Published online May 6, 2020. doi: 10.12998/wjcc.v8.i9.1580
Peer-review started: December 29, 2019
First decision: January 19, 2020
Revised: March 26, 2020
Accepted: April 24, 2020
Article in press: April 24, 2020
Published online: May 6, 2020
Processing time: 122 Days and 16.8 Hours
MicroRNAs (miRNAs) are highly deregulated in cancer and play a role in the initiation of tumorigenesis. Recently, miRNAs have attracted attention in gastrointestinal (GI) cancers. Single nucleotide polymorphisms (SNPs) could affect the genes involved in each step of miRNA biosynthesis. Several meta-analyses of case-control studies have assessed the association between miRNA “pathway” gene-SNPs (including biosynthesis regulators and binding sites) and susceptibility to GI cancers. We present in this mini-review the current knowledge on the association between miRNAs “pathway” genes and GI cancer predisposition. The interaction between miRNA/regulators/binding site-SNPs and environmental as well as genomic factors is an interesting field that should be exploited in future studies.
Core tip: We discuss in this mini-review the current knowledge on the association between microRNA-gene-single nucleotide polymorphisms as well as their regulators/binding sites and gastrointestinal cancer predisposition. They could act as tumor suppressors as well as oncogenes depending on their target. We also discuss the interaction between microRNAs and environmental factors and genomic susceptibility like microsatellite instability.
- Citation: Baz M, Ibrahim T. Role of microRNAs in the predisposition to gastrointestinal malignancies. World J Clin Cases 2020; 8(9): 1580-1585
- URL: https://www.wjgnet.com/2307-8960/full/v8/i9/1580.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i9.1580
MicroRNAs (miRNAs) are highly conserved short (nearly 20 nucleotides) non-coding RNA-molecules that control cell survival and proliferation. In their biologically active form, they induce the downregulation of mRNA. A single miRNA can regulate the expression of many mRNA-targets involved in different biological pathways[1]. Hence, even a small change in their structure and activity can have a profound effect on cell homeostasis. Studies have shown that key miRNAs are highly deregulated in cancer and play a role in the initiation of tumorigenesis by influencing the expression of oncogenic and tumor suppressor proteins[1,2].
To date, more than 2000 sequences of miRNAs have been reported in humans[3,4], and more than 4500 miRNA gene-variations have been described. Single nucleotide polymorphisms (SNPs) of miRNA-related genes have been shown to increase cancer risk and have attracted attention in recent years mainly in gastrointestinal (GI) cancers[5].
This mini-review intends to present the current knowledge on the association between SNPs affecting miRNAs as well as their machinery genes and the risk of developing GI cancers.
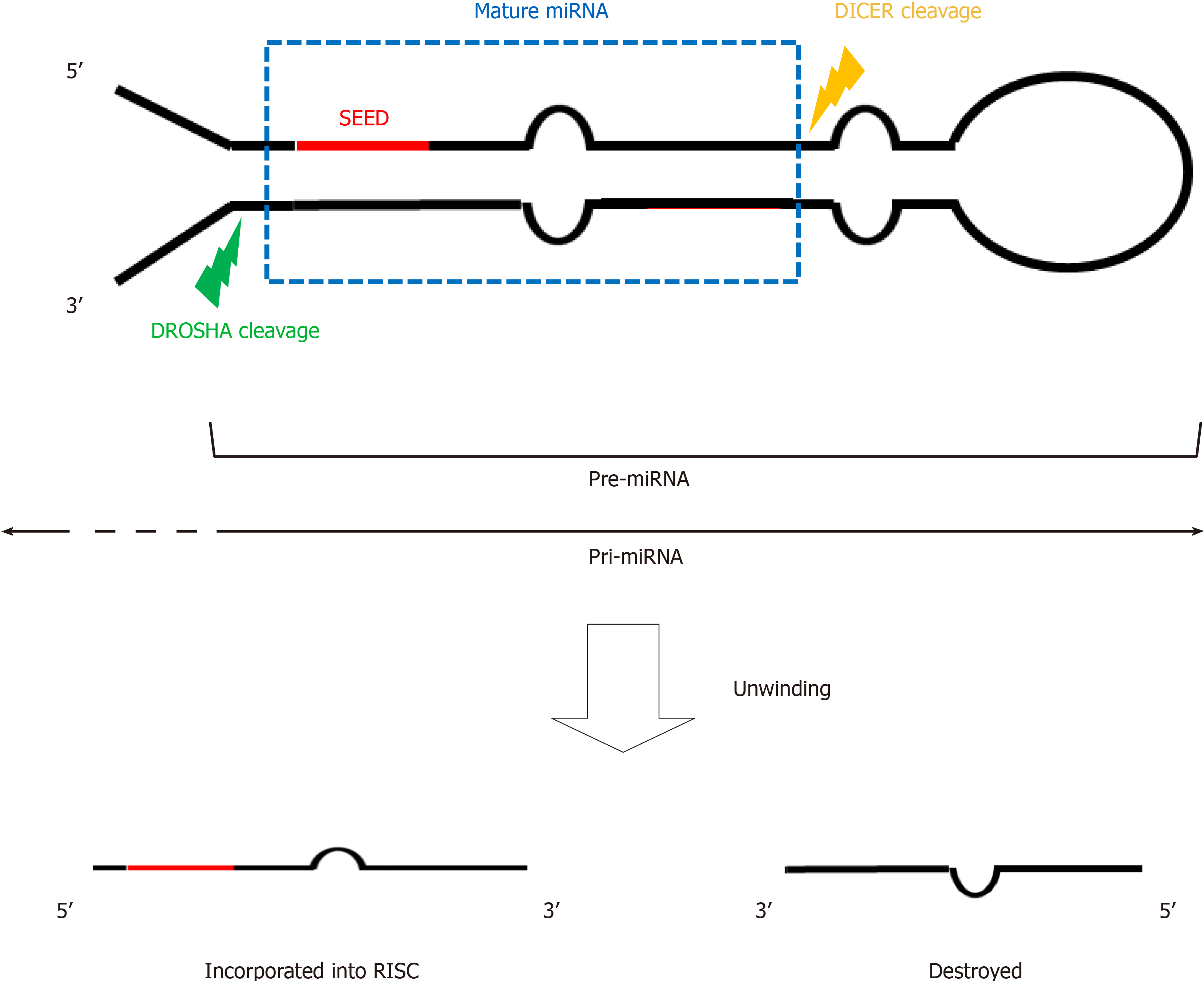
The regulation of miRNA biosynthesis is complex and under continuous investigation[6]. miRNA genes are generally transcribed by RNA polymerase II from intergenic regions of the genome, though they can be found in coding areas (intragenic) as well. Most are transcribed independently form another, but some show a tandem repeat-like cluster organization[7]. After DNA transcription, the resulting pri-miRNAs are cleaved in the nucleus into small sequences (nearly 60 nucleotides) by DROSHA ribonuclease III enzyme and its cofactor DGR8. After their export to the cytoplasm by exportin-5 (XPO5), the pre-miRNAs are processed by DICER ribonuclease into a mature duplex. The next step is the unwinding which consists in the separation of the two miRNA brands, followed by loading of the brand containing the seed region onto RNA-inducing-silencing complex (RISC). These complex guide mature miRNAs to 3’UTR region of their target mRNA-sites resulting in their cleavage and translational repression[8]. Figure 1 illustrates the structure of pri, pre, and mature miRNA.
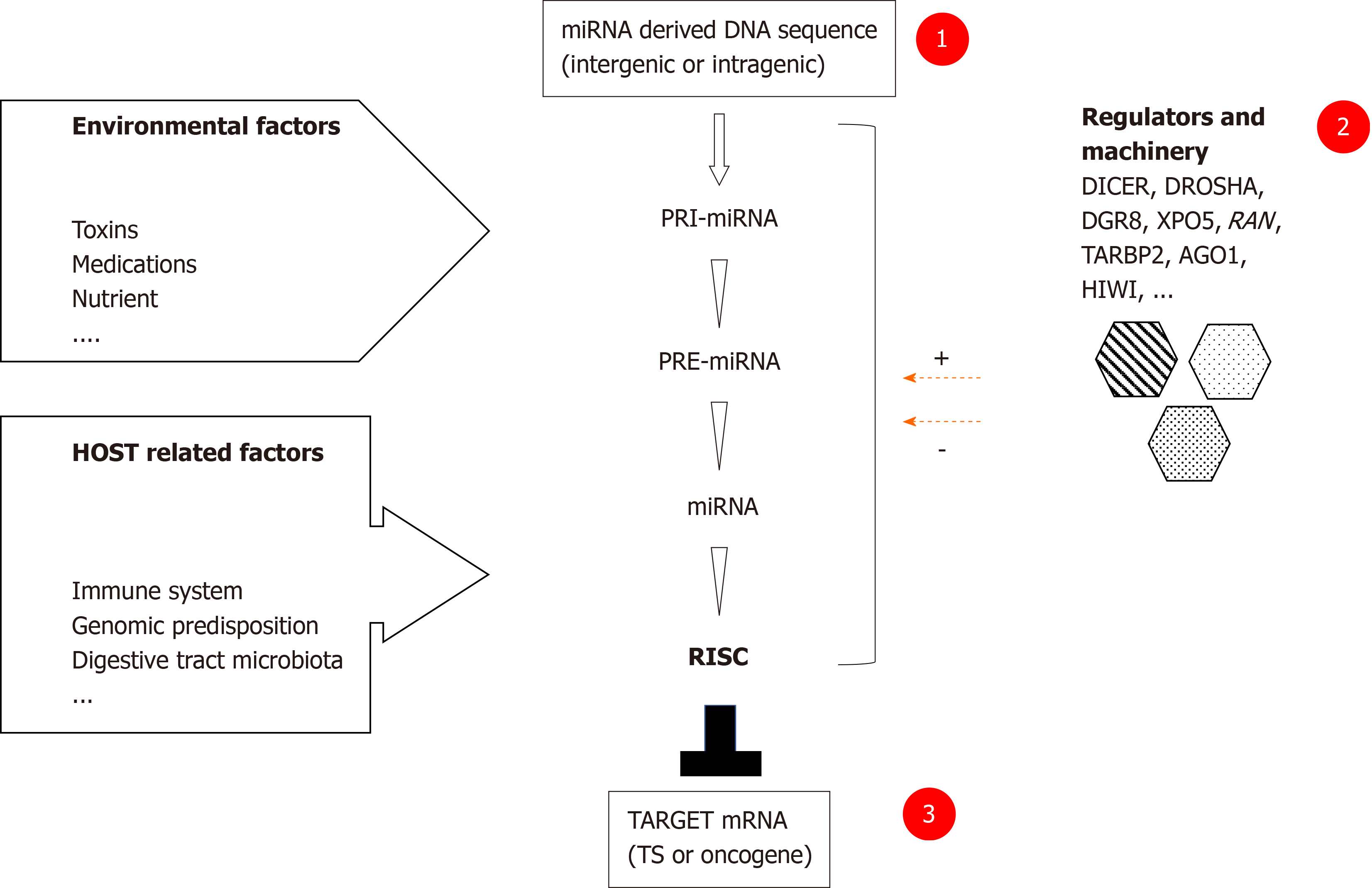
Recent studies have shown that key miRNAs could act as tumor suppressors or as oncogenes in GI cancers depending on their targets[1]. Potential tumour suppressor miRNAs include among others miRNA-15b and miRNA-16 which modulate apoptosis via targeting BCL2 in gastric cancer (GC)[9], and miRNA-34a via E2F pathway in colorectal cancer (CRC)[10]. Potential oncogenic miRNAs include among others miRNA-21 via PTEN in hepatocellular carcinoma (HCC)[11], GC and CRC[12]; miRNA-106a via RB1 in CRC[12]; miRNA-106b-25 cluster via TGF-beta in GC[13]. SNPs could affect genes involved in each step of the miRNA biosynthesis including miRNA-binding sites as well as their regulators, consequently impacting their function (gain or loss) (Figure 2)[14].
Several meta-analyses of increasing number of case-control studies have been conducted to assess the association between different miRNA-polymorphisms and GI cancer susceptibility[5]. We present in this minireview four miRNA SNPs that are, to our opinion, the most validated to date as risk factors for GI cancers. These include miRNA-146a rs2910164, miRNA-149 rs2292832, miRNA-499a rs3746444 which affect the seed region, as well as miRNA-196a2 rs11614913 affecting the premature form[5].
miRNA-146a belongs to a family of miRNAs involved in the regulation of inflammation and the innate immune system[15]. It has been associated with tissue invasion and metastases as well as independence on external growth factor signals[2]. Multiple target-sites for miRNA-146a have been identified and include the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), the CCAAT enhancer-binding-protein-β (C/EBPβ), and the interferon regulatory factor 3/7 (IRF3/7)[16]. miRNA-146a rs2910164 (C>G) polymorphism which affects the seed region[5], was closely associated with GI cancers especially GC[17,18], CRC[17], and esophageal[18] cancer as well as HCC mainly in Asian populations[19]. The odds ratio (OR) ranged between 1.1 and 1.2 (increased risk of 10% to 20%). Members of the mi-RNA 196 family are in the region of the homeobox (HOX) genes (HOXC 9-10 specifically for miR-196a-2 gene), which are transcription factors essential for embryonic development. The non-histone chromatin-binding protein HMGA2 (for High-mobility group A2) mRNA was identified as one of its most important putative targets[20]. This later has been shown to play an important role in cancer metastases and epithelial-mesenchymal transition (EMT)[21]. rs11614913 (C>T) affects the mature form of mi-RNA196a2 and potentially impact the protein-coding region of HOXC6[5]. Studies have suggested an increased risk of CRC in Asian populations (increased risk between 20%-30%)[22,23], and GC in both Asians and Caucasians[14]. A third SNP affecting the seed region is the mi-RNA-499a rs3746444 (A>G) which could impact the protein-Coding MYH7B. This SNP has been shown to increase the risk of esophageal cancer in Iranian and Chinese populations[5,24].
rs2292832 (T>C) affects the premature form of the pro-apoptotic miRNA-149 which is known to inhibit the expression of Akt1 and E2F1[25]. This SNP has been associated with CRC mainly in Asian populations with an increased risk of 20%[26].
MiRNA-machinery-gene polymorphisms could also impact the biosynthesis and activity of miRNAs. Several SNPs have been studied affecting DROSHA, DICER, DGCR8, XPO5 as well as several other regulators with conflicting results. As an example, XPO5 rs11077 (A→C) affects the activity of miR-617 and miR-4763–5p, and has been associated with CRC in Asian patients with hypertension[27] as well as esophageal cancer in Caucasian patients[28]. On the other hand, a meta-analysis including 10 case-control studies has analyzed the association between DROSHA/DGCR8 SNPs and cancer risk. Only HCC cases had been analyzed among GI cancers, and had not been associated with any of the studied SNPs[6].
SNPs affecting miRNA targets have also been suggested to affect GI cancer risk especially those located in the binding sites of miRNAs. rs12947947 (G>A) and rs28363292 (T>G) located in the 3′UTR of RAD51D have been jointly associated with an increased risk of HCC (nearly 1.5 times) mainly in Zhuang people (nearly 3 times) compared to controls[29]. Other target sites have also been studied including INSR T>C (rs1051690)[30] and CD133 rs2240688 A>C in GC[31].
The increased risk of GI cancers owing to SNPs could be potentiated by the interaction with environmental factors affecting miRNAs. As an example, it has been shown that long-term colonization of H. pylori might affect the activity of different miRNAs in gastric mucosae through epigenetic modifications[1,32,33]. miRNAs that are down-regulated in response to H. pylori include several components of the Let 7 family (which control cell cycle progression, proliferation, and invasion via RAB40C and HMGA2), miRNA-101 (proliferation, apoptosis, invasion and migration via COX-2, FOS, MCL1, EZH2), miRNA-141 (cell proliferation via FGFR2), miRNA-203 (proliferation, invasion via ABL1), miRNA-218 (proliferation, apoptosis, invasion and metastasis via ECOP and ROBO1), miRNA-375 (proliferation, apoptosis via PDK1 and JAK2), miRNA-449 (cell cycle progression and proliferation via GMNN, CCNE2, MET and SIRT1). On the other hand, up-regulated miRNAs include miRNA-17 (cell cycle progression via p21), miRNA-20a (cell cycle progression via p21), miRNA-21 (proliferation, apoptosis and invasion via PDCD4, RECK and PTEN), miRNA-146a (proliferation, apoptosis and immune response via IRAK1, TRAF6 and SMAD4), miRNA-155 (apoptosis and immune response via IKK-ε, SMAD2, FADD and PKI alpha) and miRNA-223 (invasion and metastasis via EPB41L3)[34,35].
Furthermore, accumulating evidence has supported the hypothesis of an interaction between miRNA-machinery and microsatellite instability (MSI) in the pathogenesis of GI cancers. As an example, miRNA-155 has been shown to downregulate the expression of MLH1, MSH2, and MSH6, whereas XPO5 has been found to be mutated in MSI + GI cancers[36].
In conclusion, SNPs affecting miRNA “pathway” genes (including regulators and binding sites) could have an impact on GI cancer susceptibility. However, as shown above, most meta-analyses have included only case-control studies which were mainly realized in Asian populations. To our knowledge, no prospective study has been published yet. Furthermore, even in studies where the risk of GI cancers has been found to be increased by a specific SNP, the magnitude of association was low (OR between 1.1 and 1.3) and varies according to different factors like ethnicity.
In our opinion, the interaction between miRNA/regulators/binding site SNPs and environmental factors as well as genomic susceptibility, like MSI, is an interesting field that should be exploited in future studies.
Manuscript source: Invited Manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: France
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chen XZ, Xu LB, Zhu X S-Editor: Wang YQ L-Editor: A E-Editor: Liu MY
| 1. | Saito Y, Suzuki H, Hibi T. The role of microRNAs in gastrointestinal cancers. J Gastroenterol. 2009;44 Suppl 19:18-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 2. | Babaei K, Shams S, Keymoradzadeh A, Vahidi S, Hamami P, Khaksar R, Norollahi SE, Samadani AA. An insight of microRNAs performance in carcinogenesis and tumorigenesis; an overview of cancer therapy. Life Sci. 2020;240:117077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 3. | Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu Rev Genet. 2015;49:213-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 398] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 4. | Sempere LF. Celebrating 25 Years of MicroRNA Research: From Discovery to Clinical Application. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Pipan V, Zorc M, Kunej T. MicroRNA Polymorphisms in Cancer: A Literature Analysis. Cancers (Basel). 2015;7:1806-1814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 6. | Wen J, Lv Z, Ding H, Fang X, Sun M. Association of miRNA biosynthesis genes DROSHA and DGCR8 polymorphisms with cancer susceptibility: a systematic review and meta-analysis. Biosci Rep. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | Farrell RE. miRNA. In: Farrell RE. RNA Methodologies. 5th ed. Amsterdam: Academic Press, 2017: 329-343. |
| 8. | Belter A, Gudanis D, Rolle K, Piwecka M, Gdaniec Z, Naskręt-Barciszewska MZ, Barciszewski J. Mature miRNAs form secondary structure, which suggests their function beyond RISC. PLoS One. 2014;9:e113848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 561] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 10. | Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472-15477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 699] [Cited by in RCA: 756] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 11. | Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2191] [Cited by in RCA: 2184] [Article Influence: 121.3] [Reference Citation Analysis (0)] |
| 12. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257-2261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4471] [Cited by in RCA: 4520] [Article Influence: 237.9] [Reference Citation Analysis (0)] |
| 13. | Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 678] [Cited by in RCA: 693] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 14. | Xu Q, Liu JW, Yuan Y. Comprehensive assessment of the association between miRNA polymorphisms and gastric cancer risk. Mutat Res Rev Mutat Res. 2015;763:148-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 425] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 16. | Iacona JR, Lutz CS. miR-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip Rev RNA. 2019;10:e1533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 122] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 17. | Xu X, Zhang Y, Lei Q, Wang Y, Xing C, Yang X, Zhang S, Cao J. MicroRNA-146a polymorphism and susceptibility to gastrointestinal cancer: a meta-analysis. Zhonghua Wei Chang Wai Ke Za Zhi. 2015;18:718-722. [PubMed] |
| 18. | Xie M, Li Y, Wu J, Wu J. A risk of digestive tract neoplasms susceptibility in miR-146a and miR-196a2. Fam Cancer. 2015;14:229-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 19. | Peng Q, Li S, Lao X, Chen Z, Li R, Deng Y, Qin X. The association of common functional polymorphisms in mir-146a and mir-196a2 and hepatocellular carcinoma risk: evidence from a meta-analysis. Medicine (Baltimore). 2014;93:e252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | Chen C, Zhang Y, Zhang L, Weakley SM, Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2011;15:14-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 176] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 21. | Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y, D'Armiento J, Chada K. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013;73:4289-4299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 214] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 22. | Wu Y, Hao X, Feng Z, Liu Y. Genetic polymorphisms in miRNAs and susceptibility to colorectal cancer. Cell Biochem Biophys. 2015;71:271-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Xu L, Tang W. Associations of Polymorphisms in mir-196a2, mir-146a and mir-149 with Colorectal Cancer Risk: A Meta-Analysis. Pathol Oncol Res. 2016;22:261-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Chen C, Yang S, Chaugai S, Wang Y, Wang DW. Meta-analysis of Hsa-mir-499 polymorphism (rs3746444) for cancer risk: evidence from 31 case-control studies. BMC Med Genet. 2014;15:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Alidoust M, Hamzehzadeh L, Rivandi M, Pasdar A. Polymorphisms in non-coding RNAs and risk of colorectal cancer: A systematic review and meta-analysis. Crit Rev Oncol Hematol. 2018;132:100-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | Du W, Ma XL, Zhao C, Liu T, Du YL, Kong WQ, Wei BL, Yu JY, Li YY, Huang JW, Li ZK, Liu L. Associations of single nucleotide polymorphisms in miR-146a, miR-196a, miR-149 and miR-499 with colorectal cancer susceptibility. Asian Pac J Cancer Prev. 2014;15:1047-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 27. | Cho SH, Ko JJ, Kim JO, Jeon YJ, Yoo JK, Oh J, Oh D, Kim JW, Kim NK. 3'-UTR Polymorphisms in the MiRNA Machinery Genes DROSHA, DICER1, RAN, and XPO5 Are Associated with Colorectal Cancer Risk in a Korean Population. PLoS One. 2015;10:e0131125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, Wu X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila). 2008;1:460-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 181] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 29. | Jiang YJ, Zhong JH, Zhou ZH, Qiu MQ, Zhou XG, Liu YC, Huo RR, Liang XM, Chen Z, Lin QL, Yu XY, Yu HP. Association between polymorphisms in MicroRNA target sites of RAD51D genes and risk of hepatocellular carcinoma. Cancer Med. 2019;8:2545-2552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Petkevicius V, Salteniene V, Juzenas S, Wex T, Link A, Leja M, Steponaitiene R, Skieceviciene J, Kupcinskas L, Jonaitis L, Kiudelis G, Malfertheiner P, Kupcinskas J. Polymorphisms of microRNA target genes IL12B, INSR, CCND1 and IL10 in gastric cancer. World J Gastroenterol. 2017;23:3480-3487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 18] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 31. | Wang Q, Liu H, Xiong H, Liu Z, Wang LE, Qian J, Muddasani R, Lu V, Tan D, Ajani JA, Wei Q. Polymorphisms at the microRNA binding-site of the stem cell marker gene CD133 modify susceptibility to and survival of gastric cancer. Mol Carcinog. 2015;54:449-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, Tamura G, Saito D, Sugimura T, Ichinose M, Ushijima T. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 474] [Cited by in RCA: 479] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 33. | Ushijima T, Nakajima T, Maekita T. DNA methylation as a marker for the past and future. J Gastroenterol. 2006;41:401-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 34. | Valenzuela MA, Canales J, Corvalán AH, Quest AF. Helicobacter pylori-induced inflammation and epigenetic changes during gastric carcinogenesis. World J Gastroenterol. 2015;21:12742-12756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 72] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 35. | Noto JM, Peek RM. The role of microRNAs in Helicobacter pylori pathogenesis and gastric carcinogenesis. Front Cell Infect Microbiol. 2011;1:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | Yamamoto H, Adachi Y, Taniguchi H, Kunimoto H, Nosho K, Suzuki H, Shinomura Y. Interrelationship between microsatellite instability and microRNA in gastrointestinal cancer. World J Gastroenterol. 2012;18:2745-2755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 61] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |