. Carcinogenesis. 2007;28:1241-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Pilat N, Grünberger T, Längle F, Mittlböck M, Perisanidis B, Kappel S, Wolf B, Starlinger P, Kührer I, Mühlbacher F, Kandioler D. Assessing the TP53 marker type in patients treated with or without neoadjuvant chemotherapy for resectable colorectal liver metastases: a p53 Research Group study. Eur J Surg Oncol. 2015;41:683-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | de Jong KP, Gouw AS, Peeters PM, Bulthuis M, Menkema L, Porte RJ, Slooff MJ, van Goor H, van den Berg A. P53 mutation analysis of colorectal liver metastases: relation to actual survival, angiogenic status, and p53 overexpression. Clin Cancer Res. 2005;11:4067-4073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Frankel TL, Vakiani E, Nathan H, DeMatteo RP, Kingham TP, Allen PJ, Jarnagin WR, Kemeny NE, Solit DB, D'Angelica MI. Mutation location on the RAS oncogene affects pathologic features and survival after resection of colorectal liver metastases. Cancer. 2017;123:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 32. | Chun YS, Passot G, Yamashita S, Nusrat M, Katsonis P, Loree JM, Conrad C, Tzeng CD, Xiao L, Aloia TA, Eng C, Kopetz SE, Lichtarge O, Vauthey JN. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann Surg. 2019;269:917-923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 121] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 33. | Løes IM, Immervoll H, Sorbye H, Angelsen JH, Horn A, Knappskog S, Lønning PE. Impact of KRAS, BRAF, PIK3CA, TP53 status and intraindividual mutation heterogeneity on outcome after liver resection for colorectal cancer metastases. Int J Cancer. 2016;139:647-656. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 34. | Yamashita S, Chun YS, Kopetz SE, Maru D, Conrad C, Aloia TA, Vauthey JN. APC and PIK3CA Mutational Cooperativity Predicts Pathologic Response and Survival in Patients Undergoing Resection for Colorectal Liver Metastases. Ann Surg. 2020;272:1080-1085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 35. | Mizuno T, Cloyd JM, Vicente D, Omichi K, Chun YS, Kopetz SE, Maru D, Conrad C, Tzeng CD, Wei SH, Aloia TA, Vauthey JN. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur J Surg Oncol. 2018;44:684-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 36. | Kawaguchi Y, Kopetz S, Newhook TE, De Bellis M, Chun YS, Tzeng CD, Aloia TA, Vauthey JN. Mutation Status of RAS, TP53, and SMAD4 is Superior to Mutation Status of RAS Alone for Predicting Prognosis after Resection of Colorectal Liver Metastases. Clin Cancer Res. 2019;25:5843-5851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 134] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 37. | Lin A, Zhang H, Hu X, Chen X, Wu G, Luo P, Zhang J. Age, sex, and specific gene mutations affect the effects of immune checkpoint inhibitors in colorectal cancer. Pharmacol Res. 2020;159:105028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 38. | Douglas JK, Callahan RE, Hothem ZA, Cousineau CS, Kawak S, Thibodeau BJ, Bergeron S, Li W, Peeples CE, Wasvary HJ. Genomic variation as a marker of response to neoadjuvant therapy in locally advanced rectal cancer. Mol Cell Oncol. 2020;7:1716618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 39. | Yamauchi M, Urabe Y, Ono A, Miki D, Ochi H, Chayama K. Serial profiling of circulating tumor DNA for optimization of anti-VEGF chemotherapy in metastatic colorectal cancer patients. Int J Cancer. 2018;142:1418-1426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 40. | Hühns M, Nürnberg S, Kandashwamy KK, Maletzki C, Bauer P, Prall F. High mutational burden in colorectal carcinomas with monoallelic POLE mutations: absence of allelic loss and gene promoter methylation. Mod Pathol. 2020;33:1220-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |