Published online Oct 6, 2020. doi: 10.12998/wjcc.v8.i19.4360
Peer-review started: April 19, 2020
First decision: July 25, 2020
Revised: August 8, 2020
Accepted: September 2, 2020
Article in press: September 2, 2020
Published online: October 6, 2020
Processing time: 161 Days and 9.6 Hours
The global outbreak of human severe acute respiratory syndrome coronavirus (SARS-CoV)-2 infection represents an urgent need for readily available, accurate and rapid diagnostic tests. Nucleic acid testing of respiratory tract specimens for SARS-CoV-2 is the current gold standard for diagnosis of coronavirus disease 2019 (COVID-19). However, the diagnostic accuracy of reverse transcription polymerase chain reaction (RT-PCR) tests for detecting SARS-CoV-2 nucleic acid may be lower than optimal. The detection of SARS-CoV-2-specific antibodies should be used as a serological non-invasive tool for the diagnosis and management of SARS-CoV-2 infection.
To investigate the diagnostic value of SARS-CoV-2 IgM/IgG and nucleic acid detection in COVID-19.
We retrospectively analyzed 652 suspected COVID-19 patients, and 206 non-COVID-19 patients in Wuhan Integrated TCM and Western Medicine Hospital. Data on SARS-CoV-2 nucleic acid tests and serum antibody tests were collected to investigate the diagnostic value of nucleic acid RT-PCR test kits and immunoglobulin (Ig)M/IgG antibody test kits. The χ2 test was used to compare differences between categorical variables. A 95% confidence interval (CI) was provided by the Wilson score method. All analyses were performed with IBM SPSS Statistics version 22.0 (IBM Corp., Armonk, NY, United States).
Of the 652 suspected COVID-19 patients, 237 (36.3%) had positive nucleic acid tests, 311 (47.7%) were positive for IgM, and 592 (90.8%) were positive for IgG. There was a significant difference in the positive detection rate between the IgM and IgG test groups (P < 0.001). Using the RT-PCR results as a reference, the specificity, sensitivity, and accuracy of IgM/IgG combined tests for SARS-CoV-2 infection were 98.5%, 95.8%, and 97.1%, respectively. Of the 415 suspected COVID-19 patients with negative nucleic acid test results, 366 had positive IgM/IgG tests with a positive detection rate of 88.2%.
Our data indicate that serological IgM/IgG antibody combined test had high sensitivity and specificity for the diagnosis of SARS-CoV-2 infection, and can be used in combination with RT-PCR for the diagnosis of SARS-CoV-2 infection.
Core Tip: We retrospectively analyzed 652 suspected coronavirus disease 2019 (COVID-19) patients, and 206 non-COVID-19 patients to investigate the diagnostic value of severe acute respiratory syndrome coronavirus (SARS-CoV)-2 IgM/IgG and nucleic acid detection. We found that 237/652 (36.3%) suspected COVID-19 patients had positive nucleic acid tests, 311 (47.7%) were positive for IgM, and 592 (90.8%) were positive for IgG. Using reverse transcription polymerase chain reaction (RT-PCR) results as a reference, the specificity, sensitivity, and accuracy of IgM/IgG combined tests for SARS-CoV-2 infection were 98.5%, 95.8%, and 97.1%, respectively. Our data indicate that the serological IgM/IgG combined test can be used in combination with RT-PCR for the diagnosis of SARS-CoV-2 infection.
- Citation: Meng QB, Peng JJ, Wei X, Yang JY, Li PC, Qu ZW, Xiong YF, Wu GJ, Hu ZM, Yu JC, Su W. Clinical application of combined detection of SARS-CoV-2-specific antibody and nucleic acid. World J Clin Cases 2020; 8(19): 4360-4369
- URL: https://www.wjgnet.com/2307-8960/full/v8/i19/4360.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i19.4360
Coronavirus disease 2019 (COVID-19) was first reported in Wuhan, Hubei Province, China and has spread worldwide[1,2]. COVID-19 is a highly transmissible disease caused by severe acute respiratory syndrome coronavirus (SARS-CoV)-2, which was also named 2019 novel coronavirus (2019-nCoV)[2,3]. As of April 19, 2020, 2160207 confirmed COVID-19 cases were reported worldwide, causing 146088 deaths[4]. The World Health Organization (WHO) declared COVID-19 a global pandemic on March 11, 2020[5,6]. The global outbreak of human SARS-CoV-2 infection has highlighted the necessity for readily available, accurate and rapid diagnostic tests.
Nucleic acid testing or genetic sequencing of respiratory tract specimens for SARS-CoV-2 is the current gold standard for the diagnosis of COVID-19[7-9]. However, according to recent evidence, the diagnostic accuracy of reverse transcription polymerase chain reaction (RT-PCR) tests for detecting SARS-CoV-2 nucleic acid may be lower than optimal. Liu et al[10] analyzed the RT-PCR results of throat swab samples from 4880 cases of suspected SARS-CoV-2 infection, and found that only 38.42% were positive. Another important concern is the number of false-negative RT-PCR results for COVID-19[11]. RT-PCR has some other limitations, including potential biological safety hazards due to handling of patient samples and long waiting time for results.
Given the limitations of the currently used nucleic acid detection for diagnosis of COVID-19, clinical laboratories should apply sensitive and accurate assays such as immunological detection kits that target viral antigens or antibodies for diagnosing SARS-CoV-2 infection as quickly as possible[12]. Therefore, SARS-CoV-2 serum IgM and IgG antibody positivity was added to the diagnostic criteria in the Novel Coronavirus Pneumonia Diagnosis and Treatment Guidelines (Trial Version 7)[13]. Dong et al[14] reported a COVID-19 case without detectable virus in oropharyngeal specimens and suggested testing for serum IgM and IgG antibodies to SARS-CoV-2 as an alternative for diagnosis. Li et al[15] demonstrated a rapid and simple point-of-care lateral flow immunoassay that can detect SARS-CoV-2 IgM and IgG antibodies in the blood of patients at different stages of infection. The overall testing sensitivity was 88.66% and specificity was 90.63%. However, there is limited clinical information on the SARS-CoV-2 antibody test (colloidal gold).
In the present study, we collected clinical data from 652 suspected COVID-19 patients and 206 non-COVID-19 patients to investigate the diagnostic value of SARS-CoV-2 IgM/IgG antibody test kits with colloidal gold immunoassays and nucleic acid RT-PCR test kits.
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Wuhan Integrated TCM and Western Medicine Hospital, Huazhong University of Science and Technology (No. [2020]8). The requirement for written informed consent was waived given the context of emerging infectious diseases.
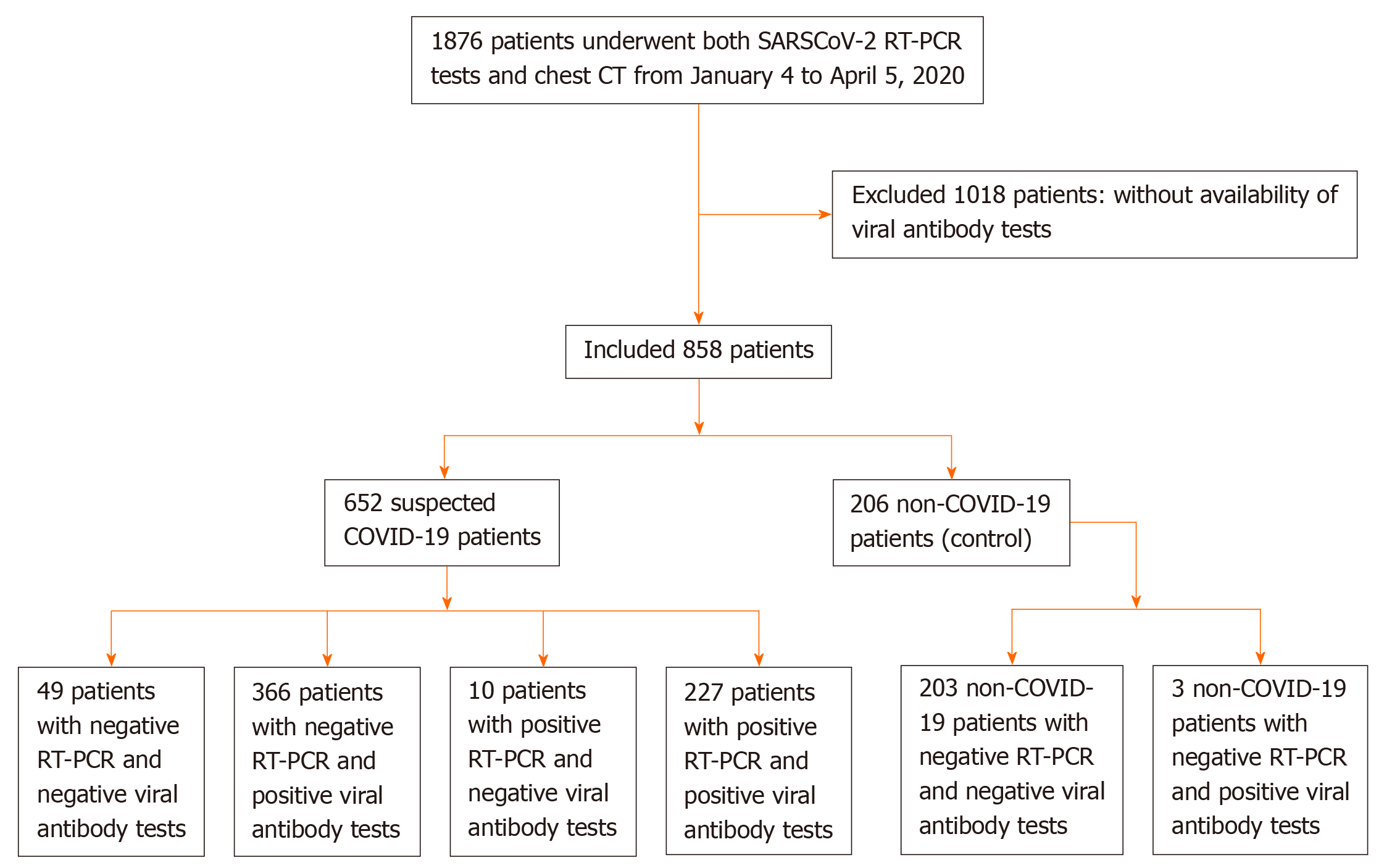
From January 4 to April 5, 2020, data on 1876 consecutive patients who underwent SARS-CoV-2 nucleic acid tests and chest computed tomography were retrospectively collected in Wuhan Integrated TCM and Western Medicine Hospital. A total of 1018 patients were excluded because they did not have SARS-CoV-2 IgM/IgG antibody tests. We included 652 suspected COVID-19 patients and 206 non-COVID-19 patients (Figure 1). RT-PCR, SARS-CoV-2 IgM/IgG antibody tests, and pulmonary imaging features were extracted from patients’ electronic medical records in our hospital information system. The patients were clinically diagnosed with COVID-19 according to the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7)[13]. Suspected COVID-19 patients met the following criteria: Clear history of epidemiological contact, typical clinical symptoms and pulmonary imaging features.
The SARS-CoV-2 Antibody Test Kit (catalog No. 20203400177, colloidal gold) was obtained from Innovita (Tangshan) Biotechnology Co., Ltd. (Tangshan, China), with the recombinant SARS-CoV-2 antigen coated on the surface of the colloid gold particles. The SARS-CoV-2 nucleic acid detection kit (catalog No. 20203400057, fluorescent PCR) was purchased from Shanghai Zhijiang Biotechnology Co., Ltd. (Shanghai, China)[16].
SARS-CoV-2 antibody test: Peripheral blood (4 mL) was collected from each patient with a yellow top collection tube and sent to the laboratory for serum SARS-CoV-2 IgM/IgG antibody test. Detection of serum IgM/IgG antibody was performed using the SARS-CoV-2 Antibody Test Kit (colloidal gold). The presence of two purple bands (M and C) indicated the presence of SARS-CoV-2 IgM antibodies. The presence of two purple bands (G and C) indicated the presence of SARS-CoV-2 IgG antibodies. For negative results, only one purple band appeared at the control line (C). If the control line (C) failed to appear, regardless of whether the G/M line was visible, the test was invalid.
Fluorescent PCR for SARS-CoV-2 nucleic acid detection: Pharyngeal swabs were used to collect secretions from the lateral and posterior pharyngeal walls and placed in sterile tubes (containing 1 mL sterile normal saline). Fluorescence PCR was performed using the SARS-CoV-2 Nucleic Acid Detection Kit. The results were divided into positive and negative according to the manufacturer’s protocol.
The statistical methods used in this study were reviewed by Guang-Jiang Wu from Beijing Shijitan Hospital, Capital Medical University. Categorical variables were displayed as counts and percentages. Continuous variables were presented as median (interquartile range; IQR). The χ2 test was used to compare differences between categorical variables. The specificity, sensitivity, positive predictive value (PPV) and negative predictive value (NPV) of the SARS-CoV-2 Antibody Test Kit (colloidal gold) were calculated according to the following formulas. Specificity (%) = 100 × [true negative/(true negative + false positive)]; Sensitivity (%) = 100 × [true positive/(true positive + false negative); PPV (%) = 100 × [true positive/(true positive + false positive); NPV (%) = 100 × [true negative/(true negative + false negative)]; and Accuracy (%) = 100 × (true positive + true negative)/( true positive + false positive + true negative + false negative). A 95% confidence interval (CI) was provided by the Wilson score method. All P values were two-sided, and P < 0.05 was considered statistically significant. All analyses were performed with IBM SPSS Statistics version 22.0 (IBM Corp., Armonk, NY, United States).
Of the 652 suspected COVID-19 patients, 237 had positive and 415 had negative SARS-CoV-2 nucleic acid tests with a positive detection rate of 36.3% (95%CI: 32.6%–40.1%); therefore, 237 patients were confirmed to have COVID-19 by the SARS-CoV-2 nucleic acid RT-PCR test.
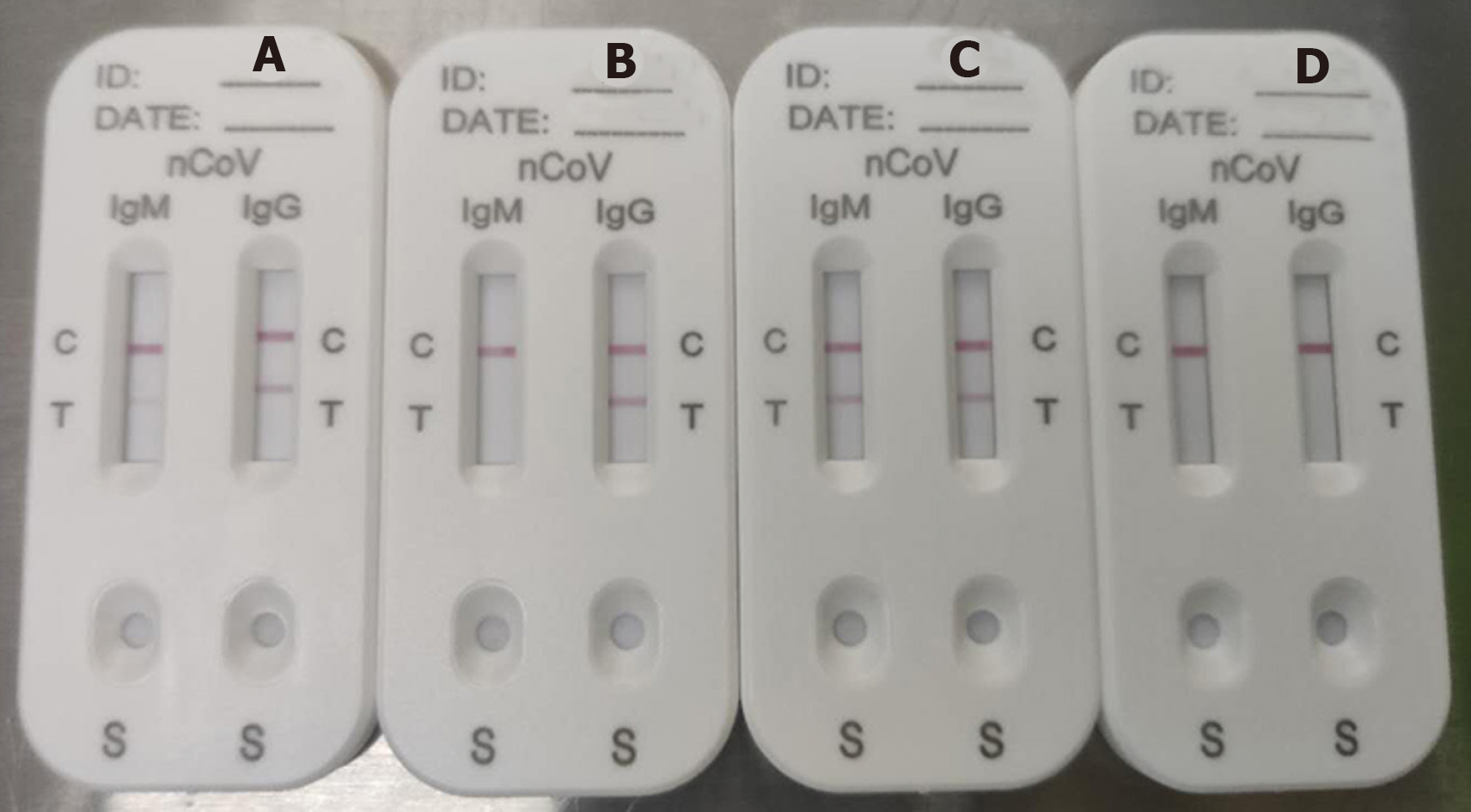
The median time from illness onset to IgM/IgG antibody tests was 34 d (IQR 28–39 d). A representative photograph of SARS-CoV-2 IgM and IgG antibody tests is shown in Figure 2. Figure 2A shows detection of IgM in low concentration (Score 1) and IgG in high concentration (Score 2); Figure 2B shows IgG only in high concentration (Score 2); Figure 2C shows both IgM and IgG in high concentration (Score 2); and Figure 2D shows no IgM and IgG (Score 0). Score ≥ 1 was defined as positive.
Of 206 non-COVID-19 patients, one was positive for IgM antibody against SARS-CoV-2, and two were positive for IgG antibody against SARS-CoV-2. Of the 652 suspected COVID-19 patients, 311 were positive for SARS-CoV-2-specific IgM antibody with a positive detection rate of 47.7% (95%CI: 43.9%–51.5%); 592 patients were positive for SARS-CoV-2-specific IgG antibody with a positive detection rate of 90.8% (95CI: 88.6%–93.0%); and 593 patients were positive for SARS-CoV-2-specific IgM and/or IgG antibody combined tests with a positive detection rate of 91.0% (95%CI: 88.7%–93.2%). There was a significant difference regarding the positive detection rate between the IgM and IgG test groups (P < 0.001) (Table 1).
Of the 237 patients who were positive for SARS-CoV-2 nucleic acid tests, 109 were positive for IgM, 227 patients were positive for IgG, and 227 patients were positive for IgM and/or IgG. Using the RT-PCR results as a reference, the specificity, sensitivity, and accuracy of SARS-CoV-2-specific IgM, IgG and IgM/IgG combined tests for detecting SARS-CoV-2 infection are shown in Table 2.
| Results of IgM/IgG test (n) | IgM/IgG antibody test performance (%) | ||||||||
| TP | TN | FP | FN | Specificity (95%CI) | Sensitivity (95%CI) | PPV (95%CI) | NPV (95%CI) | Accuracy (95%CI) | |
| IgM | 109 | 205 | 1 | 128 | 99.5 (98.6-100.0) | 46.0 (39.6-52.4) | 99.1 (97.3-100.0) | 61.6 (56.3-66.8) | 70.9 (66.6-75.1) |
| IgG | 227 | 204 | 2 | 10 | 99.0 (97.7-100.0) | 95.8 (93.2-98.4) | 99.1 (97.9-100.0) | 95.3 (92.5-98.2) | 97.3 (95.8-98.8) |
| M/G | 227 | 203 | 3 | 10 | 98.5 (96.9-100.0) | 95.8 (93.2-98.4) | 98.7 (97.2-100.0) | 95.3 (92.4-98.2) | 97.1 (95.5-98.6) |
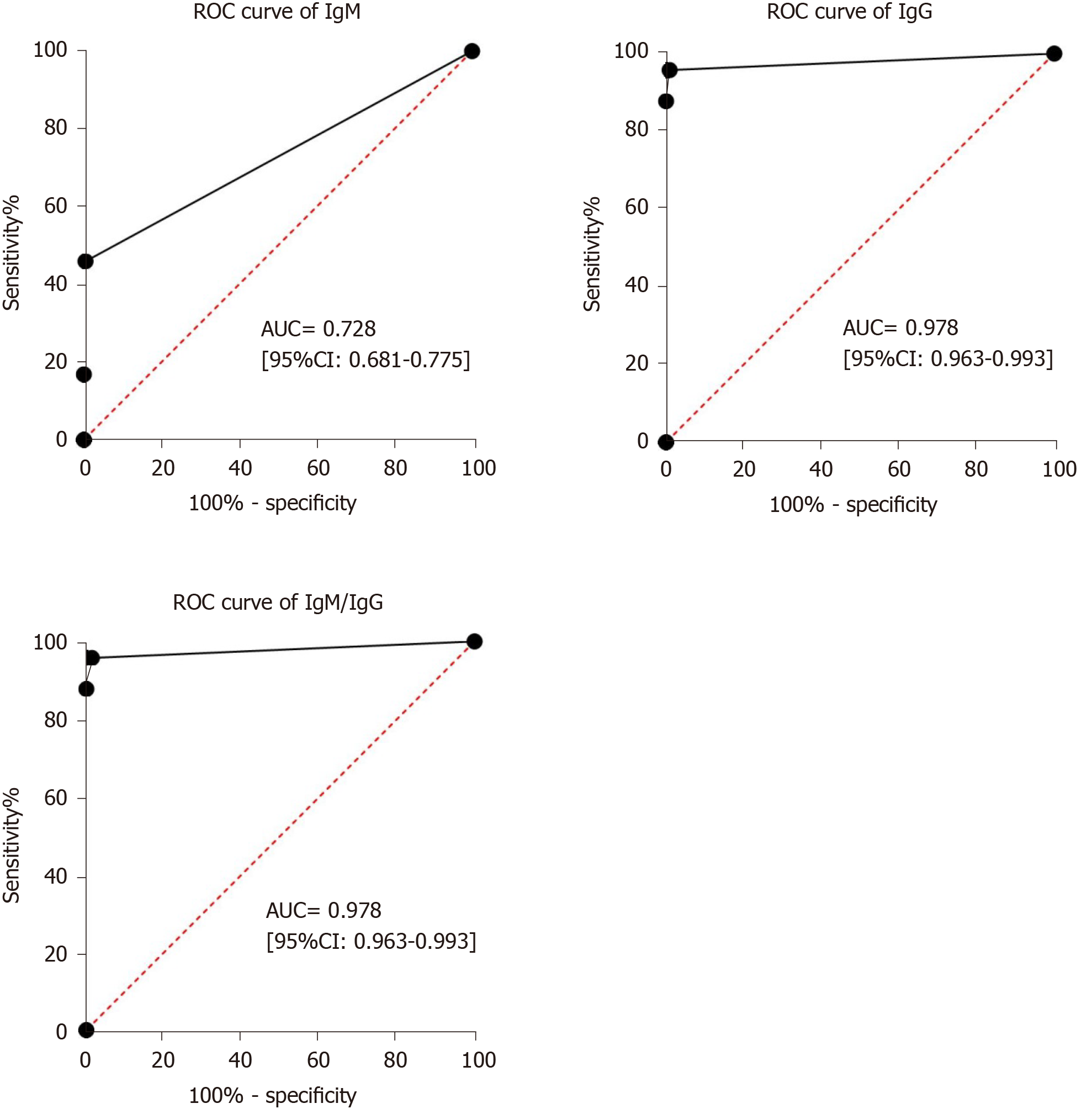
Receiver operating characteristic curve analysis showed that the area under the curve of IgM tests, IgG tests and IgM/IgG combined tests for diagnosing COVID-19 were 0.728 (95%CI: 0.681–0.775), 0.978 (95%CI: 0.963–0.993) and 0.978 (95%CI: 0.963–0.993) (Figure 3).
Of the 415 suspected COVID-19 patients who were negative for SARS-CoV-2 nucleic acid tests, 366 patients were positive for SARS-CoV-2-specific IgM and/or IgG antibody tests with a positive detection rate of 88.2% (95%CI: 85.1–91.3%).
In the current retrospective study, we included 652 suspected COVID-19 patients with a clear history of epidemiological contact, typical clinical symptoms and pulmonary imaging features, to investigate the positive detection rate of nucleic acid and antibody tests. The results showed that of 652 patients, 237 (36.3%) were confirmed to have COVID-19 by the SARS-CoV-2 nucleic acid test, 311 (47.7%) were positive for SARS-CoV-2-specific IgM antibodies, 592 (90.8%) were positive for SARS-CoV-2-specific IgG antibodies, and 593 (91.0%) were positive for SARS-CoV-2 specific IgM and/or IgG antibodies. We included 237 confirmed COVID-19 patients with positive SARS-CoV-2 nucleic acid tests and 206 confirmed non-COVID-19 patients to evaluate the performance of the SARS-CoV-2-specific IgM and IgG antibody test kit.
Liu et al[10] reported that the positive rate of RT-PCR detection of SARS-CoV-2 infection was 38.4% (1875/4880) and an increased positive percentage was found in male and older patients. Ai et al[17] reported that 59% (601/1014) of patients had positive SARS-CoV-2 RT-PCR results. In the current study, the positive percentage of SARS-CoV-2 RT-PCR tests was lower than that of previous studies. According to the Novel Coronavirus Pneumonia Diagnosis and Treatment Guidelines, SARS-CoV-2 nucleic acid test by RT-PCR assay on respiratory tract specimens plays an important role in the etiological diagnosis of SARS-CoV-2 infection and discharge evaluation[13]. However, the accuracy of the laboratory diagnosis of COVID-19 using RT-PCR is affected by many potential factors, including preanalytical problems such as improper collection, storage and transport of swabs; sample contamination and testing patients receiving antiretroviral therapy; as well as analytical problems such as active viral recombination, use of inadequately validated assays, instrument malfunctioning, along with other specific technical issues[9]. Therefore, it is necessary to develop a more sensitive, accurate and simple detection method for the diagnosis of SARS-CoV-2 infection.
Dong et al[14] reported that COVID-19 was confirmed with positive IgM and positive IgG antibodies tests against SARS-CoV-2. As recently reported, a rapid IgM/IgG combined antibody test was used for the diagnosis of SARS-CoV-2 infection, showing 88.66% sensitivity and 90.63% specificity[15]. In the current study, we found that the specificity, sensitivity, and accuracy of IgM and/or IgG antibody combined detection for the diagnosis of SARS-CoV-2 infection were 98.5% (203/206), 95.8% (227/237), and 97.1% (430/443), respectively. Of the 415 suspected COVID-19 patients who were negative for SARS-CoV-2 nucleic acid tests, 88.2% (366) of patients were positive for SARS-CoV-2 specific IgM and/or IgG antibody tests. Therefore, 366 patients were considered to have COVID-19 with SARS-CoV-2 IgM and/or IgG antibody tests. All the results confirmed that IgM and/or IgG antibody tests can be used as an effective method for serological diagnosis of SARS-CoV-2 infection.
According to the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7), double positive results of IgM and IgG antibody tests can be used as serological evidence for the diagnosis of SARS-CoV-2 infection[13]. However, the fact that IgM testing may be negative in convalescent patients with COVID-19 is not surprising considering the probable kinetics of SARS-CoV-2-specific IgM antibody[18-20]. Therefore, IgM and/or IgG positivity should be used as serological evidence for the diagnosis of SARS-CoV-2 infection.
There were several notable limitations in the present study. Firstly, the main weaknesses were its single center retrospective nature and small sample size. Secondly, due to the retrospective nature of the study, most patients did not complete the dynamic monitoring of SARS-CoV-2-specific IgM and IgG by the end of the study. Thirdly, the median time from symptom onset to the IgM and IgG test was long due to late availability of the SARS-CoV-2-specific IgM and IgG test kits.
In summary, this retrospective study indicated that serum specific IgM and IgG antibody combined test has high sensitivity, specificity and accuracy for the diagnosis of SARS-CoV-2 infection. Our data indicate that the antibody-based test can be used as a detection tool in combination with RT-PCR in the diagnosis of SARS-CoV-2 infection in epidemic areas.
Coronavirus disease 2019 (COVID-19) is a highly transmissible disease caused by severe acute respiratory syndrome coronavirus (SARS-CoV)-2. The global outbreak of human SARS-CoV-2 infection has highlighted the necessity for readily available, accurate and rapid diagnostic tests. SARS-CoV-2 serum IgM and IgG antibody positivity was added to the diagnostic criteria in the Novel Coronavirus Pneumonia Diagnosis and Treatment Guidelines (Trial Version 7). However, there is limited clinical information on the SARS-CoV-2 antibody test (colloidal gold).
According to recent evidence, the diagnostic accuracy of reverse transcription polymerase chain reaction (RT-PCR) tests for detecting SARS-CoV-2 nucleic acid may be lower than optimal. Given the limitations of RT-PCR tests for the diagnosis of COVID-19, clinical laboratories should apply sensitive and accurate assays such as immunological detection kits that target viral antigens or antibodies for diagnosing SARS-CoV-2 infection as quickly as possible. We are very interested in this issue and hope that we can present a new antibody test adopted in our hospital.
The objectives were to report the diagnostic value of SARS-CoV-2 IgM/IgG and nucleic acid detection in COVID-19.
We retrospectively analyzed data on 652 suspected COVID-19 patients, and 206 non-COVID-19 patients in Wuhan Integrated TCM and Western Medicine Hospital. RT-PCR, SARS-CoV-2 IgM/IgG antibody tests, and pulmonary imaging features were extracted from patients’ electronic medical records in our hospital information system. The specificity, sensitivity, PPV and NPV of the SARS-CoV-2 Antibody Test Kit were calculated. A 95% confidence interval (CI) was provided by the Wilson score method. All analyses were performed with IBM SPSS Statistics version 22.0 (IBM Corp., Armonk, NY, United States), and two-tailed P values less than 0.05 were considered to be statistically significant.
Of the 652 suspected COVID-19 patients, 237 (36.3%) patients were confirmed to have COVID-19 by the SARS-CoV-2 nucleic acid RT-PCR test. Using RT-PCR results as a reference, the specificity, sensitivity, and accuracy of the SARS-CoV-2-specific IgM/IgG combined tests for detecting SARS-CoV-2 infection were 98.5%, 95.8%, and 97.1%, respectively. Of the 415 suspected COVID-19 patients who were negative for the SARS-CoV-2 nucleic acid tests, 366 patients were positive for the SARS-CoV-2-specific IgM and/or IgG antibody tests with a positive detection rate of 88.2%.
Our data indicate that the serological IgM/IgG antibody combined test had high specificity, sensitivity, and accuracy for the diagnosis of SARS-CoV-2 infection, and can be used in combination with RT-PCR for the diagnosis of SARS-CoV-2 infection.
For COVID-19 patients, it is worth further completing the dynamic monitoring of SARS-CoV-2-specific IgM and IgG.
The authors thank Professor Ling-Qian Chang (Beijing Advanced Innovation Center for Biomedical Engineering, School of Biological Science and Medical Engineering, Beihang University), Associate Professor Feng Chen (College of Materials Science and Engineering, Zhejiang University of Technology) and Jie Qiao (Hubei College of Traditional Chinese Medicine, Wuhan, Hubei Province, China) for their guidance in study design and interpretation of results, and review of the manuscript.
Manuscript source: Invited manuscript
Specialty type: Infectious diseases
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sukocheva O S-Editor: Ma YJ L-Editor: Webster JR P-Editor: Li JH
| 1. | Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14113] [Cited by in RCA: 14751] [Article Influence: 2950.2] [Reference Citation Analysis (0)] |
| 2. | Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18987] [Cited by in RCA: 17613] [Article Influence: 3522.6] [Reference Citation Analysis (0)] |
| 3. | Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536-544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5202] [Cited by in RCA: 4624] [Article Influence: 924.8] [Reference Citation Analysis (0)] |
| 4. | World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report-89. 2020. [18 April 2020]. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200418-sitrep-89-covid-19.pdf?sfvrsn=3643dd38_2. |
| 5. | Cucinotta D, Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91:157-160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2616] [Reference Citation Analysis (0)] |
| 6. | Mahase E. Covid-19: WHO declares pandemic because of "alarming levels" of spread, severity, and inaction. BMJ. 2020;368:m1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 387] [Article Influence: 77.4] [Reference Citation Analysis (0)] |
| 7. | Pang J, Wang MX, Ang IYH, Tan SHX, Lewis RF, Chen JI, Gutierrez RA, Gwee SXW, Chua PEY, Yang Q, Ng XY, Yap RK, Tan HY, Teo YY, Tan CC, Cook AR, Yap JC, Hsu LY. Potential Rapid Diagnostics, Vaccine and Therapeutics for 2019 Novel Coronavirus (2019-nCoV): A Systematic Review. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 337] [Cited by in RCA: 305] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 8. | Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, Fan YP, Fang C, Huang D, Huang LQ, Huang Q, Han Y, Hu B, Hu F, Li BH, Li YR, Liang K, Lin LK, Luo LS, Ma J, Ma LL, Peng ZY, Pan YB, Pan ZY, Ren XQ, Sun HM, Wang Y, Wang YY, Weng H, Wei CJ, Wu DF, Xia J, Xiong Y, Xu HB, Yao XM, Yuan YF, Ye TS, Zhang XC, Zhang YW, Zhang YG, Zhang HM, Zhao Y, Zhao MJ, Zi H, Zeng XT, Wang YY, Wang XH; for the Zhongnan Hospital of Wuhan University Novel Coronavirus Management and Research Team, Evidence-Based Medicine Chapter of China International Exchange and Promotive Association for Medical and Health Care (CPAM). A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil Med Res. 2020;7:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 881] [Cited by in RCA: 1136] [Article Influence: 227.2] [Reference Citation Analysis (0)] |
| 9. | Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med. 2020;58:1070-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 371] [Cited by in RCA: 411] [Article Influence: 82.2] [Reference Citation Analysis (0)] |
| 10. | Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, Feng Y, Zhu C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172-175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 404] [Cited by in RCA: 386] [Article Influence: 77.2] [Reference Citation Analysis (0)] |
| 11. | Li D, Wang D, Dong J, Wang N, Huang H, Xu H, Xia C. False-Negative Results of Real-Time Reverse-Transcriptase Polymerase Chain Reaction for Severe Acute Respiratory Syndrome Coronavirus 2: Role of Deep-Learning-Based CT Diagnosis and Insights from Two Cases. Korean J Radiol. 2020;21:505-508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 250] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 12. | Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1000] [Cited by in RCA: 935] [Article Influence: 187.0] [Reference Citation Analysis (0)] |
| 13. | National Health Commission of the people's Republic of China. Novel coronavirus pneumonia diagnosis and treatment guideline (trial version 7). 2020. Available from: http://www.nhc.gov.cn/xcs/zhengcwj/202003/202046c209294a202007dfe202004cef202080dc202007f205912eb201989.shtml. |
| 14. | Dong X, Cao YY, Lu XX, Zhang JJ, Du H, Yan YQ, Akdis CA, Gao YD. Eleven faces of coronavirus disease 2019. Allergy. 2020;75:1699-1709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 218] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 15. | Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 901] [Cited by in RCA: 1087] [Article Influence: 217.4] [Reference Citation Analysis (0)] |
| 16. | Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg Microbes Infect. 2020;9:747-756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 545] [Cited by in RCA: 500] [Article Influence: 100.0] [Reference Citation Analysis (1)] |
| 17. | Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020;296:E32-E40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3614] [Cited by in RCA: 3283] [Article Influence: 656.6] [Reference Citation Analysis (0)] |
| 18. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14097] [Article Influence: 2819.4] [Reference Citation Analysis (1)] |
| 19. | Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, Wang YY, Xiao GF, Yan B, Shi ZL, Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1192] [Cited by in RCA: 1219] [Article Influence: 243.8] [Reference Citation Analysis (0)] |
| 20. | Jin Y, Wang M, Zuo Z, Fan C, Ye F, Cai Z, Wang Y, Cui H, Pan K, Xu A. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 203] [Article Influence: 40.6] [Reference Citation Analysis (0)] |