Published online Sep 6, 2020. doi: 10.12998/wjcc.v8.i17.3859
Peer-review started: March 3, 2020
First decision: April 28, 2020
Revised: July 8, 2020
Accepted: July 18, 2020
Article in press: July 18, 2020
Published online: September 6, 2020
Processing time: 185 Days and 2.6 Hours
X-linked agammaglobulinemia is a primary immunodeficiency disease caused by gene mutations of Bruton’s tyrosine kinase (BTK). We found a new mutation point and summarized the correlation analysis and performed a literature review.
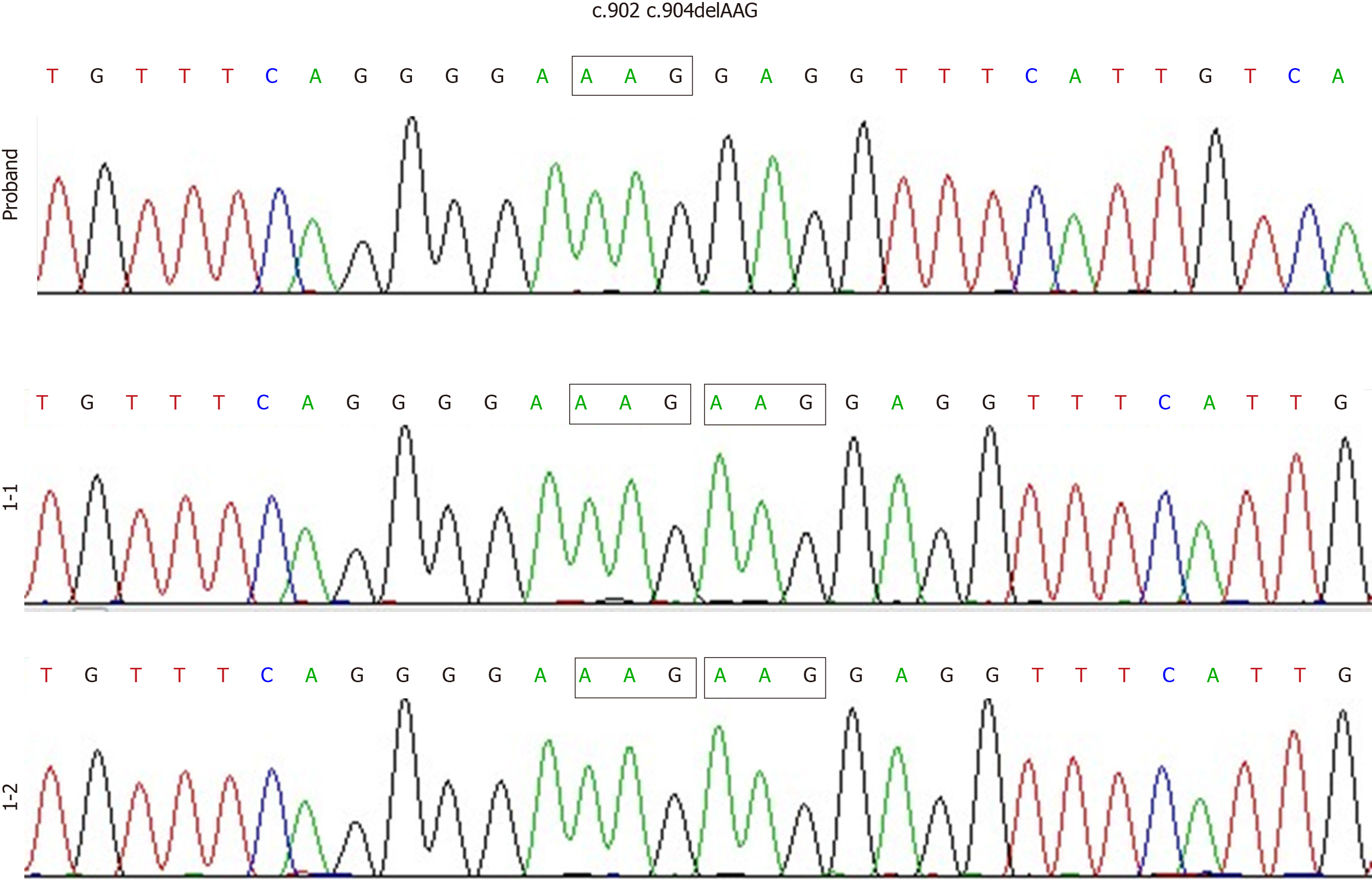
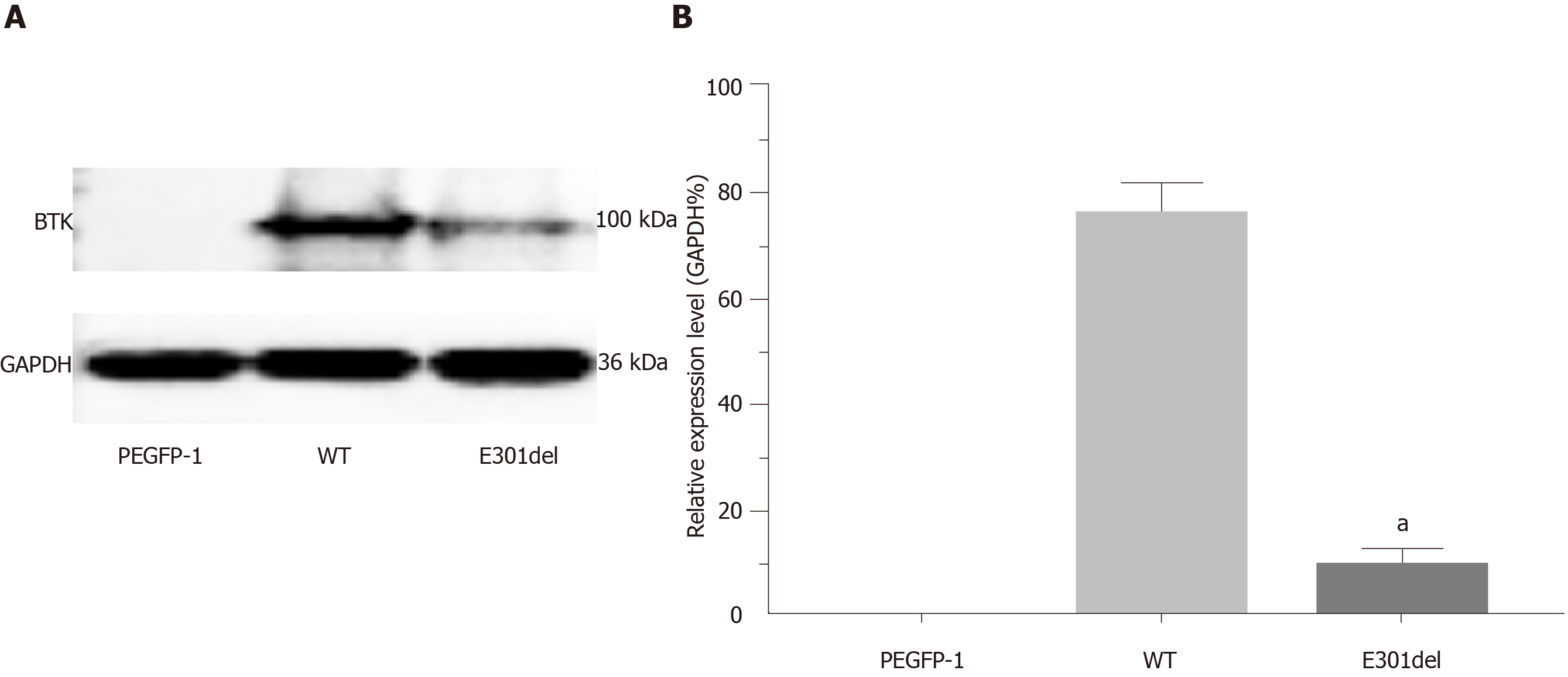
The proband was a 5-year-old boy. He was admitted to our hospital due to a recurrent cough and a fever that had persisted for a month. He had a history of multiple respiratory infections and sinusitis. There was no immunodeficiency or recurrent infection history among his family members. Agammaglobulinemia was characterized as follows: Immunoglobulin (Ig) A, 90.0 mg/dL (90-450 mg/dL); IgG, 20.0 mg/dL (800-1800 mg/dL); and IgM, 18.0 mg/dL (60-280 mg/dL). Notably, the assessment of IgG subtypes revealed the following very low levels: Subtype 1, 0.26 g/L (3.62-12.28 g/L); subtype 2, 0.10 g/L (0.57-2.9 g/L); subtype 3, 0.009 g/L (0.129-0.789 g/L); and subtype 4, 0.003 g/L (0.013-1.446 g/L). Cellular immunological test results were as follows: CD3, 74.6% (50%-84.0%); CD4, 47.3% (27.0%-51.0%); and CD8, 24.9% (15.0%-44.0%). A de novo hemizygous deletion in BTK was detected: c.902_c.904delAAG/p.E301del. Transcript levels of the mutant BTK were similar to those of the wild-type gene, though overexpression resulted in markedly reduced levels of mutant BTK (9.49% ± 1.58%), relative to the wild-type BTK (75.8% ± 2.98%, P < 0.01).
This case of X-linked agammaglobulinemia was attributed to a de novo hemizygous deletion mutation in BTK (c.902_c.904delAAG/p.E301del). The mutation resulted in markedly reduced BTK protein stability in vitro.
Core tip: This report introduces the diagnosis and treatment of X-linked agammaglobulinemia caused by a new Bruton’s tyrosine kinase gene mutation, and includes a detailed clinical and laboratory analysis of its pathogenic principle, which provided the diagnosis and led to the treatment of X-linked agammaglobulinemia.
- Citation: Hu XM, Yuan K, Chen H, Chen C, Fang YL, Zhu JF, Liang L, Wang CL. Novel deletion mutation in Bruton’s tyrosine kinase results in X-linked agammaglobulinemia: A case report. World J Clin Cases 2020; 8(17): 3859-3866
- URL: https://www.wjgnet.com/2307-8960/full/v8/i17/3859.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i17.3859
X-linked agammaglobulinemia (XLA, OMIM 300755; also known as Bruton's disease) is a primary immunodeficiency disease caused by mutations of the Bruton’s tyrosine kinase (BTK) gene. The BTK protein, discovered in 1993, is named after Ogden Bruton, who first described XLA in 1952. XLA is a developmental disorder characterized by severely reduced mature peripheral B lymphocyte counts and serum immunoglobulin (Ig) levels[1]. Typically, patients with XLA suffer from various bacterial infections caused by many extracellular bacteria. They are also vulnerable to viruses and parasites. The presentation and severity of XLA differ among patients, with some BTK mutations being associated with severe and complex disease processes[2-4]. However, the relationship between in vitro findings and clinical phenotypes of BTK mutations remains poorly understood.
In the present study, we report the case of a Chinese boy with a typical clinical XLA phenotype carrying a de novo hemizygous deletion in BTK. We identified the mutation (c.902_c.904delAAG/p. E301del) and conducted a predictive analysis of its effect on the tertiary structure of BTK. The present study was approved by the Ethics Committee of The First Affiliated Hospital, College of Medicine, Zhejiang University, and conducted in accordance with the Declaration of Helsinki Principles. Furthermore, written informed consents were obtained from the patient’s parents.
A 5-year-old boy was admitted to our hospital due to recurrent cough and fever that had persisted for more than a month.
The patient’s symptoms started over a month ago with recurrent cough and fever, which had worsened over the previous week.
The patient had a history of multiple respiratory infections and sinusitis episodes since the age of 1 year, but no peripheral lymphadenopathy, tonsil enlargement, or hepatosplenomegaly were observed.
The boy was the first child of Han Chinese nonconsanguineous parents. He was born full term with a birth weight of 4000 g and a length of 50 cm. There was no family history of immunodeficiency or recurrent infections.
The patient’s temperature was 36.6°C, heart rate was 90 bpm, respiratory rate was 25 breaths per minute, blood pressure was 121/60 mmHg and oxygen saturation in room air was 97%. His mind was clear, he had shortness of breath, middle trachea, hyperemia of the pharynx, no obvious enlargement of the tonsils, thick breath sounds in both lungs, obvious moist rale, homogeneous rhythm, no obvious pathological Mur, soft abdomen, normal bowel sound, no obvious abnormalities during nervous system examination.
The patient’s routine blood results were as follows: White blood cell count, 21.5 × 109/L (4.0-12.0 × 109/L); neutrophils, 67.9% (50%-70%); lymphocytes, 18.1% (20%-40%); hemoglobin, 131 g/L (120-140 g/L); platelet count, 401 × 109/L (100-300 × 109/L); and high-sensitivity C-reactive protein, 112.7 mg/dL (0–8 mg/dL). Agammaglobulinemia was characterized as follows: IgG, 20.0 mg/dL (800-1800 mg/dL); IgG subtype 1, 0.26 g/L (3.62-12.28 g/L); IgG subtype 2, 0.10 g/L (0.57-2.9 g/L); IgG subtype 3, 0.009 g/L (0.129-0.789 g/L); IgG subtype 4, 0.003 g/L (0.013-1.446 g/L); IgA, 90.0 mg/dL (90-450 mg/dL); and IgM, 18.0 mg/dL (60-280 mg/dL) (determined by enzyme-linked immunosorbent assay kits from Shanghai Langton Biotechnology Co., Ltd.). Complement C4 and C3 levels were found to be 18.0 mg/dL (7-149 mg/dL) and 106.0 mg/dL (58-160 mg/dL), respectively. CD3+, CD4+, and CD8+ T lymphocytes were 74.6% (50%-84.0%), 47.3% (27.0%-51.0%), and 24.9% (15.0%-44.0%), respectively.
A chest spiral computed tomography scan revealed a bilateral lung infection.
Due to the patient’s history of repeated respiratory tract infections, his very low serum IgG levels (< 2.5 g/L), and general agammaglobulinemia, he was treated with Ig infusions every four weeks. This treatment markedly reduced the patient’s respiratory infection incidence.
Genetic analysis: Genomic deoxyribonucleic acid (DNA) was extracted from peripheral blood samples obtained from the proband and his parents. Sequence variants detected in BTK were described according to the National Center of Biotechnology Information entry NM_000061.2. An analysis of pathogenicity and mutation conservation was performed with the bioinformatics Mutation Taster (http://www.mutationtaster.org) and University of California Santa Cruz Genome Browser (http://genome.UCSC.edu) programs.
Molecular modeling: The three-dimensional structure of the mutation was generated in the Swiss-Model program (https://swissmodel.expasy.org/interactive).
BTK expression: The wild-type (WT) full-length human BTK messenger ribonucleic acid (mRNA) (NM_000061.2), which includes NheI and BamH1 restriction enzyme sites, was synthesized (Tsingke, China), and cloned into the plasmid vector pEGFP-N1. The mutation 902-904delAAG was introduced into the BTK sequence via two polymerase chain reaction (PCR) rounds with mutagenic primers. The two PCR products were connected by homologous recombination[5] with a TreliefTM SoSoo cloning kit (Tsingke, China). The connection product was transformed into Escherichia coli DH5α. Positive clones were selected, and the full-length mutant cDNA was sequenced to confirm the site-directed mutation.
For transient transfection, HEK-293T cells were cultured in 24-well plates to 70%-80% confluence and transfected with 500 ng of pEGFP-N1 containing no insert, pEGFP-N1-WT-BTK (WT), or pEGFP-N1-Mutation-BTK (Mut) with Lipofectamine™ 3000 transfection reagent (Thermo Fisher Scientific). Cells were collected 24 h later.
Quantitative real-time PCR: Cells were collected from 24-well plates and total RNA preparations were obtained with a MiniBEST Universal RNA Extraction Kit (Takara). cDNA was produced with a Primescript RT reagent kit with gDNA Eraser, applied prior to reverse transcription to remove genomic DNA (Takara, AK3920). Glyceraldehyde 3-phosphate dehydrogenase gene fragment was used as an internal reference, and quantitative PCR was performed with a real-time fluorescence quantitative PCR instrument (BIO-RAD CFX Connect). The forward and reverse primers used for the PCR amplification were 5’ GAA AGG TTC CCT TAT CCC TTC C 3’ and 5’ GAA TCC ACC GCT TCC TTA GTT 3’, respectively.
Western blotting: Cell protein was extracted with a whole cell extraction kit (Abcam), and 10-μg protein aliquots were subjected to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis. Protein bands were electro transferred onto a polyvinylidene difluoride membrane and detected with monoclonal anti-BTK antibody (Abcam) and horseradish peroxidase-conjugated goat anti-rabbit secondary antibody. Immunoreactive bands were visualized with enhanced chemiluminescence reagent (Super Signal, Pierce) and observed with a gel imaging system.
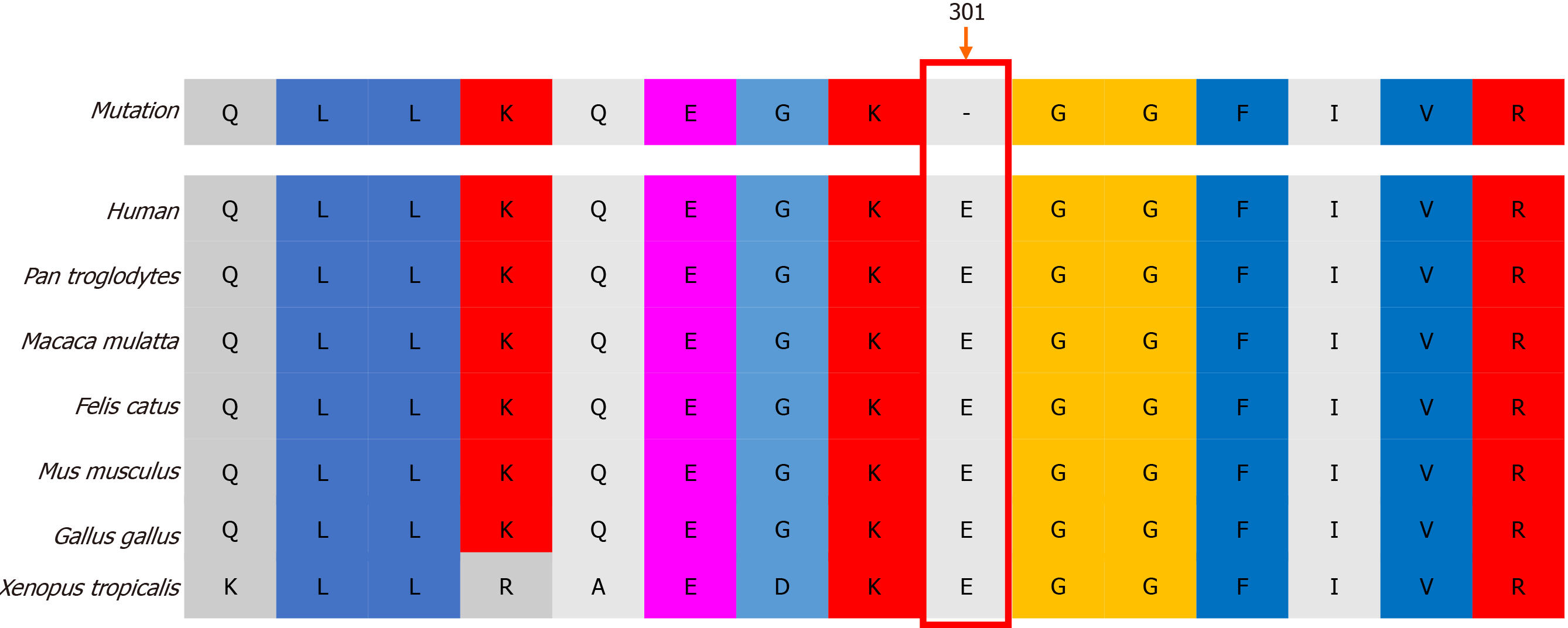
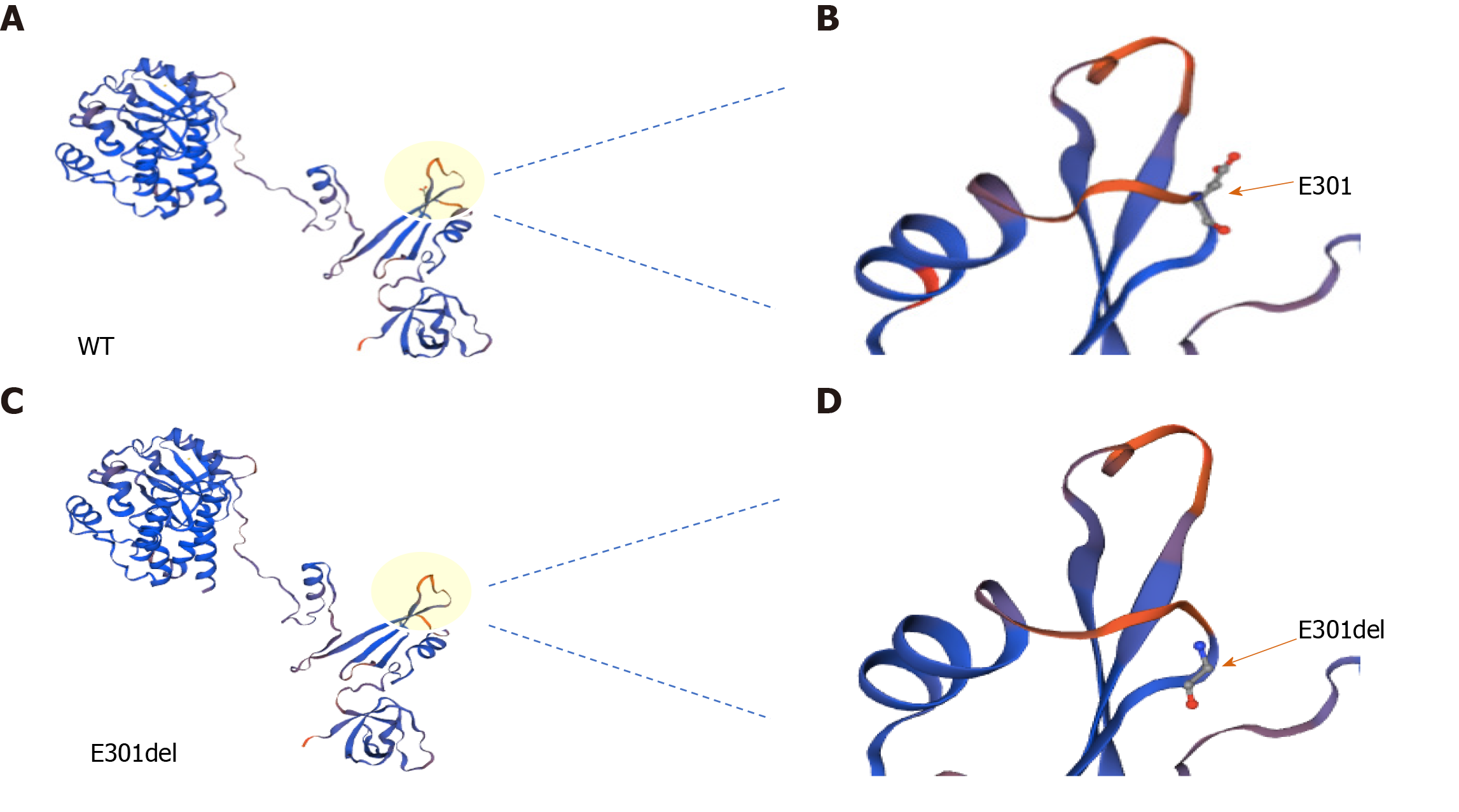
Genetic diagnosis and protein structure modeling: Genetic analysis revealed that the proband had a de novo hemizygous deletion in BTK: c.902_c.904delAAG/p.E301del. No BTK variants were detected in the proband’s parents’ genomes (Figure 1). Bioinformatics analysis in Mutation Taster indicated that BTK c.902_c.904delAAG /p.E301del would be highly pathogenic. Furthermore, sequence alignment revealed that the E301 residue lost in this deletion is conserved across diverse vertebrates (e.g. Homo sapiens, Pan troglodytes, Macaca mulatta, Felis catus, Mus musculus, Gallus gallus, and Xenopus tropicalis), a finding consistent with it being functionally important (Figure 2). The deletion did not produce a frameshift. However, three-dimensional structural modeling indicated that the deletion altered the Src homology (SH) 2 domain of the mutant BTK protein relative to WT BTK (Figure 3).
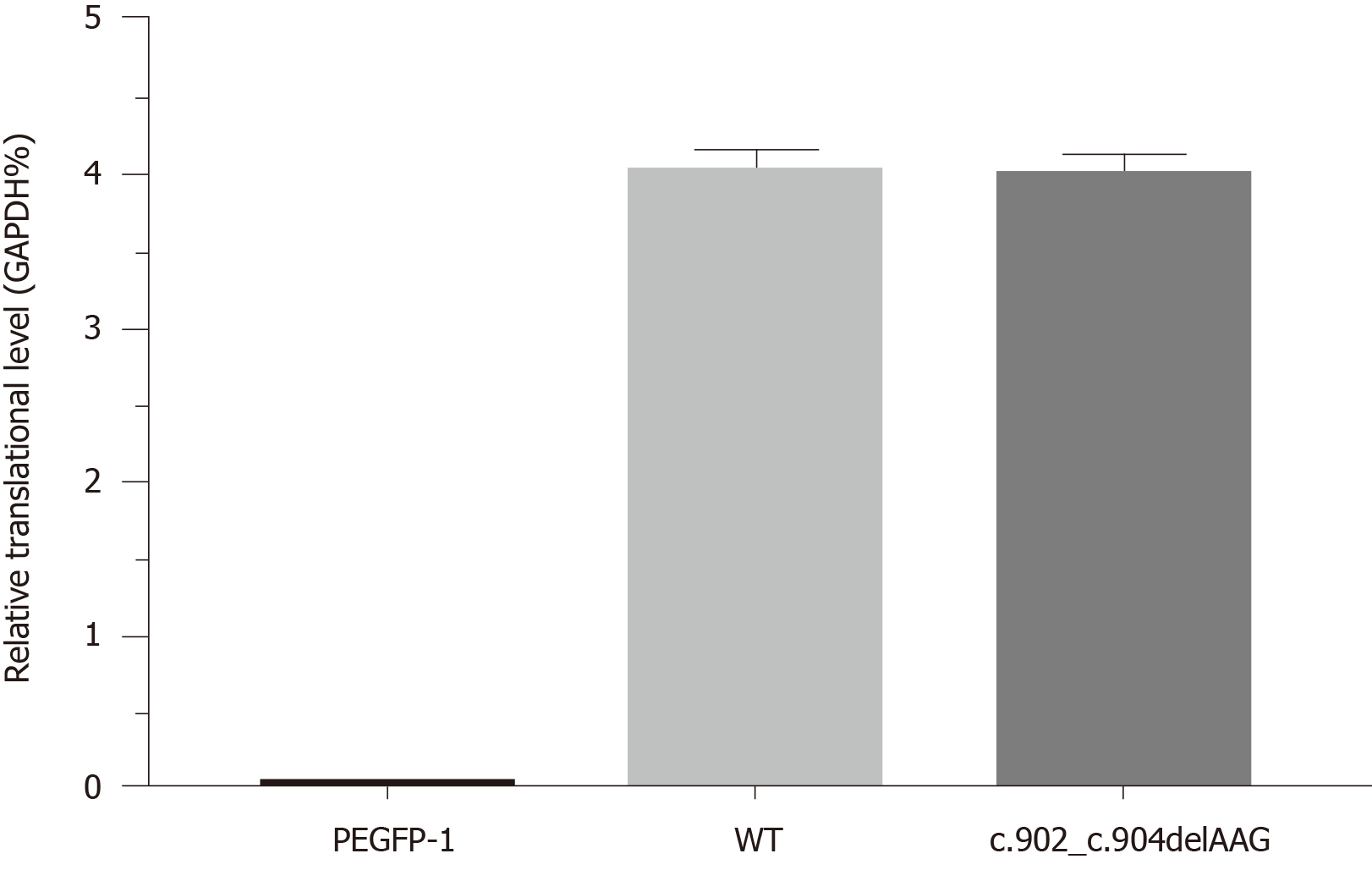
Reduction in steady-state levels of BTK: Quantitative PCR results revealed similar BTK mRNA levels between WT and mutant cDNAs, indicating that the mutation was not disruptive to transcription (Figure 4). Western blotting revealed plentiful overexpression of WT BTK (75.8% ± 2.98%) and markedly reduced overexpression of the BTK c.902_c.904delAAG/p.E301del deletion mutant (9.49% ± 1.58%, P < 0.01) (Figure 5). Hence, mutant BTK protein levels were depressed despite the deletion mutation not being disruptive to transcription, providing evidence of a posttranslational loss.
X-linked agammaglobulinemia.
Regular injections of intravenous immunoglobulin.
The child’s immune function was basically normal, and there were no obvious abnormalities during long-term follow-up.
The history of repeated infections and reduced serum Ig levels in the proband in the present case led to a clear diagnosis of Ig deficiency. This phenotype together with the identified BTK mutation led to the final diagnosis of XLA. As BTK-mediated signaling plays an important role in the differentiation, development, proliferation, and apoptosis of B lymphocytes, mutations in BTK that disrupt BTK function result in the severe Ig deficiency characteristic of XLA[1,3]. Affected patients exhibit typical development in infancy while they are protected by maternal IgGs, after which patients become susceptible to repeated bacterial infections. The disrupted B-cell development in bone marrow in patients with XLA leads to severe insufficiencies of B cells and antibodies in the circulation.
Human BTK is located on the X chromosome at q21.33-22 (total length, 37.5 kb) and expressed in all hematopoietic cells. It contains 21 exons.
Approximately 900 different BTK mutations have been identified in patients with XLA, most of which are missense mutations, followed by nonsense, deletion, insertion, and splice mutations. Some two-thirds of patients with XLA have a family history of XLA, with the remainder having a de novo mutation.
The BTK protein consists of five functional domains: A pleckstrin-homology domain [amino acids (aa) 1-138], a Tec homology domain (aa 139-215), an SH3 domain (aa 216-280), an SH2 domain (aa 281-377), and the tyrosine kinase-SH1 domain (aa 375-659)[14].
BTK mutations, such as L295P, G302E, G302R and G302del, have been reported to severely disrupt protein activity, leading to XLA[6-8]. Although the deletion mutation in our patient (c.902_c.904 delAAG/p.E301del) did not result in a frameshift and did not alter the structure of the whole protein (Figure 3), it is located in the important functional SH2 domain. The evolutionary conservation of the residue lost in our patient’s mutant BTK (E301) across diverse vertebrates suggests that it is functionally critical.
BTK genotypes have been related, to some extent, with disease phenotypes[9,10]. Patients with some mild missense mutations causing just a single change have relatively intact Ig levels and mild clinical manifestations. However, patients with more severe mutations, including frameshift and nonsense mutations, may exhibit a wide range of severe phenotypes. Notwithstanding, the same mutation may result in different clinical manifestations in different individuals[11-14]. In the present case, we found that BTK mutation was associated with a clinically typical presentation of XLA. The pathogenesis appears to be attributable to a loss of stability of the mutant protein, whose stability level upon overexpression in vitro was only about that of the WT protein.
In summary, a de novo hemizygous deletion mutation in BTK was identified as the cause of XLA in the present case. The mutation was shown to cause a significant reduction in protein stability in vitro.
The authors would like to thank the proband, his family, and the control subjects for providing blood samples and agreeing to participate in this research.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territoryof origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Rangel-Corona R S-Editor: Zhang L L-Editor: Webster JR P-Editor: Wang LL
| 1. | Tsukada S, Saffran DC, Rawlings DJ, Parolini O, Allen RC, Klisak I, Sparkes RS, Kubagawa H, Mohandas T, Quan S. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993;72:279-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 998] [Cited by in RCA: 981] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 2. | Aghamohammadi A, Fiorini M, Moin M, Parvaneh N, Teimourian S, Yeganeh M, Goffi F, Kanegane H, Amirzargar AA, Pourpak Z, Rezaei N, Salavati A, Pouladi N, Abdollahzade S, Notarangelo LD, Miyawaki T, Plebani A. Clinical, immunological and molecular characteristics of 37 Iranian patients with X-linked agammaglobulinemia. Int Arch Allergy Immunol. 2006;141:408-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 3. | Broides A, Yang W, Conley ME. Genotype/phenotype correlations in X-linked agammaglobulinemia. Clin Immunol. 2006;118:195-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 69] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | Conley ME, Broides A, Hernandez-Trujillo V, Howard V, Kanegane H, Miyawaki T, Shurtleff SA. Genetic analysis of patients with defects in early B-cell development. Immunol Rev. 2005;203:216-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 129] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 5. | Esenboga S, Cagdas D, Ozgur TT, Gur Cetinkaya P, Turkdemir LM, Sanal O, VanDerBurg M, Tezcan I. Clinical and genetic features of the patients with X-Linked agammaglobulinemia from Turkey: Single-centre experience. Scand J Immunol. 2018;87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Gaspar HB, Bradley LA, Katz F, Lovering RC, Roifman CM, Morgan G, Levinsky RJ, Kinnon C. Mutation analysis in Bruton's tyrosine kinase, the X-linked agammaglobulinaemia gene, including identification of an insertional hotspot. Hum Mol Genet. 1995;4:755-757. [PubMed] [DOI] [Full Text] |
| 7. | Hagemann TL, Rosen FS, Kwan SP. Characterization of germline mutations of the gene encoding Bruton's tyrosine kinase in families with X-linked agammaglobulinemia. Hum Mutat. 1995;5:296-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Schuster V, Seidenspinner S, Kreth HW. Detection of a novel mutation in the SRC homology domain 2 (SH2) of Bruton's tyrosine kinase and direct female carrier evaluation in a family with X-linked agammaglobulinemia. Am J Med Genet. 1996;63:318-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 9. | Lappalainen I, Thusberg J, Shen B, Vihinen M. Genome wide analysis of pathogenic SH2 domain mutations. Proteins. 2008;72:779-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Vetrie D, Vořechovský I, Sideras P, Holland J, Davies A, Flinter F, Hammarström L, Kinnon C, Levinsky R, Bobrow M, Smith CI, Bentley DR. The gene involved in X-linked agammaglobulinaemia is a member of the Src family of protein-tyrosine kinases. 1993. J Immunol. 2012;188:2948-2955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1093] [Cited by in RCA: 1073] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 11. | Lee PP, Chen TX, Jiang LP, Chan KW, Yang W, Lee BW, Chiang WC, Chen XY, Fok SF, Lee TL, Ho MH, Yang XQ, Lau YL. Clinical characteristics and genotype-phenotype correlation in 62 patients with X-linked agammaglobulinemia. J Clin Immunol. 2010;30:121-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 12. | López-Granados E, Pérez de Diego R, Ferreira Cerdán A, Fontán Casariego G, García Rodríguez MC. A genotype-phenotype correlation study in a group of 54 patients with X-linked agammaglobulinemia. J Allergy Clin Immunol. 2005;116:690-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 72] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Singh S, Rawat A, Suri D, Gupta A, Garg R, Saikia B, Minz RW, Sehgal S, Chan KW, Lau YL, Kamae C, Honma K, Nakagawa N, Imai K, Nonoyama S, Oshima K, Mitsuiki N, Ohara O. X-linked agammaglobulinemia: Twenty years of single-center experience from North West India. Ann Allergy Asthma Immunol. 2016;117:405-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Winkelstein JA, Marino MC, Lederman HM, Jones SM, Sullivan K, Burks AW, Conley ME, Cunningham-Rundles C, Ochs HD. X-linked agammaglobulinemia: report on a United States registry of 201 patients. Medicine (Baltimore). 2006;85:193-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 370] [Article Influence: 19.5] [Reference Citation Analysis (0)] |