Published online Jul 16, 2025. doi: 10.12998/wjcc.v13.i20.102651
Revised: January 21, 2025
Accepted: March 4, 2025
Published online: July 16, 2025
Processing time: 166 Days and 22.8 Hours
Floating-harbor syndrome (FHS) is a rare genetic disorder caused by pathogenic variants in the SRCAP gene. Most individuals with FHS have short stature, delayed speech and language development, and dysmorphic facial features. However, the patients with FHS are not easy to diagnose due to the overlap of clinical phenotypes with other disorders.
We reported a 10-year-old boy who presented with severe short stature, developmental delay and distinctive facial features. Exome sequencing was provided for the proband and his parents. We identified a novel frameshift variant c.7235de
This case confirms that the c.7235delinsGT (p.Thr2412fs) variant in the SRCAP gene is associated with FHS and expands the spectrum of SRCAP variants.
Core Tip: In conclusion, a de novo heterozygous variant in SRCAP (c.7235delinsGT; p.Thr2412fs) was identified in a Chinese male patient with floating-harbor syndrome (FHS) using ES. This proband exhibited classical clinical features of FHS, including severe short stature, developmental delay, characteristic facial features, and additional malformations. This novel SRCAP variant expands the known variant spectrum in FHS and supports the clinical homogeneity of this disorder. Fortunately, we provided genetic counseling and prenatal genetic diagnosis for this family based on the known variant in the proband.
- Citation: Xiao X, Wang P, Wang H, Xie HB, Liu SL. Identifying a novel SRCAP variant in floating-harbor syndrome and prenatal genetic diagnosis in this Chinese family: A case report. World J Clin Cases 2025; 13(20): 102651
- URL: https://www.wjgnet.com/2307-8960/full/v13/i20/102651.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v13.i20.102651
Floating-harbor syndrome (FHS; OMIM #136140) is a rare autosomal dominant genetic disorder characterized by short stature, delayed speech and language development, and dysmorphic facial features[1-4]. Typical characteristic facial features include a triangular face, a short philtrum, long eyelashes, a narrow nasal root, a broad nasal tip, a prominent nose, a broad mouth and dental malocclusion[5,6]. Other manifestations have been reported such as skeletal features, genital anomalies and cardiac malformations[3,7]. Some authors have highlighted the phenotypic overlap between FHS and Rubinstein-Taybi syndrome (RSTS; OMIM #180849, #613684), which is characterized by distinctive facial features, anomalous thumbs and halluces, short stature, and moderate-to-severe intellectual disability.
The genetic cause of FHS are heterozygous variants in the SRCAP gene (OMIM: 611421)[6]. SRCAP encodes an ATPase that functions as the core catalytic subunit of the multiprotein SRCAP complex, which regulates transcription of various target genes by chromatin remodeling[8]. SRCAP contains several discrete functional domains, which include a N-terminal HSA (Helicase-SANT-associated) domain, SNF2-like ATPase domains, a CBP-binding domain and three C-terminal A/T-hook DNA-binding motifs[9-11]. Pathogenic variants in SRCAP were found to be located mostly within a small region of exon 34, but also in exon 33[7,12,13]. In the vast majority of cases, the variants were either nonsense or frameshift and occurred de novo.
Here, we report on a case of FHS confirmed by exome sequencing, and describe the clinical and molecular findings in this patient. We also performed prenatal genetic diagnosis for this family whose next generation was at increased risk of FHS.
Growth delay and speech impairment.
Following irregular prenatal examinations on his mother, he was born at 39 weeks of gestation via vaginal delivery with a birth weight of 2.3 kg (< 3rd percentile) and a length of 45 cm (< 3rd percentile).
The proband had a history of significant developmental delays. Bone age was of 0.4 years at the age of 1 year. He started to walk when he was 1 year and 4 months old, and had no word at age two years. At the age of 1.5 years, inguinal hernia corrective surgery was performed, with good results. Later, when he was 2 years and 11 months old, he was referred to clinic because of growth delay and speech impairment. The proband showed short stature with a weight of 11 kg (< 3rd percentile) and height of 83 cm (< 3rd percentile). At this age, his bone age was 1 year. His language production is limited with no more than two words. Echocardiography showed mild aortic and pulmonary valve regurgitation. On clinical examination, he presented a distinctive facial appearance with short philtrum, long eyelashes, prominent nose, narrow nasal root, broad nasal tip, dental malocclusion and mild clubbing of fingers (Figure 1A). At the age of 5 years and 11 months, bone age development had markedly accelerated, and his bone age was 7 years. Pituitary magnetic resonance imaging showed a normal pituitary morphology. Upon comprehensive assessment no dysfunction of thyroid or the low level of growth hormone was found. He had behavioural problems with hyperactivity and attention deficit. He attended a normal school, but had the lowest score in the class. At the latest evaluation of proband at age of 10 years, weight was 24 kg (3rd-10th percentile), height was 124 cm (< 3rd percentile), head circumference was 48 cm (< -1SDS), and was able to speak full sentences with simple sentence structures (Table 1).
| Years | Weight (kg) | Height (cm) | Bone age (years) | Features |
| At birth (39 weeks) | 2.3 (< 3rd) | 45 (< 3rd) | NA | NA |
| 1 year | NA | NA | 0.4 | NA |
| 1 year and 4 months | NA | NA | NA | Started to walk |
| 1 year and 6 months | NA | NA | NA | Inguinal hernia corrective surgery |
| 2 years | NA | NA | NA | No word |
| 2 years and 11 months | 11 (< 3rd) | 83 (< 3rd) | 1 | No more than two words; mild aortic and pulmonary valve regurgitation; a distinctive facial appearance |
| 5 years and 11 months | NA | NA | 7 years | Hyperactivity and attention deficit |
| 10 years | 24 (3rd-10th) | 124 (< 3rd) | NA | Speak simple sentences |
There is no significant history of past illness.
The family history was unremarkable. The child’s mother was 157 cm tall, and his father was 173 cm tall (Figure 1B).
He was born at 39 weeks of gestation with a birth weight of 2.3 kg (< 3rd percentile) and a length of 45 cm (< 3rd percentile).
When he was 2 years and 11 months old, he weighed 11 kg (< 3rd percentile) and was 83cm tall (< 3rd percentile).
At the age of 10 years, weight was 24 kg (3rd-10th percentile), height was 124 cm (< 3rd percentile), head circumference was 48 cm (< -1SDS).
There was no dysfunction of the thyroid, nor was a low level of growth hormone found. We identified a novel frameshift variant c.7235delinsGT (p.Thr2412fs) in SRCAP gene through exome sequencing[14]. The variant in our study is highly conserved across species and substitutes the encoded threonine for serine, causing premature termination at a downstream amino acid (Figure 1C). We attempted to predict the 3D structure of the SRCAP protein of the wild-type and which with the variant through AlphaFold3. When the highest rank model of p.Thr2412fs aligned to the wild-type, it changed the protein twists direction, concurrently obtained a TM-score of 0.23 and a RMSD value of 8.06. The predicted SRCAP protein structure of the p.Thr2412fs variant with low TM-score and high RMSD value showed it was highly variable, compared to the wild-type (Figure 1D and E). According to ACMG criteria, the variant was classified as likely pathogenic[15]. This variant was validated by Sanger sequencing (Figure 1F).
Bone age was of 0.4 years at the age of 1 year. When he was 2 years and 11 months old, his bone age was 1 year. At the age of 5 years and 11 months, his bone age was 7 years. Echocardiography showed mild aortic and pulmonary valve regurgitation. Pituitary magnetic resonance imaging showed a normal pituitary morphology.
The proband was diagnosed with FHS.
The proband did not receive any special treatment. His mother had a second pregnancy. No fetal abnormalities were found through ultrasound examination till amniocentesis. At 21 weeks of gestation, amniocentesis was performed to analyze the specific variant of the target gene and fetal chromosomal abnormalities.
The prenatal genetic diagnosis for c.7235delinsGT (p.Thr2412fs) of SRCAP gene showed that the fetus had no target variant. Further, the fetus had no chromosomal abnormality using chromosomal microarray analysis and no abnormal finding through ultrasound scan throughout pregnancy. The fetus has been born successfully and is one month old now. The newborn does not show any similar symptom to the proband.
We describe a detailed clinical phenotype and genetic analysis of a patient with FHS. The patient had severe short stature, developmental delay, a distinctive facial appearance, clubbing of fingers and inguinal hernia.
In general, the clinical features of our patient with SRCAP variant (c.7235delinsGT; p.Thr2412fs) were concordant with earlier descriptions of FHS. One of the typical phenotypes of FHS is delayed bone maturation[16]. The patient’s bone age was severely delayed in early childhood and markedly accelerated around 6 years. These findings are consistent with previous cases showing significantly delayed bone age in most FHS patients before the age of 6 years, then markedly accelerated and reach normal level[17-19]. The phenotypes of short stature, language impairment, and a distinctive facial appearance in the proband were remarkably similar to those of nearly all individuals with FHS. The patient had additional systemic malformations: Aortic and pulmonary valve regurgitation, clubbing of fingers and inguinal hernia, these also had been reported in some patients with FHS[6].
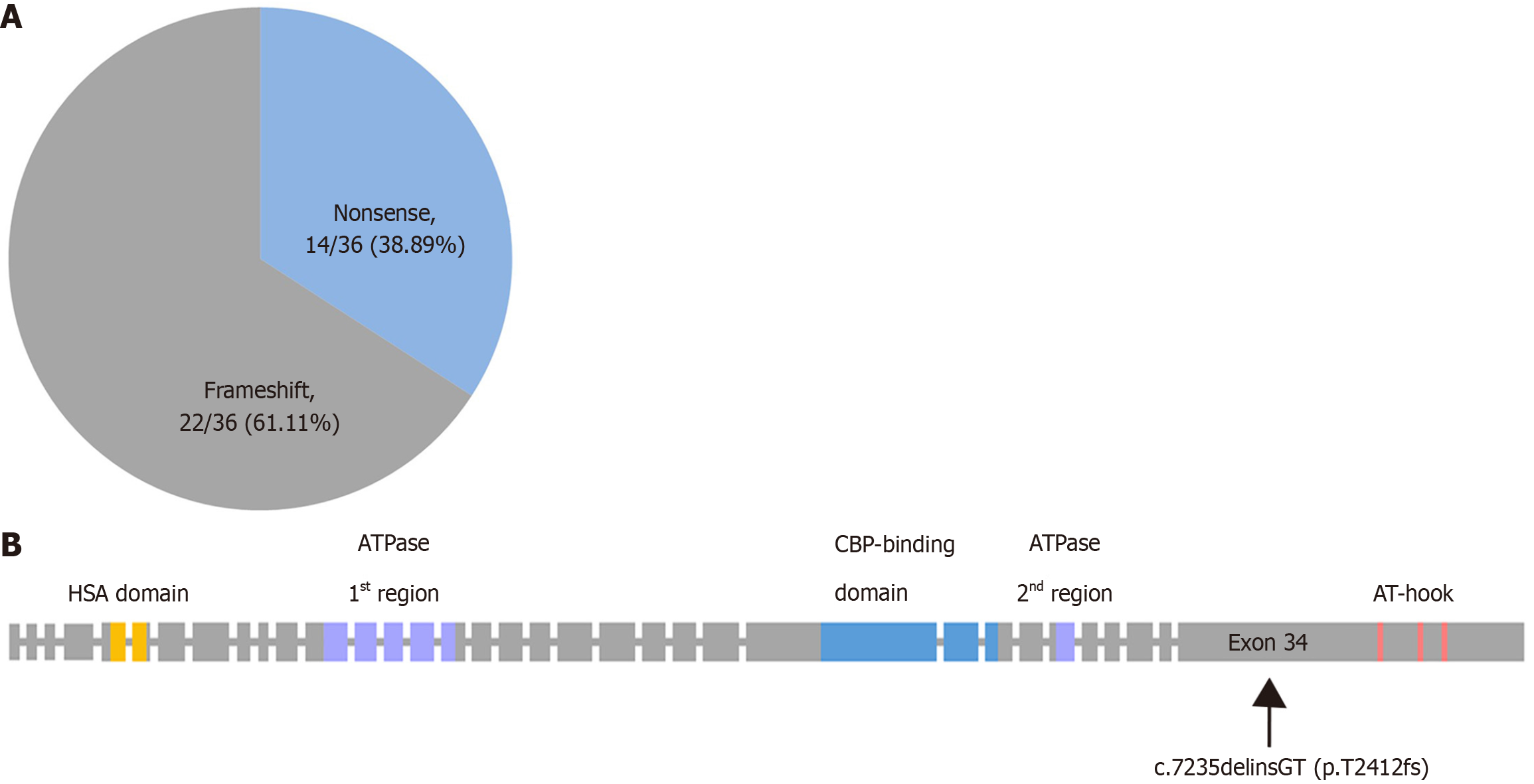
FHS is caused by heterozygous variants in the SRCAP gene. So far 36 pathogenic SRCAP gene variants associated with FHS had been reported in Human Gene Mutation Database (HGMD, accessed by Sep. 2023, Figure 2A). Currently, most of the pathogenic and likely pathogenic SRCAP variants associated with FHS reported in the literature fall inside the hot spots of exon 34, the final exon of the SRCAP gene, and the majority of these variants occur between codons 2407 and 2517. Only two studies have shown that SRCAP gene variants are located in exon 33[7,13]. Additionally, all of them were nonsense or frameshift variants in the terminal portion of the SRCAP gene predicting to result in the formation of C-terminally truncated SRCAP proteins. It has also been noted that the complete heterozygous deletion of the SRCAP gene has no features of FHS[6]. It has been suggested that the truncated SRCAP protein escaping nonsense-mediated mRNA decay, may disrupt the binding to the DNA and its chromatin targets, in a dominant negative manner[20]. The heterozygous SRCAP variant (c.7235delinsGT; p.Thr2412fs) of our patient was a de novo frameshift variant in exon 34 (Figure 2B), situated between codons 2407 and 2517, which is consistent with the previous reports[3]. In addition, this variant is not recorded in the HGMD, expanding the variant spectrum of FHS. On the other hand, some SRCAP missense variants could result in phenotypes distinct from FHS[21]. A similar phenomenon has been reported on NOTCH2, where truncating variants in the last exon are associated with Hajdu-Cheney syndrome, while missense variants in other exons result in Alagille syndrome[22]. Functional studies of biological mechanisms will eventually lead to a better under
The SRCAP gene is located at chromosome 16p11.2, which contains 3230 amino acids composed of a N-terminal HSA structural domain (helicase-SANT-associated domain), a SNF2-like ATPase structural domain, a CREBBP-binding domain and three C-terminal AT-hook structural domains. The SRCAP protein can participate in various aspects of transcriptional regulation through activating CREBBP, which plays a crucial role in essential cellular pathways[20]. Variants in SRCAP gene may result in an altered protein that destroys the normal activation of CREBBP, leading to FHS. Some authors have highlighted the phenotypic overlap between FHS and RSTS, which is caused by variants in the gene encoding CREBBP[23,24]. In addition to the CREBBP-binding domain, SRCAP carries three C-terminal AT-hook structural domains, which exert transcription independently of CREBBP. The AT-hook structural domains are responsible for chromatin organization and genes expression that control essential cellular processes. These variants of the SRCAP gene producing a truncated protein that result in loss of AT-hook structural domains. With these variants of the SRCAP gene, the expression levels of the genes controlling the onset of differentiation and developmental processes of the embryo are perturbed[20]. Taken together, the multiple functions of SRCAP may drive the phenotypic manifestation of FHS.
The discovery of the SRCAP variant (c.7235delinsGT; p.Thr2412fs) has far-reaching implications for public and global health. It may facilitate prenatal diagnosis, allowing for precise determination of whether a fetus carries this variant, thereby effectively preventing the birth of affected children. Additionally, genetic testing may help identify patients early, creating favorable conditions for early intervention and treatment, which significantly enhances the quality of life and recovery chances for patients.
In conclusion, a de novo heterozygous variant in SRCAP (c.7235delinsGT; p.Thr2412fs) was identified in a Chinese male patient with FHS using ES. This proband exhibited classical clinical features of FHS, including severe short stature, developmental delay, characteristic facial features, and additional malformations. This novel SRCAP variant expands the known variant spectrum in FHS and supports the clinical homogeneity of this disorder. Fortunately, we provided genetic counseling and prenatal genetic diagnosis for this family based on the known variant in the proband.
| 1. | Robinson PL, Shohat M, Winter RM, Conte WJ, Gordon-Nesbitt D, Feingold M, Laron Z, Rimoin DL. A unique association of short stature, dysmorphic features, and speech impairment (Floating-Harbor syndrome). J Pediatr. 1988;113:703-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 2. | White SM, Morgan A, Da Costa A, Lacombe D, Knight SJ, Houlston R, Whiteford ML, Newbury-Ecob RA, Hurst JA. The phenotype of Floating-Harbor syndrome in 10 patients. Am J Med Genet A. 2010;152A:821-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Nikkel SM, Dauber A, de Munnik S, Connolly M, Hood RL, Caluseriu O, Hurst J, Kini U, Nowaczyk MJ, Afenjar A, Albrecht B, Allanson JE, Balestri P, Ben-Omran T, Brancati F, Cordeiro I, da Cunha BS, Delaney LA, Destrée A, Fitzpatrick D, Forzano F, Ghali N, Gillies G, Harwood K, Hendriks YM, Héron D, Hoischen A, Honey EM, Hoefsloot LH, Ibrahim J, Jacob CM, Kant SG, Kim CA, Kirk EP, Knoers NV, Lacombe D, Lee C, Lo IF, Lucas LS, Mari F, Mericq V, Moilanen JS, Møller ST, Moortgat S, Pilz DT, Pope K, Price S, Renieri A, Sá J, Schoots J, Silveira EL, Simon ME, Slavotinek A, Temple IK, van der Burgt I, de Vries BB, Weisfeld-Adams JD, Whiteford ML, Wierczorek D, Wit JM, Yee CF, Beaulieu CL; FORGE Canada Consortium, White SM, Bulman DE, Bongers E, Brunner H, Feingold M, Boycott KM. The phenotype of Floating-Harbor syndrome: clinical characterization of 52 individuals with mutations in exon 34 of SRCAP. Orphanet J Rare Dis. 2013;8:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 4. | Amita M, Srivastava P, Agarwal D, Phadke SR. Floating Harbor Syndrome. Indian J Pediatr. 2016;83:896-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Feingold M. Thirty-two year follow-up of the first patient reported with the Floating-Harbor syndrome. Am J Med Genet A. 2006;140:782-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 6. | Hood RL, Lines MA, Nikkel SM, Schwartzentruber J, Beaulieu C, Nowaczyk MJ, Allanson J, Kim CA, Wieczorek D, Moilanen JS, Lacombe D, Gillessen-Kaesbach G, Whiteford ML, Quaio CR, Gomy I, Bertola DR, Albrecht B, Platzer K, McGillivray G, Zou R, McLeod DR, Chudley AE, Chodirker BN, Marcadier J; FORGE Canada Consortium, Majewski J, Bulman DE, White SM, Boycott KM. Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am J Hum Genet. 2012;90:308-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 136] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Kehrer M, Beckmann A, Wyduba J, Finckh U, Dufke A, Gaiser U, Tzschach A. Floating-Harbor syndrome: SRCAP mutations are not restricted to exon 34. Clin Genet. 2014;85:498-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Dong S, Han J, Chen H, Liu T, Huen MSY, Yang Y, Guo C, Huang J. The human SRCAP chromatin remodeling complex promotes DNA-end resection. Curr Biol. 2014;24:2097-2110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 9. | Johnston H, Kneer J, Chackalaparampil I, Yaciuk P, Chrivia J. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J Biol Chem. 1999;274:16370-16376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 67] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Monroy MA, Ruhl DD, Xu X, Granner DK, Yaciuk P, Chrivia JC. Regulation of cAMP-responsive element-binding protein-mediated transcription by the SNF2/SWI-related protein, SRCAP. J Biol Chem. 2001;276:40721-40726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Eissenberg JC, Wong M, Chrivia JC. Human SRCAP and Drosophila melanogaster DOM are homologs that function in the notch signaling pathway. Mol Cell Biol. 2005;25:6559-6569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Zhao B, Madden JA, Lin J, Berry GT, Wojcik MH, Zhao X, Brand H, Talkowski M, Lee EA, Agrawal PB. A neurodevelopmental disorder caused by a novel de novo SVA insertion in exon 13 of the SRCAP gene. Eur J Hum Genet. 2022;30:1083-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Seifert W, Meinecke P, Krüger G, Rossier E, Heinritz W, Wüsthof A, Horn D. Expanded spectrum of exon 33 and 34 mutations in SRCAP and follow-up in patients with Floating-Harbor syndrome. BMC Med Genet. 2014;15:127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754-1760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29052] [Cited by in RCA: 34455] [Article Influence: 2153.4] [Reference Citation Analysis (0)] |
| 15. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19696] [Cited by in RCA: 22507] [Article Influence: 2250.7] [Reference Citation Analysis (0)] |
| 16. | Rots D, Chater-Diehl E, Dingemans AJM, Goodman SJ, Siu MT, Cytrynbaum C, Choufani S, Hoang N, Walker S, Awamleh Z, Charkow J, Meyn S, Pfundt R, Rinne T, Gardeitchik T, de Vries BBA, Deden AC, Leenders E, Kwint M, Stumpel CTRM, Stevens SJC, Vermeulen JR, van Harssel JVT, Bosch DGM, van Gassen KLI, van Binsbergen E, de Geus CM, Brackel H, Hempel M, Lessel D, Denecke J, Slavotinek A, Strober J, Crunk A, Folk L, Wentzensen IM, Yang H, Zou F, Millan F, Person R, Xie Y, Liu S, Ousager LB, Larsen M, Schultz-Rogers L, Morava E, Klee EW, Berry IR, Campbell J, Lindstrom K, Pruniski B, Neumeyer AM, Radley JA, Phornphutkul C, Schmidt B, Wilson WG, Õunap K, Reinson K, Pajusalu S, van Haeringen A, Ruivenkamp C, Cuperus R, Santos-Simarro F, Palomares-Bralo M, Pacio-Míguez M, Ritter A, Bhoj E, Tønne E, Tveten K, Cappuccio G, Brunetti-Pierri N, Rowe L, Bunn J, Saenz M, Platzer K, Mertens M, Caluseriu O, Nowaczyk MJM, Cohn RD, Kannu P, Alkhunaizi E, Chitayat D, Scherer SW, Brunner HG, Vissers LELM, Kleefstra T, Koolen DA, Weksberg R. Truncating SRCAP variants outside the Floating-Harbor syndrome locus cause a distinct neurodevelopmental disorder with a specific DNA methylation signature. Am J Hum Genet. 2021;108:1053-1068. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 17. | Lacombe D, Patton MA, Elleau C, Battin J. Floating-Harbor syndrome: description of a further patient, review of the literature, and suggestion of autosomal dominant inheritance. Eur J Pediatr. 1995;154:658-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Wiltshire E, Wickremesekera A, Dixon J. Floating-Harbor syndrome complicated by tethered cord: a new association and potential contribution from growth hormone therapy. Am J Med Genet A. 2005;136:81-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 19. | Nagasaki K, Asami T, Sato H, Ogawa Y, Kikuchi T, Saitoh A, Ogata T, Fukami M. Long-term follow-up study for a patient with Floating-Harbor syndrome due to a hotspot SRCAP mutation. Am J Med Genet A. 2014;164A:731-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Messina G, Atterrato MT, Dimitri P. When chromatin organisation floats astray: the Srcap gene and Floating-Harbor syndrome. J Med Genet. 2016;53:793-797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | White-Brown A, Choufani S; Care4Rare Canada Consortium, Weksberg R, Dyment D. Missense variant in SRCAP with distinct DNA methylation signature associated with non-FLHS SRCAP-related neurodevelopmental disorder. Am J Med Genet A. 2023;191:2640-2646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 22. | Kamath BM, Bauer RC, Loomes KM, Chao G, Gerfen J, Hutchinson A, Hardikar W, Hirschfield G, Jara P, Krantz ID, Lapunzina P, Leonard L, Ling S, Ng VL, Hoang PL, Piccoli DA, Spinner NB. NOTCH2 mutations in Alagille syndrome. J Med Genet. 2012;49:138-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 23. | Spena S, Gervasini C, Milani D. Ultra-Rare Syndromes: The Example of Rubinstein-Taybi Syndrome. J Pediatr Genet. 2015;4:177-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 24. | Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 882] [Cited by in RCA: 855] [Article Influence: 28.5] [Reference Citation Analysis (0)] |