Published online Aug 16, 2014. doi: 10.12998/wjcc.v2.i8.395
Revised: May 11, 2014
Accepted: June 10, 2014
Published online: August 16, 2014
Processing time: 147 Days and 17.8 Hours
Cystic fibrosis, a common autosomal recessive genetic disorder among Caucasians, is caused by defects in the transmembrane conductance regulatory (CFTR) gene. The analysis of CFTR gene mutations is useful to better characterize the disease, and for preconceptional screening, prenatal and preimplantation genetic diagnosis. Here we report the results of a genetic analysis in a 16-year-old boy from southwestern Iran diagnosed as having cystic fibrosis in infancy based on gastrointestinal and pulmonary manifestations, with positive sweat chloride tests. He lacked both normal and mutant forms of the fragment corresponding to the ∆F508 allele in initial genetic studies. Multiplex ligation-dependent probe amplification-based testing revealed a homozygous deletion spanning exons 4 to 10 of the CFTR gene. We predict an in-frame deletion removing 373 amino acids based on our sequencing results. Determining CFTR gene mutations in patients and their family members would be helpful to prevent the occurrence of new cases, especially in populations in which consanguinity is common.
Core tip: Genetic analysis of the transmembrane conductance regulatory (CFTR) gene is helpful to characterize patients with cystic fibrosis, but sequencing and multiplex ligation-dependent probe amplification-based testing are only done to diagnose rare or unknown variants. Here we report a 16-year-old boy, the son of consanguineous healthy parents, who lacked both the normal and mutant forms of the ∆F508 alleles in initial molecular tests. Further analysis disclosed a rare large homozygous CFTR gene deletion in this patient.
-
Citation: Farjadian S, Moghtaderi M, Zuntini R, Ferrari S. Rare large homozygous
CFTR gene deletion in an Iranian patient with cystic fibrosis. World J Clin Cases 2014; 2(8): 395-397 - URL: https://www.wjgnet.com/2307-8960/full/v2/i8/395.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v2.i8.395
Cystic fibrosis (CF), a common autosomal recessive genetic disorder among Caucasians, is caused by defects in the transmembrane conductance regulatory (CFTR) gene. This gene spans more than 250 kb on chromosome 7q31.2 and comprises 27 exons encoding a 170 kDa chloride channel expressed exclusively in secretory epithelial cells[1]. To date, more than 1969 sequence variations have been identified in the CFTR gene, including mutations that are involved in disease expression and polymorphisms which have no effect on the phenotype[2]. The rate of CFTR gene mutations varies greatly among different populations. Although the prevalence of CF in Iran is not known, current data suggest that the disease is not rare in this country. The most common mutation is ΔF508 with a frequency of 16% to 24% in different parts of Iran; these rates are much lower than in European countries[3].
The clinical presentations of CF varies widely from atypical mild disease to the classical form characterized by multiorgan involvement. The highly variable presentation depends on specific mutations, gene penetrance, the presence of genetic modifiers and environmental factors[4]. The diagnosis of classical CF is straightforward and based on specific clinical features, family history and positive sweat chloride tests, whereas the diagnosis of nonclassical CF is often delayed because of its unusual presentation or the late onset of symptoms. Delays in the diagnosis usually lead to progressive disease and even irreversible multiorgan damage[5]. The analysis of CFTR gene mutations is useful to better characterize the disease, especially when the results of sweat chloride tests are uncertain or variable. DNA-based testing is also useful for preconceptional screening, prenatal diagnosis for couples with a family history of CF, and preimplantation genetic diagnosis for couples with known CFTR genetic mutations who hope to have a healthy child by in vitro fertilization[5,6]. These tests are usually performed with a panel of known CFTR mutations for the ethnic group of interest. Sequencing the CFTR gene and multiplex ligation-dependent probe amplification (MLPA)-based testing are only done to diagnose rare or unknown variants[4].
A 16-year-old boy from Southwestern Iran with chronic productive cough and dyspnea was diagnosed as having CF in infancy based on typical findings of gastrointestinal and pulmonary manifestations with a positive sweat chloride test. He was the sixth child of healthy consanguineous parents and had two healthy older sisters and one healthy brother. The results of sweat chloride tests were normal for the parents and siblings, and none of them reported any symptoms or problems related with CF. Two of the patient’s older brothers had died at the age of 6 mo; their medical history was unremarkable.
This patient had been hospitalized several times during infancy due to severe dehydration. He suffered from numerous recurrent pulmonary infections and greasy stools, which required frequent visits to his physician. Physical examination showed scattered bilateral coarse crackles, increased anteroposterior diameter of chest and digital clubbing.
At his most recent visit his bone age was estimated at about 12-year-old based on left-hand X-ray, and he also had symptoms compatible with delayed sexual maturation and delayed puberty. Laboratory parameters including blood cell count, fasting blood glucose, blood urea nitrogen, serum creatinine, calcium, phosphorus, erythrocyte sedimentation rate, C-reactive protein levels and liver function tests were normal at this visit, but his sweat chloride test results were higher than normal (> 100 mEq/L). Chest X-ray revealed bilateral infiltration and bronchiectasis in both lung fields. Abdominal and pelvic ultrasound examination disclosed no abnormal findings. Because of his abnormal heart sounds, echocardiography was performed which showed mild pulmonary artery hypertension. The patient was advised to continue treatment with antibiotics, chest physiotherapy, pancreatic enzyme replacement and vitamin supplementation.
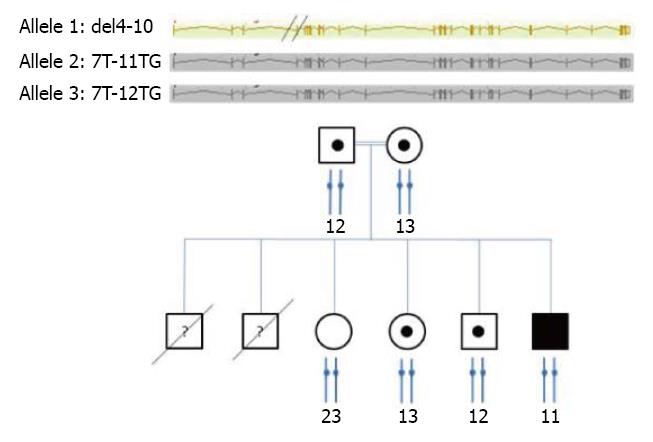
An initial genetic study was done with the Elucigene CF29 v.2 kit (Tepnel, Oxfordshire, United Kingdom). Our patient lacked of both the normal and mutant forms of the fragment corresponding to the ∆F508 allele, whereas all his first-degree relatives carried the normal allele. This test was repeated three times with new blood samples, and the results were consistent across tests. Genetic analysis was then performed with the Elucigene CF-EU2 v.1 kit (Gen-Probe Life Science Ltd., Manchester, United Kingdom), which is designed to identify 50 mutations. This kit is also able to identify the number of TG repeats associated to the polythymidine tract at the junction of intron 8 and exon 9, which affects the splicing efficiency of exon 9 and influences the gene transcription rate. This analysis showed the absence of PCR amplification products for all fragments mapping to exons 4-10, suggesting that he was homozygous for a deletion spanning exons 4 to 10 of the CFTR gene (CFTR del 4-10), as a result of first-degree consanguinity between his parents. This homozygous deletion was confirmed by MLPA and was detected in the heterozygous state in both parents (Figure 1), in one of the sisters and in his brother. The 40-kb del 4-10 CF mutation was previously reported in compound heterozygous patterns in two patients with CF: an 8-year-old French girl with the ΔF508/CF 40-kb del 4-10 genotype combination[7] and a 19-year-old Caucasian female with the c.1220del20/CF 40-kb del 4-10 genotype combination[8]. In contrast to the latter patient with a frameshift mutation in the CFTR gene because of a 40-kb deletion, in our patient we predict an in-frame deletion removing 373 amino acids based on our sequencing results.
In conclusion, although there is no evidence to prove the relationship between CFTR gene mutations and disease severity or response to therapy, determining CFTR gene mutations in patients and their family members would be helpful to prevent the occurrence of new cases, especially in populations in which consanguinity is common.
The authors thank Professor Romeo G for coordinating this collaboration, the Institute of Advanced Studies, University of Bologna, Italy for providing accommodations for Dr. Farjadian S while she was in Bologna for the molecular analysis and Shashok K (Author AID in the Eastern Mediterranean) for improving the use of English in the manuscript.
A 16-year-old boy with chronic productive cough and dyspnea was diagnosed as having cystic fibrosis (CF) in infancy based on gastrointestinal and pulmonary manifestations with a positive sweat chloride test.
Hospitalization during infancy due to severe dehydration and recurrent pulmonary infections and greasy stools.
Celiac disease, primary immunodeficiency disorders.
Positive sweat chloride test and lack of both normal and mutant forms of the fragment corresponding to the ∆F508 allele in molecular analysis.
Left-hand X-ray: bone age about 12-year-old based on. Chest X-ray: bilateral infiltration and bronchiectasis in both lung fields. Echocardiography: mild pulmonary artery hypertension.
Antibiotics therapy, chest physiotherapy, pancreatic enzyme replacement and vitamin supplementation.
Homozygous 40-kb del 4-10 in cystic fibrosis transmembrane regulatory (CFTR) gene was detected in this patient by multiplex ligation-dependent probe amplification (MLPA).
Determining CFTR gene mutations in CF patients and their family members would be helpful to prevent the occurrence of new cases, especially in populations in which consanguinity is common.
MLPA is a technique for detecting deletions or duplications of one or more parts of a gene.
The manuscript reports a patient with homozygous exon 4-10 CFTR gene deletion mutation. Overall, this manuscript is well written and suitable as a case-report.
P- Reviewer: Bener A S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Tousson A, Van Tine BA, Naren AP, Shaw GM, Schwiebert LM. Characterization of CFTR expression and chloride channel activity in human endothelia. Am J Physiol. 1998;275:C1555-1564. [PubMed] |
| 2. | Available from: http://www.genet.sickkids.on.ca/cftr/Statistics Page.html. |
| 3. | Farjadian S, Moghtaderi M, Kashef S, Alyasin S, Najib K, Saki F. Clinical and genetic features in patients with cystic fibrosis in southwestern Iran. Iran J Pediatr. 2013;23:212-215. [PubMed] |
| 4. | Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet. 2013;132:1077-1130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 493] [Cited by in RCA: 440] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 5. | Karczeski B, Cutting GR. Diagnosis of Cystic Fibrosis, CFTR-Related Disease and Screening. In: Bush A, Alton EWFW, Davies JC, Griesenbach U, Jaffe A. Cystic Fibrosis in the 21st Century. Basel: Karger. 2006;69-76. [PubMed] |
| 6. | Rechitsky S, Verlinsky O, Kuliev A. PGD for cystic fibrosis patients and couples at risk of an additional genetic disorder combined with 24-chromosome aneuploidy testing. Reprod Biomed Online. 2013;26:420-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Chevalier-Porst F, Bonardot AM, Chazalette JP, Mathieu M, Bozon D. 40 kilobase deletion (CF 40 kb del 4-10) removes exons 4 to 10 of the Cystic Fibrosis Transmembrane Conductance Regulator gene. Hum Mutat. 1998;Suppl 1:S291-294. [PubMed] |
| 8. | Hantash FM, Rebuyon A, Peng M, Redman JB, Sun W, Strom CM. Apparent homozygosity of a novel frame shift mutation in the CFTR gene because of a large deletion. J Mol Diagn. 2009;11:253-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |