Published online Mar 26, 2024. doi: 10.12998/wjcc.v12.i9.1622
Peer-review started: January 7, 2024
First decision: January 16, 2024
Revised: January 23, 2024
Accepted: February 27, 2024
Article in press: February 27, 2024
Published online: March 26, 2024
Processing time: 78 Days and 1 Hours
The pathogenesis of ulcerative colitis (UC) is complex, and recent therapeutic advances remain unable to fully alleviate the condition.
To inform the development of novel UC treatments, bioinformatics was used to explore the autophagy-related pathogenesis associated with the active phase of UC.
The GEO database was searched for UC-related datasets that included healthy controls who met the screening criteria. Differential analysis was conducted to obtain differentially expressed genes (DEGs). Au
A total of 4822 DEGs were obtained, of which 58 were classified as DEARGs. SERPINA1, BAG3, HSPA5, CASP1, and CX3CL1 were identified as core targets. GO enrichment analysis revealed that DEARGs were primarily enriched in processes related to autophagy regulation and macroautophagy. KEGG enrichment analysis showed that DEARGs were predominantly associated with NOD-like receptor signaling and other signaling pathways. Disease enrichment analysis indicated that DEARGs were significantly linked to diseases such as malignant glioma and middle cerebral artery occlusion. Immune infiltration analysis demonstrated a higher presence of immune cells like activated memory CD4 T cells and follicular helper T cells in active UC pa
Autophagy is closely related to the active phase of UC and the potential targets obtained from the analysis in this study may provide new insight into the treatment of active UC patients.
Core Tip: This study used bioinformatics to explore the autophagy-related pathogenesis of ulcerative colitis (UC) during its active phase. A total of 58 differentially expressed autophagy-related genes (DEARGs) were found in gene expression datasets from UC patients and healthy controls. Of these, SERPINA1, BAG3, HSPA5, CASP1, and CX3CL1 were identified as core targets. Enrichment analysis highlighted the involvement of DEARGs in autophagy regulation, and macroautophagy, in addition to NOD-like receptor signaling and other pathways. These DEARGs were also shown to be associated with diseases like malignant glioma and middle cerebral artery occlusion. Immune infiltration analysis revealed an increased presence of immune cells, including activated memory CD4 T cells and follicular helper T cells in active UC pa
- Citation: Gong ZZ, Li T, Yan H, Xu MH, Lian Y, Yang YX, Wei W, Liu T. Exploring the autophagy-related pathogenesis of active ulcerative colitis. World J Clin Cases 2024; 12(9): 1622-1633
- URL: https://www.wjgnet.com/2307-8960/full/v12/i9/1622.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i9.1622
Ulcerative colitis (UC) is a chronic, recurrent inflammatory disease in humans that profoundly affects normal functioning[1]. It has both active and remission phases that are classified according to disease severity. UC is characterized by sym
Autophagy is a finely coordinated process that segregates misfolded proteins, damaged or aged organelles, and mutated proteins into double-membrane vesicles called autophagosomes. The autophagosomes later merge with lyso
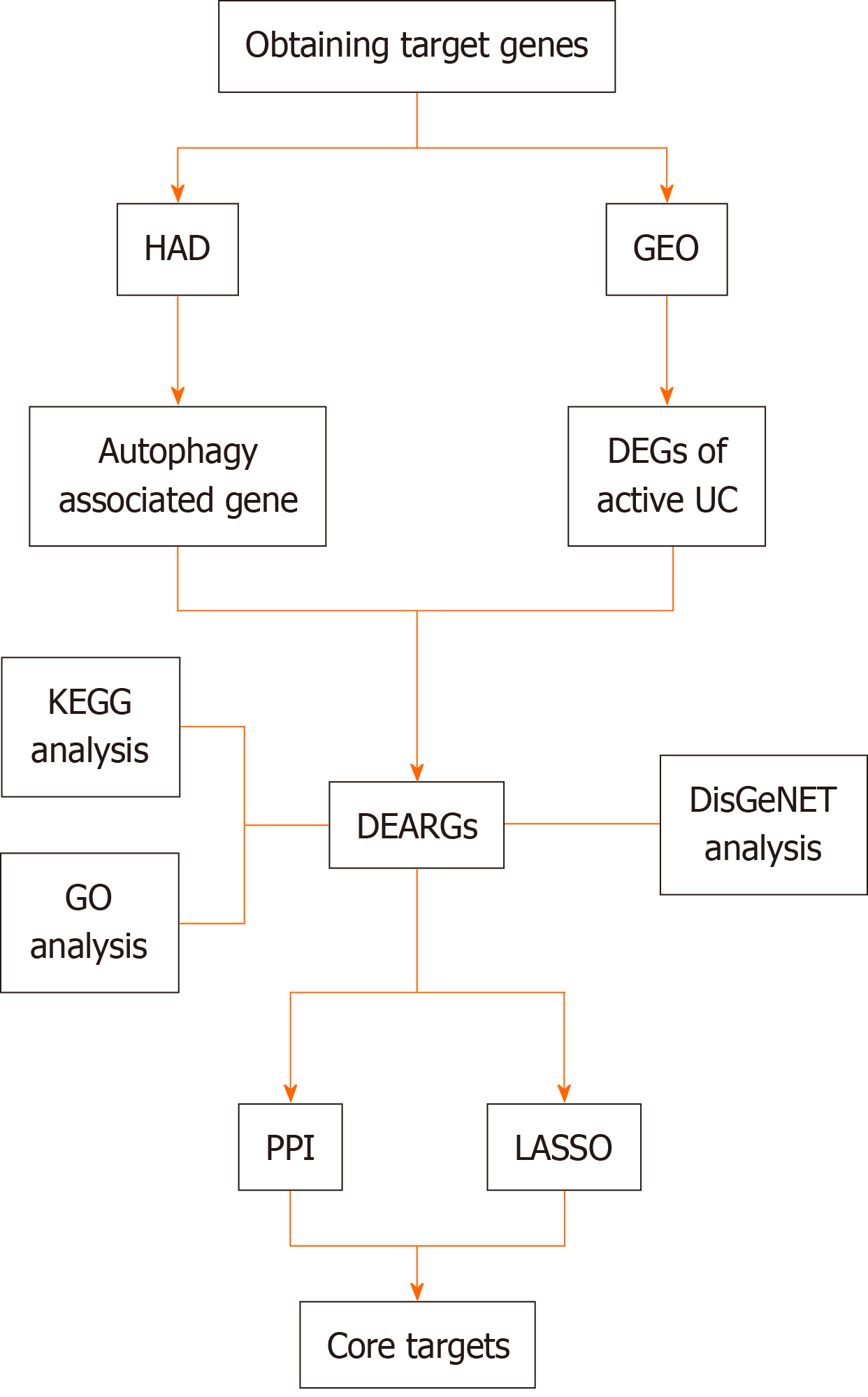
Given the importance of autophagy in preserving intestinal balance and the role of autophagy dysfunction in UC development, identifying autophagy-related disease predictors is essential for the design of new UC treatments. The current study uses bioinformatics to define gene expression patterns associated with the autophagy-related pathogenesis of active UC (Figure 1).
Datasets related to active UC which included normal control and active UC samples and had a sample size > 30 were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/gds/). The selected data set was normalized and the “limma” package was downloaded using “Bioconductor.” R 4.3.1 software was then used to perform differential gene analysis on targets identified in the dataset. A |log FC| ≥ 0.585 and an adj. P < 0.05 were used to obtain differentially expressed genes (DEGs).
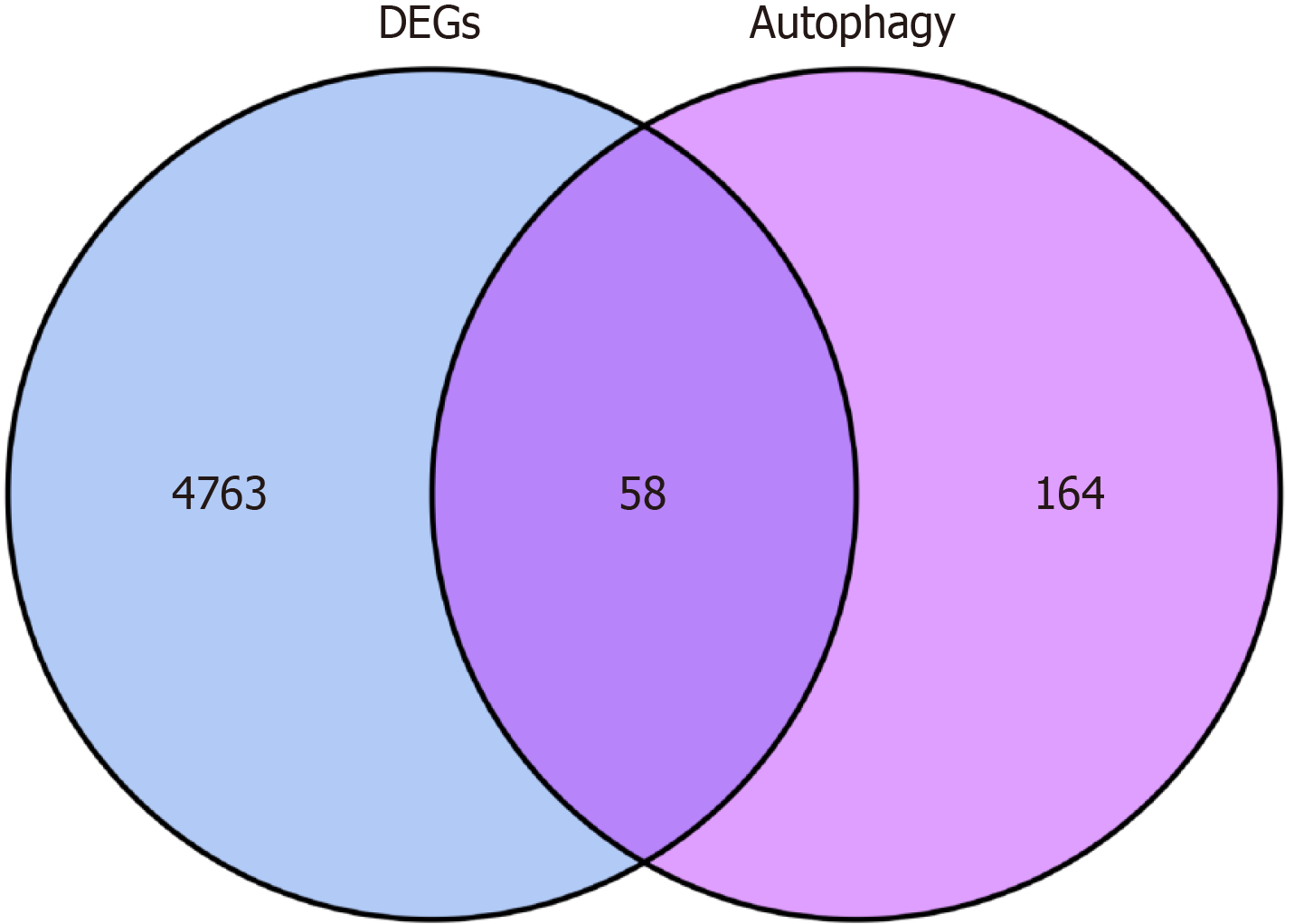
To obtain differentially expressed autophagy-related genes (DEARGs), autophagy-related genes were downloaded from the Human Autophagy Database (http://www.autophagy.lu/). Using the “Venn Diagram” package in R, autophagy-related targets were intersected with the DEGs, identifying DEARGs as the central targets for further analysis. DEARG heat maps were generated using the “limma” and “pheatmap” packages.
The immune microenvironment is typically composed of immune cells, inflammatory cells, fibroblasts, and mesenchymal stem cells, along with various cytokines and chemokines. Assessing immune cell infiltration is vital for predicting disease progression and treatment response. Several methods exist to analyze immune cell infiltration, including CiberSort, an inverse convolution algorithm developed by BinderG. This method calculates the cellular composition of complex tissues based on normalized gene expression data, allowing specific cell types to be quantified. The CiberSort deconvolution algorithm was used with 100 simulations and subsequent analyses were conducted with a significance threshold of P < 0.05 to determine the proportion of immune cells in different samples. The results were visualized using the “ggpubr” package in R.
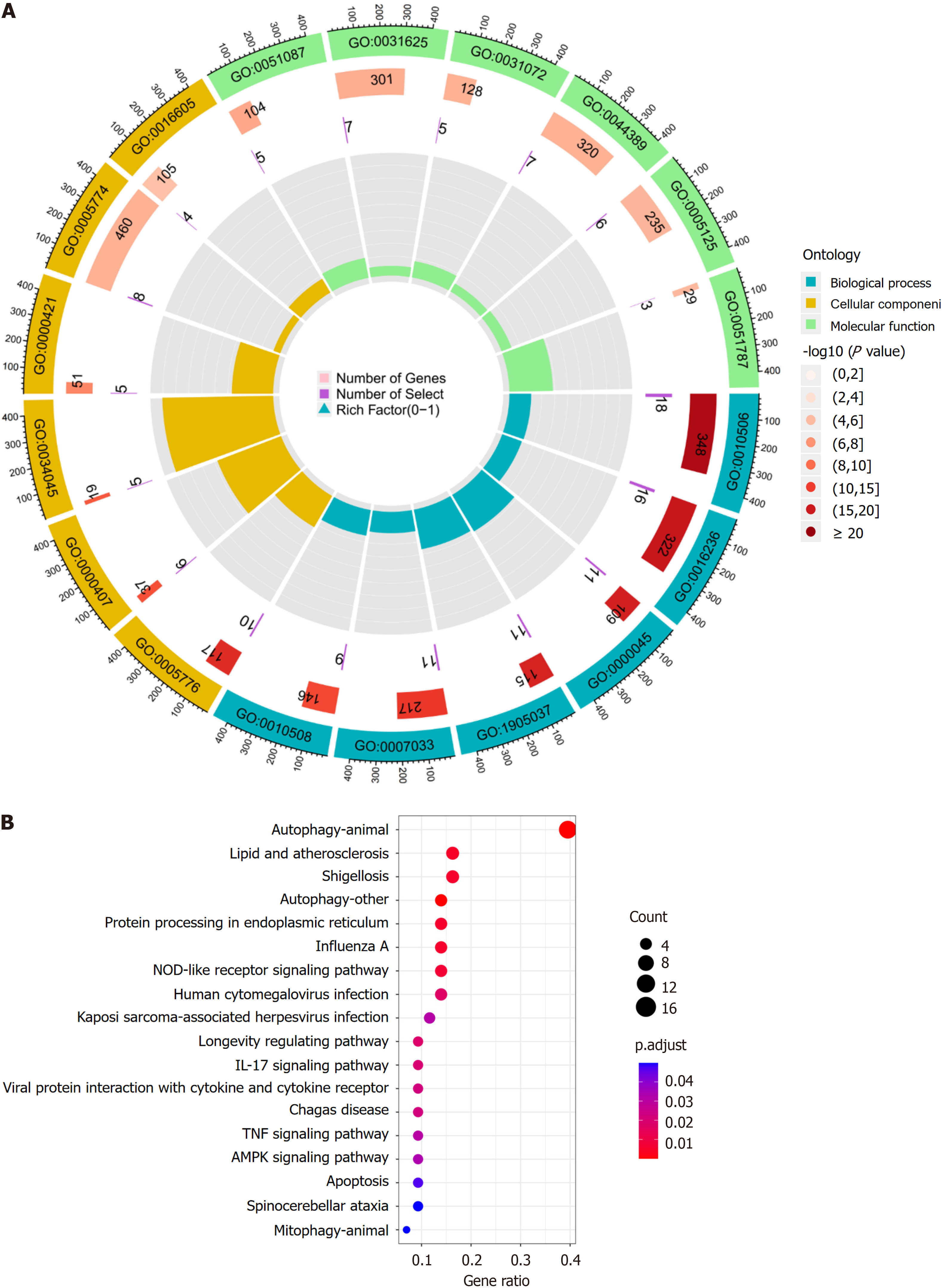
Gene ontology (GO) analysis categorizes genes into biological processes (BP), molecular functions (MF), and cellular components (CC), which help to inform their biological functions. The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database that integrates genomic, chemical, and systemic information. It is often used for the functional annotation of genes to understand their associated activities and pathways of action. To further understand the target functions of autophagy in patients with active UC and the associated signaling pathways, the “clusterProfiler” package was downloaded from Bioconductor, and GO and KEGG enrichment analysis of the DEARGs was conducted using R. The “clusterProfiler” package was downloaded from “Bioconductor” and the DEARGs were analyzed by GO and KEGG enrichment analysis using R with a threshold value of P < 0.05.
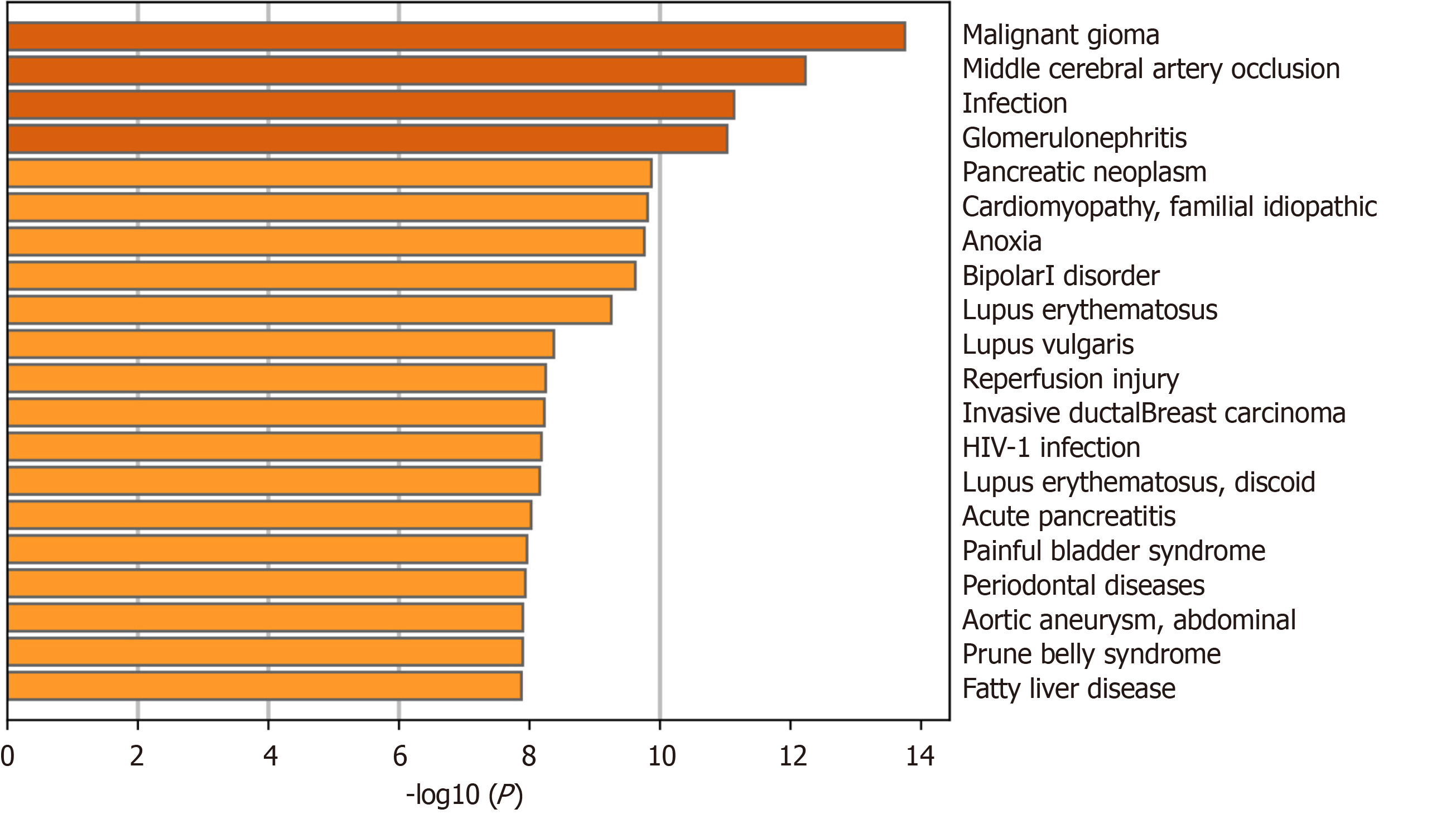
To explore the role of autophagy in UC-related diseases, DEARGs were input into the Metascape platform (https://metascape.org/) using “H. sapiens” as the species setting for both “Input” and “Analysis.” The “Summary of enrichment analysis in DisGeNET” was then exported.
For more precise identification of the core targets, the least absolute shrinkage and selection operator (LASSO) algorithm was used along with the construction of a protein-protein interaction (PPI) network to refine DEARG selection and pre
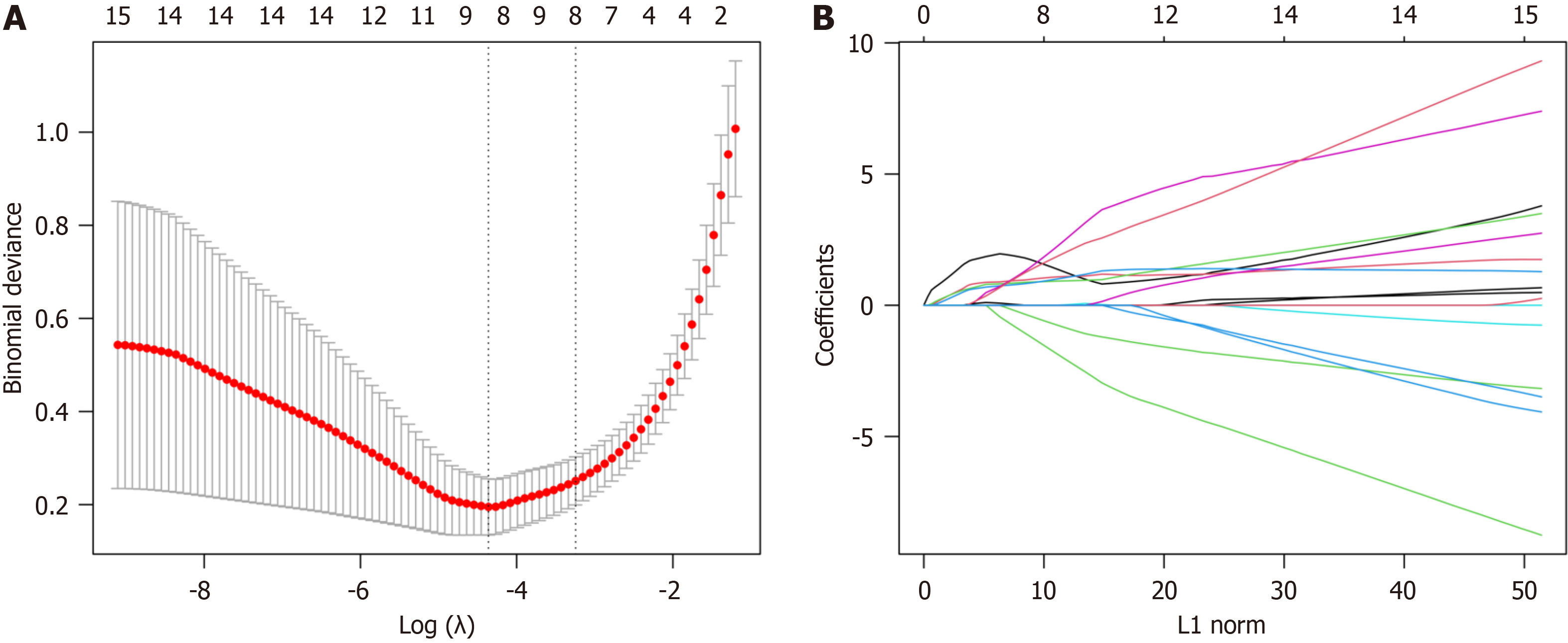
The LASSO algorithm was used for DEARG validation and feature gene selection using the “glmnet” package in R. The identified genes were then uploaded to the String database (https://cn.string-db.org/) with a “minimum required interaction score” of 0.15 and the results were imported into Cytoscape 3.9.1. To further refine the selection, the “Cyto
GEO database screening identified two UC-related datasets: GSE87466 and GSE53306. GSE53306 includes data on differential gene expression between the active and quiescent stages of UC, providing insight into the disease characteristics. The dataset, which has information on 40 individuals, including 16 active UC cases and 12 normal controls, was pub
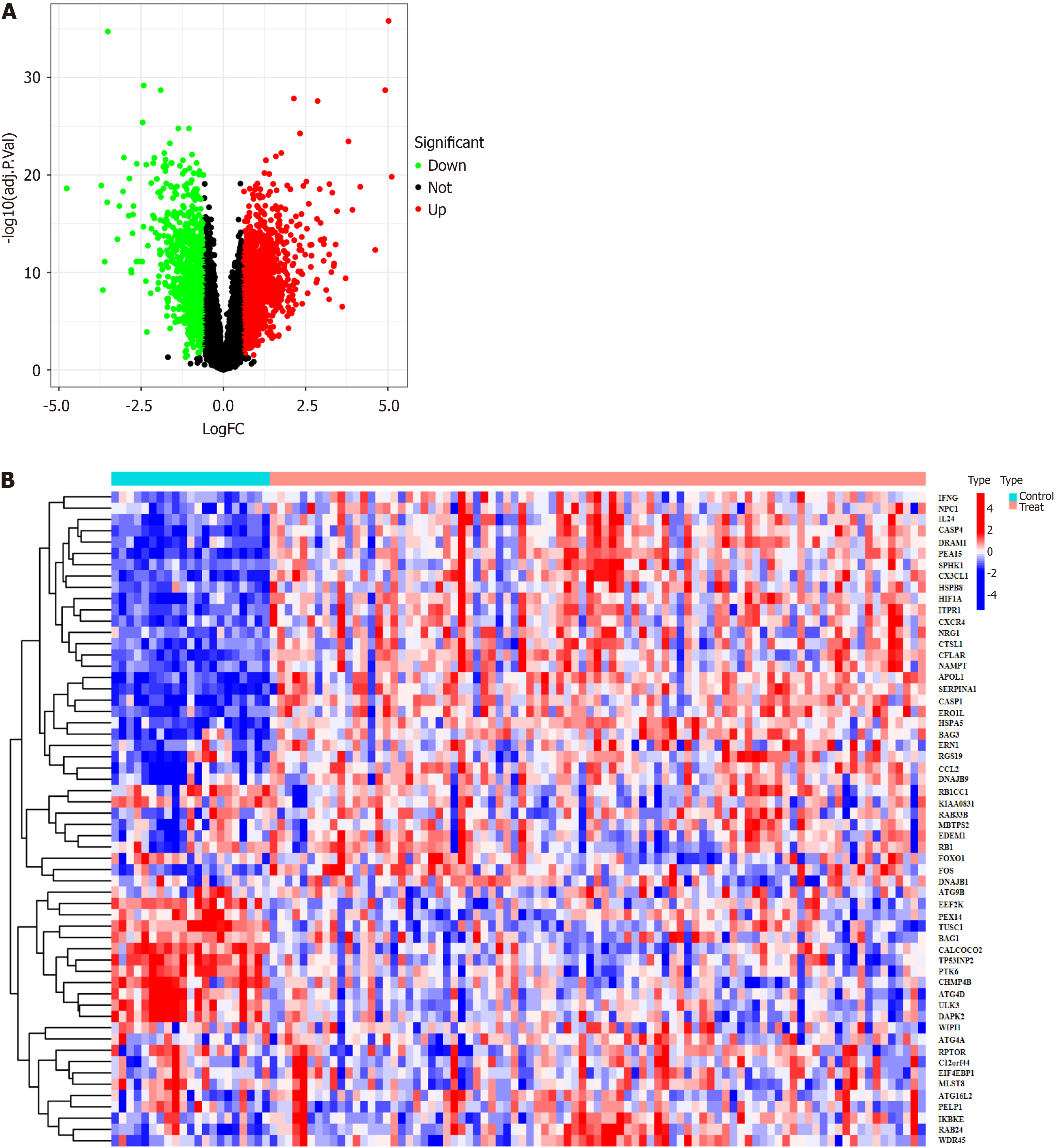
A total of 232 autophagy-related genes were obtained from the Human Autophagy Database (http://www.autophagy.lu/). These autophagy-related genes were intersected with the DEGs, resulting in 58 DEARGs (Figure 2). R was then used to analyze the DEARGs and generate heat map and volcano map (Figure 3).
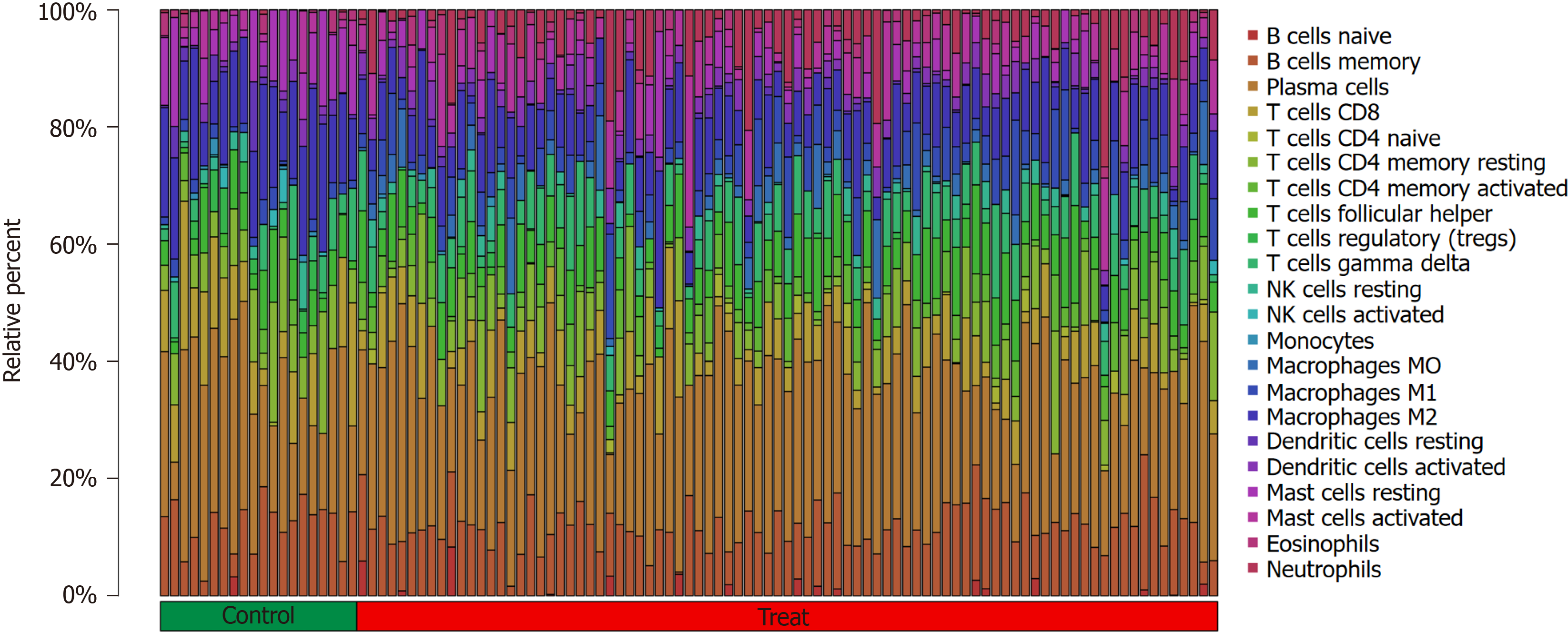
The Cibersort algorithm was used to evaluate immune cell infiltration in two distinct immune states. The following immune cell types were more abundant in active UC cases than in healthy controls: activated memory CD4 T cells, fo
The following immune cell types were significantly higher in the UC group than in the healthy control group: activated memory CD4 T cells (P < 0.001), follicular helper T cells (P < 0.05), gamma delta T cells (P < 0.05), M0 macrophages (P < 0.001), M1 macrophages (P < 0.001), activated dendritic cells (P < 0.001), activated mast cells (P < 0.001), and neutrophils (P <0.001). Meanwhile, CD8 T cells (P < 0.05), resting memory CD4 T cells (P < 0.05), regulatory T cells (Tregs) (P < 0.001), activated NK cells (P < 0.01), monocytes (P < 0.01), M2 macrophages (P < 0.001), resting dendritic cells (P < 0.05), and resting mast cells (P < 0.001) were significantly higher in the healthy control group than in the active UC group. No sig
BP analysis revealed that the DEARGs were primarily associated with the regulation of autophagy, macroautophagy, autophagosome assembly, autophagosome organization, and vacuole organization. CC analysis showed that the DE
The Metascape “Summary of enrichment analysis in DisGeNET” revealed that the DEARGs were mainly enriched in malignant glioma, middle cerebral artery occlusion, infection, glomerulonephritis, and other diseases (Figure 6).
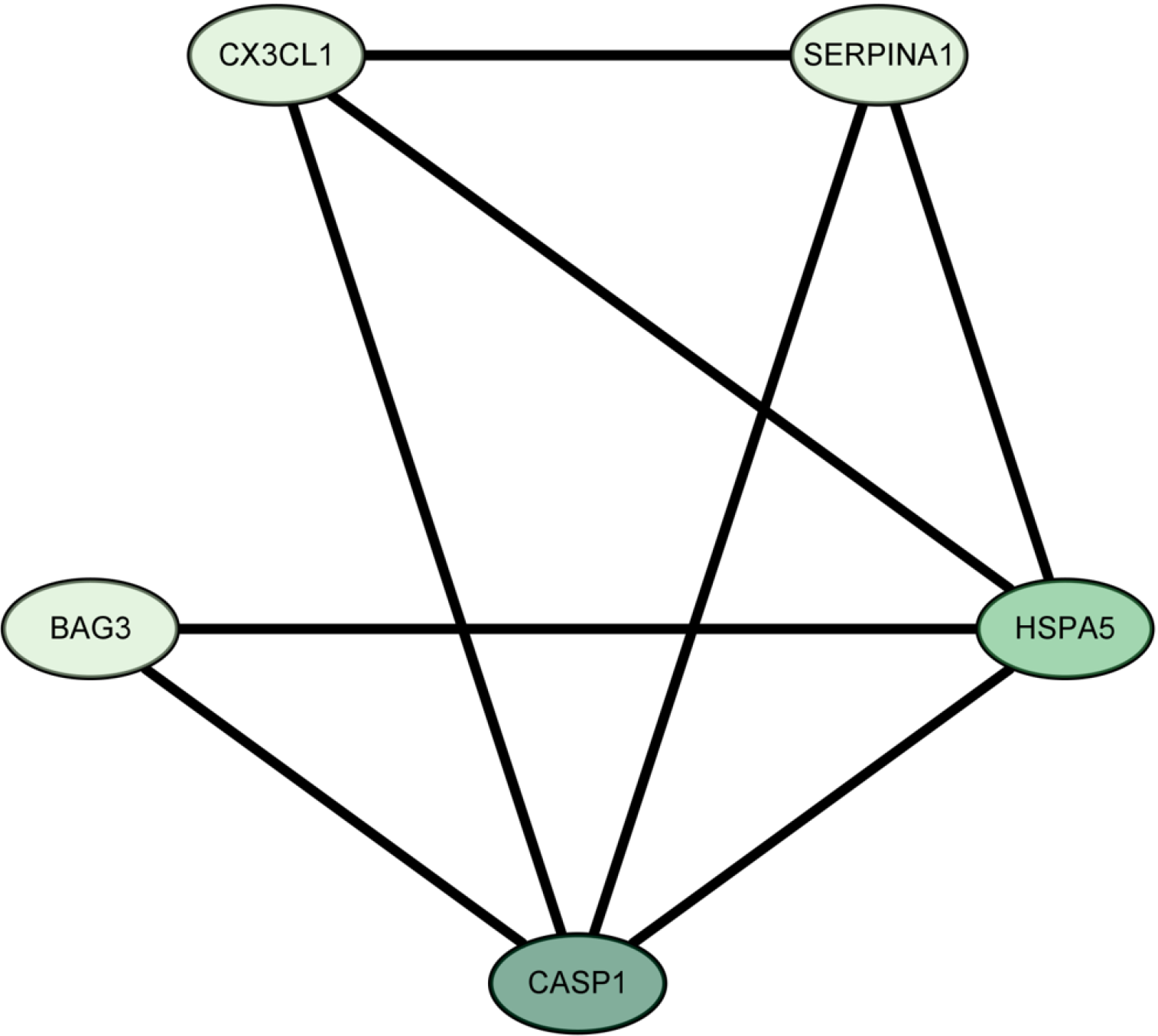
The LASSO algorithm narrowed the range of DEARGs and identified 13 targets: proliferation and apoptosis adaptor protein 15 (PEA15), heat shock 70-kDa protein 5 (HSPA5), caspase 1 (CASP1), serine protease inhibitor A1 (SERPINA1), C-X3-C chemokine ligand 1 (CX3CL1), Bcl2-associated athanogene 3 (BAG3), tumor protein p53 inducible nuclear protein 2 (TP53INP2), and peroxisomal biogenesis factor 14 (PEX14) (Figure 7). Their relationships were further established using the String database. The Cytoscape 3.9.1 software “CytoNCA” plug-in was used to sort the 13 targets according to their degree values, and the top five were selected as the core targets. The Fold Change (logFC) of these targets was obtained from the difference analysis results. All five were up-regulated and had the following parameters: SERPINA1 (logFC = 1.051), BAG3 (logFC = 0.661), HSPA5 (logFC = 0.790), CASP1 (logFC = 1.231), and CX3CL1 (logFC = 0.837) (Figure 8).
The current study identified HSPA5, CASP1, SERPINA1, CX3CL1, and BAG3 as core autophagy-related targets in active UC, all of which were upregulated during the disease. Key signaling pathways linked to these targets included auto
The results, including those predicted using core targeting and immune infiltration analysis, are supported by existing literature. R-HSPA5 is a specific form of HSPA5 that is localized in the endoplasmic reticulum (ER) and shown to play a critical role in autophagy-mediated lysosomal protein hydrolysis. Significant overexpression of HSPA5 mRNA and pro
The disease enrichment analysis results discussed here are confirmed by prior studies. Different stages of IBD are linked to the development of thrombosis, with IBD episodes or activity serving as a primary risk factor[33]. The predicted core genes are critical to the pathogenesis+ADs- hSPA5, for example, offering neuroprotection in ischemic strokes[34]. BAG3 overexpression is shown to improve neurological outcomes associated with middle cerebral artery embolism in mice, reducing infarct volume and enhancing cell survival by activating autophagy and inhibiting apoptosis[35]. Ische
In summary, HSPA5, CASP1, SERPINA1, CX3CL1, and BAG3 were identified as core autophagy-related targets that are upregulated in active UC patients. These targets are associated with key signaling pathways, including autophagy in ani
The etiology of ulcerative colitis (UC), a chronic inflammatory bowel disease (IBD), remains poorly understood. The pathogenesis of UC is complex and is influenced by genetic, environmental, and immune-related factors. While some recent progress has been made in the development of effective UC treatments, few patients experience complete relief of their symptoms. Thus, finding new therapeutic avenues to improve UC patient quality of life remains an urgent need. Autophagy is a cellular self-degradation and repair process that can help remove harmful proteins and organelles from cells and maintain intracellular homeostasis. Recent studies suggest that autophagy may play a key role in the patho
The motivation of this study was to provide an in-depth investigation of the autophagy-related pathogenesis of active phase UC. Bioinformatics analysis was used to better understand whether autophagy plays a key role in active UC and which autophagy-related genes may contribute to the disease process.
This study sought to provide new ideas and potential therapeutic targets for the treatment of active UC to better understand the pathogenesis of the disease and improve clinical symptoms.
A bioinformatics approach was used to compare gene expression data between patients with active UC and healthy controls to identify core genes associated with autophagy and to obtain more information about the role of autophagy in this disease.
HSPA5, CASP1, SERPINA1, CX3CL1, and BAG3 were identified as core targets associated with autophagy-related pathogenesis in active UC, all of which were upregulated. Key signaling pathways linked to these targets include autophagy in animals, other autophagy pathways, and lipids and atherosclerosis pathways. DisGeNET enrichment ana
HSPA5, CASP1, SERPINA1, CX3CL1, and BAG3 were identified as core autophagy-related targets in active UC patients, all of which were upregulated. These targets are associated with key signaling pathways, including autophagy in animals, other autophagy pathways, and lipid and atherosclerosis pathways. DisGeNET enrichment analysis revealed a significant connection between middle cerebral artery occlusion, glomerulonephritis, and the autophagy-related patho
Future research in this field should focus on better understanding the molecular mechanisms by which HSPA5, CASP1, SERPINA1, CX3CL1, and BAG3 contribute to autophagy in patients with active UC. Investigating the specific roles of these core targets in UC pathogenesis and their interactions with the identified key signaling molecules should be a priority. Interventions that target the core autophagy-related genes and pathways could offer promising treatment op
We thank the GEO database for providing their platforms and contributors for their valuable datasets.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lin SR, Taiwan S-Editor: Gong ZM L-Editor: A P-Editor: Xu ZH
| 1. | Mandlik DS, Mandlik SK, Patel S. Protective effect of sarsasapogenin in TNBS induced ulcerative colitis in rats associated with downregulation of pro-inflammatory mediators and oxidative stress. Immunopharmacol Immunotoxicol. 2021;43:571-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 2. | Kobayashi T, Siegmund B, Le Berre C, Wei SC, Ferrante M, Shen B, Bernstein CN, Danese S, Peyrin-Biroulet L, Hibi T. Ulcerative colitis. Nat Rev Dis Primers. 2020;6:74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 988] [Article Influence: 197.6] [Reference Citation Analysis (0)] |
| 3. | Le Berre C, Honap S, Peyrin-Biroulet L. Ulcerative colitis. Lancet. 2023;402:571-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 576] [Reference Citation Analysis (104)] |
| 4. | Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1338] [Cited by in RCA: 1274] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 5. | Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20:521-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 509] [Article Influence: 72.7] [Reference Citation Analysis (0)] |
| 6. | Cao SS. Endoplasmic reticulum stress and unfolded protein response in inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:636-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 7. | Kim S, Lee JY, Shin SG, Kim JK, Silwal P, Kim YJ, Shin NR, Kim PS, Won M, Lee SH, Kim SY, Sasai M, Yamamoto M, Kim JM, Bae JW, Jo EK. ESRRA (estrogen related receptor alpha) is a critical regulator of intestinal homeostasis through activation of autophagic flux via gut microbiota. Autophagy. 2021;17:2856-2875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 8. | Shi W, Peng K, Yu H, Wang Z, Xia S, Xiao S, Tian D, Vallance BA, Yu Q. Autotaxin (ATX) inhibits autophagy leading to exaggerated disruption of intestinal epithelial barrier in colitis. Biochim Biophys Acta Mol Basis Dis. 2023;1869:166647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 9. | Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, Barnich N, Ng SC, Colombel JF. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut. 2018;67:574-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 370] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 10. | Shao BZ, Yao Y, Zhai JS, Zhu JH, Li JP, Wu K. The Role of Autophagy in Inflammatory Bowel Disease. Front Physiol. 2021;12:621132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 11. | Shen T, Li S, Cai LD, Liu JL, Wang CY, Gan WJ, Li XM, Wang JR, Sun LN, Deng M, Liu YH, Li JM. Erbin exerts a protective effect against inflammatory bowel disease by suppressing autophagic cell death. Oncotarget. 2018;9:12035-12049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Cha-Molstad H, Sung KS, Hwang J, Kim KA, Yu JE, Yoo YD, Jang JM, Han DH, Molstad M, Kim JG, Lee YJ, Zakrzewska A, Kim SH, Kim ST, Kim SY, Lee HG, Soung NK, Ahn JS, Ciechanover A, Kim BY, Kwon YT. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol. 2015;17:917-929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 203] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 13. | Bogaert S, De Vos M, Olievier K, Peeters H, Elewaut D, Lambrecht B, Pouliot P, Laukens D. Involvement of endoplasmic reticulum stress in inflammatory bowel disease: a different implication for colonic and ileal disease? PLoS One. 2011;6:e25589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Lie MRKL, van der Giessen J, Fuhler GM, de Lima A, Peppelenbosch MP, van der Ent C, van der Woude CJ. Low dose Naltrexone for induction of remission in inflammatory bowel disease patients. J Transl Med. 2018;16:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 15. | Chiu HW, Chen CH, Chang JN, Hsu YH. Erratum to: Far-infrared promotes burn wound healing by suppressing NLRP3 inflammasome caused by enhanced autophagy. J Mol Med (Berl). 2016;94:821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Siegmund B, Lehr HA, Fantuzzi G, Dinarello CA. IL-1 beta -converting enzyme (caspase-1) in intestinal inflammation. Proc Natl Acad Sci U S A. 2001;98:13249-13254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 350] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 17. | Bauer C, Duewell P, Lehr HA, Endres S, Schnurr M. Protective and aggravating effects of Nlrp3 inflammasome activation in IBD models: influence of genetic and environmental factors. Dig Dis. 2012;30 Suppl 1:82-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 18. | Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, Tschopp J, Endres S, Latz E, Schnurr M. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59:1192-1199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 719] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 19. | Soendergaard C, Nielsen OH, Seidelin JB, Kvist PH, Bjerrum JT. Alpha-1 antitrypsin and granulocyte colony-stimulating factor as serum biomarkers of disease severity in ulcerative colitis. Inflamm Bowel Dis. 2015;21:1077-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 20. | Li Y, Chen Y, Xiao X, Deng S, Kuang J, Wang Y. CX3CL1 represses autophagy via CX3CR1/ CaMKIIδ/HDAC4/Rubicon axis and exacerbates chronic intermittent hypoxia induced Kupffer cell apoptosis. Cell Signal. 2023;111:110873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 21. | Kostadinova FI, Baba T, Ishida Y, Kondo T, Popivanova BK, Mukaida N. Crucial involvement of the CX3CR1-CX3CL1 axis in dextran sulfate sodium-mediated acute colitis in mice. J Leukoc Biol. 2010;88:133-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Ishida Y, Hayashi T, Goto T, Kimura A, Akimoto S, Mukaida N, Kondo T. Essential involvement of CX3CR1-mediated signals in the bactericidal host defense during septic peritonitis. J Immunol. 2008;181:4208-4218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 23. | Nemoto Y, Kanai T, Takahara M, Oshima S, Nakamura T, Okamoto R, Tsuchiya K, Watanabe M. Bone marrow-mesenchymal stem cells are a major source of interleukin-7 and sustain colitis by forming the niche for colitogenic CD4 memory T cells. Gut. 2013;62:1142-1152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 24. | Wang X, Zhu Y, Zhang M, Hou J, Wang H, Jiang Y, Gao P. The shifted balance between circulating follicular regulatory T cells and follicular helper T cells in patients with ulcerative colitis. Clin Sci (Lond). 2017;131:2933-2945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Inagaki-Ohara K, Chinen T, Matsuzaki G, Sasaki A, Sakamoto Y, Hiromatsu K, Nakamura-Uchiyama F, Nawa Y, Yoshimura A. Mucosal T cells bearing TCRgammadelta play a protective role in intestinal inflammation. J Immunol. 2004;173:1390-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 130] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 26. | Pastorelli L, Garg RR, Hoang SB, Spina L, Mattioli B, Scarpa M, Fiocchi C, Vecchi M, Pizarro TT. Epithelial-derived IL-33 and its receptor ST2 are dysregulated in ulcerative colitis and in experimental Th1/Th2 driven enteritis. Proc Natl Acad Sci U S A. 2010;107:8017-8022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 347] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 27. | Chu HQ, Li J, Huang HP, Hao WD, Duan LP, Wei XT. Protective effects of tranilast on oxazolone-induced rat colitis through a mast cell-dependent pathway. Dig Liver Dis. 2016;48:162-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Lopetuso LR, De Salvo C, Pastorelli L, Rana N, Senkfor HN, Petito V, Di Martino L, Scaldaferri F, Gasbarrini A, Cominelli F, Abbott DW, Goodman WA, Pizarro TT. IL-33 promotes recovery from acute colitis by inducing miR-320 to stimulate epithelial restitution and repair. Proc Natl Acad Sci U S A. 2018;115:E9362-E9370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 29. | Lai LJ, Shen J, Ran ZH. Natural killer T cells and ulcerative colitis. Cell Immunol. 2019;335:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 30. | Cherfane CE, Gessel L, Cirillo D, Zimmerman MB, Polyak S. Monocytosis and a Low Lymphocyte to Monocyte Ratio Are Effective Biomarkers of Ulcerative Colitis Disease Activity. Inflamm Bowel Dis. 2015;21:1769-1775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 31. | Pinelli DF, Ford ML. Novel insights into anti-CD40/CD154 immunotherapy in transplant tolerance. Immunotherapy. 2015;7:399-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 32. | Rescigno M. Dendritic cell-epithelial cell crosstalk in the gut. Immunol Rev. 2014;260:118-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 33. | Barclay AR, Keightley JM, Horrocks I, Garrick V, McGrogan P, Russell RK. Cerebral thromboembolic events in pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:677-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 34. | Peng ZF, Zhang NB, Meng J, Zhang JH. Early Aerobic Exercise Promotes Neurological Function Recovery of Rats after Cerebral Ischemia/Reperfusion by Upregulating the Expression of Heat Shock Protein A5. Curr Med Sci. 2022;42:267-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 35. | Liu X, Ye Q, Huang Z, Li X, Zhang L, Liu X, Wu YC, Brockmeier U, Hermann DM, Wang YC, Ren L. BAG3 Overexpression Attenuates Ischemic Stroke Injury by Activating Autophagy and Inhibiting Apoptosis. Stroke. 2023;54:2114-2125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 37] [Reference Citation Analysis (0)] |
| 36. | He HY, Ren L, Guo T, Deng YH. Neuronal autophagy aggravates microglial inflammatory injury by downregulating CX3CL1/fractalkine after ischemic stroke. Neural Regen Res. 2019;14:280-288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 37. | Choi ME. Autophagy in Kidney Disease. Annu Rev Physiol. 2020;82:297-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 112] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 38. | Minami S, Yamamoto T, Yamamoto-Imoto H, Isaka Y, Hamasaki M. Autophagy and kidney aging. Prog Biophys Mol Biol. 2023;179:10-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 39. | Oikonomou K, Kapsoritakis A, Eleftheriadis T, Stefanidis I, Potamianos S. Renal manifestations and complications of inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:1034-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 40. | Yoshimoto S, Nakatani K, Iwano M, Asai O, Samejima K, Sakan H, Terada M, Harada K, Akai Y, Shiiki H, Nose M, Saito Y. Elevated levels of fractalkine expression and accumulation of CD16+ monocytes in glomeruli of active lupus nephritis. Am J Kidney Dis. 2007;50:47-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 90] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 41. | Hirono K, Imaizumi T, Aizawa T, Watanabe S, Tsugawa K, Shiratori T, Kawaguchi S, Seya K, Matsumiya T, Ito E, Tanaka H. Endothelial expression of fractalkine (CX3CL1) is induced by Toll-like receptor 3 signaling in cultured human glomerular endothelial cells. Mod Rheumatol. 2020;30:1074-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 42. | Ding X, Ren Y, He X. IFN-I Mediates Lupus Nephritis From the Beginning to Renal Fibrosis. Front Immunol. 2021;12:676082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 43. | Xiong J, Wang Y, Shao N, Gao P, Tang H, Su H, Zhang C, Meng XF. The Expression and Significance of NLRP3 Inflammasome in Patients with Primary Glomerular Diseases. Kidney Blood Press Res. 2015;40:344-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 44. | Zhang C, Boini KM, Xia M, Abais JM, Li X, Liu Q, Li PL. Activation of Nod-like receptor protein 3 inflammasomes turns on podocyte injury and glomerular sclerosis in hyperhomocysteinemia. Hypertension. 2012;60:154-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |