Published online Mar 26, 2024. doi: 10.12998/wjcc.v12.i9.1606
Peer-review started: October 19, 2023
First decision: January 25, 2024
Revised: February 2, 2024
Accepted: March 4, 2024
Article in press: March 4, 2024
Published online: March 26, 2024
Processing time: 157 Days and 17.6 Hours
Circular RNAs (circRNAs) are involved in the pathogenesis of many diseases through competing endogenous RNA (ceRNA) regulatory mechanisms.
To investigate a circRNA-related ceRNA regulatory network and a new predictive model by circRNA to understand the diagnostic mechanism of circRNAs in ulcerative colitis (UC).
We obtained gene expression profiles of circRNAs, miRNAs, and mRNAs in UC from the Gene Expression Omnibus dataset. The circRNA-miRNA-mRNA net
A circRNA-miRNA-mRNA regulatory network was obtained, containing 12 circRNAs, three miRNAs, and 38 mRNAs. Two optimal prognostic-related differentially expressed circRNAs, hsa_circ_0085323 and hsa_circ_0036906, were included to construct a predictive nomogram. The model showed good discrimination, with a C-index of 1(> 0.9, high accuracy). ROC and DCA suggested that the nomogram had a beneficial diagnostic ability.
This novel predictive nomogram incorporating hsa_circ_0085323 and hsa_circ_0036906 can be conveniently used to predict the risk of UC. The circRNa-miRNA-mRNA network in UC could be more clinically significant.
Core Tip: In this study, we constructed a circRNA-miRNA-mRNA regulatory network to investigate the probable mechanism of circRNAs in ulcerative colitis. Initially, differentially expressed circRNAs (DEcircRNAs), differentially expressed miRNAs (DEmiRNAs), and differentially expressed mRNAs (DEmRNAs) were retrieved using the RNA expression spectrum of circRNA, miRNA, and mRNA in the Gene Expression Omnibus dataset. The competing endogenous RNA (ceRNA) network was then established after determining the DEcircRNA-DEmiRNA and DEmiRNA-DEmRNA interactions using bioinformatics analysis methods. Functional enrichment analysis was performed to evaluate the biological functions of DEmRNAs in the ceRNA network. Ultimately, DEcircRNAs in the circRNA-miRNA-mRNA network were used to construct a new diagnostic model.
- Citation: Yuan YY, Wu H, Chen QY, Fan H, Shuai B. Construction of the underlying circRNA-miRNA-mRNA regulatory network and a new diagnostic model in ulcerative colitis by bioinformatics analysis. World J Clin Cases 2024; 12(9): 1606-1621
- URL: https://www.wjgnet.com/2307-8960/full/v12/i9/1606.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i9.1606
Ulcerative colitis (UC) is an inflammatory bowel disease (IBD) that affects the mucosa and submucosa of the colon and rectum and sometimes a small segment of the terminal ileum. Severe UC is characterised by fever, abnormal C-reactive protein and haemoglobin levels, and other signs of intestinal inflammation[1]. In addition to these clinical symptoms, the diagnosis of UC involves fibreoptic colonoscopy, double-contrast angiography with barium enema, and characteristic biomarkers. Following the expansion of treatment facilities for ulcerative colitis, the number of drugs with new targets is expected to increase rapidly in future[2,3]. However, despite the progress in biology, the information regarding the pathogenesis of UC continues to be limited. Therefore, exploring the molecular mechanism of UC to formulate appropriate therapeutic strategies and diagnose the disease is important.
Circular RNAs (circRNAs), a type of ncRNA, are formed from a covalently closed loop by 50-30 back-splicing or specific splicing[4]. CircRNAs regulate mRNA translation by sponging miRNAs as competing endogenous RNA (ceRNA) or acting as mRNA traps[5,6]. The biological mechanisms of circRNAs fall into six categories: (1) Acting as circRNA sponge miRNA; (2) acting as circRNA sponge RNA-binding protein RBP; (3) acting as a protein-RNA complex member; (4) acting as a protein scaffold; (5) recruiting trans-regulatory factors to regulate transcription; and (6) translation of peptides[7,8]. Previous studies have shown that circRNAs are closely related to many human diseases such as tumours, ischaemic heart disease, rheumatoid arthritis, and degenerative diseases[9]. However, studies on the relationship between UC and circRNAs, especially the expression matrix of circRNAs in UC species, are few[10]. Therefore, whether circRNAs can serve as clinical biomarkers of UC needs to be addressed. We believe that the contributions of circRNAs to UC will become a hotspot in future studies[11]. Commonly, miRNAs recognise the miRNA response elements (MREs) on RNAs that mediate their interaction and binding. These RNAs containing circRNA, lncRNAs, and mRNAs act as competing endogenous RNAs (ceRNAs). They combine with miRNAs through MREs to regulate a series of subsequent life activities[12,13]. Through clinical samples and cell experiments, Li et al[14] showed that hsa_circ_0001021 is expressed in epithelial cells and is associated with ZO-1, occludin, and CLDN-2. Furthermore, hsa_circ_0001021 sponges miR-224-5p to upregulate smad4 and increase ZO-1 and occludin to regulate UC epithelial barrier function[14]. However, the potential role of circRNAs in UC remains to be investigated.
In this study, we constructed a circRNA-miRNA-mRNA regulatory network to investigate the probable mechanism of circRNAs in UC. Initially, differentially expressed circRNAs (DEcircRNAs), differentially expressed miRNAs (DEmiRNAs), and differentially expressed mRNAs (DEmRNAs) were retrieved using the RNA expression spectrum of circRNA, miRNA, and mRNA in the Gene Expression Omnibus (GEO) dataset. The ceRNA network was then established after determining the DEcircRNA-DEmiRNA and DEmiRNA-DEmRNA interactions using bioinformatics analysis methods. Functional enrichment analysis was performed to evaluate the biological functions of DEmRNAs in the ceRNA network. Ultimately, DEcircRNAs in the circRNA-miRNA-mRNA network were used to construct a new diagnostic model. The circRNA-miRNA-mRNA network regulatory network and new diagnostic model play an important role in the comprehensive analysis of gene interactions and identification of potential biomarkers that can be used for disease diagnosis, therapy, and prognosis of UC[15,16].
The NCBI GEO (http://www.ncbi.nlm.nih.gov/gds/) public database was used to obtain microarray data. The ctiteria for the selected dataset were as follows: (1) The grouping had biological replication significance; and (2) Colon tissue belonging to UC human or mouse. The circRNA expression profile of UC was derived from GSE131911, which constituted four patients with ulcerative colitis and four healthy controls. The miRNA expression data of UC were retrieved from GSE43009 (including five UC patients and five controls), and the mRNA expression data of UC were acquired from GSE48958 (including nine UC patients and 10 controls). The basics of the three microarray datasets are presented in Table 1.
After downloading the raw microarray data, the data were normalised and logarithmically expressed. The associated probe ID was converted to an international uniform name. When multiple probes corresponded to the same gene symbol, they were averaged, and probes that did not match the gene symbol were eliminated. DEcircRNAs, DEmiRNAs, and DEmRNAs between UC and control samples were screened using Bioconductor limma R. The thresholds were the absolute value of |log2(fold-change)| > 1.0, and the adjusted P-value < 0.05.
The circRNA interactome[20] (https://circinteractome.nia.nih.gov/), a comprehensive database of circRNAs, was used to predict the target circRNAs of the DEmiRNAs. We then deployed the online database Venny 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html) to evaluate the intersection of target circRNAs of DEmiRNAs and DEcircRNAs. The target mRNAs of DEmiRNAs were derived from three different miRNA target gene databases, miRTarBase, TargetScan, and miRDB, which intersect with DEmRNAs[21]. In addition, target genes appearing in at least two databases were selected[22].
Because circRNA can competitively bind miRNA to regulate miRNA expression, only those who conform to circRNA-miRNA and miRNA-mRNA targeting rules can be selected for the construction of ceRNA network. By combining the circRNA-miRNA and miRNA-mRNA pairs, we constructed a circRNA-miRNA-mRNA regulatory network, which was visualised using Cytoscape (version 3.9.1).
To assess functional enrichment, Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed on DEmRNAs using the R package (corrected P-value < 0.05, show category = 5)[23].
The least absolute shrinkage and selection operator (LASSO) algorithm was used to screen statistically significant model features for the subsequent establishment of a risk model for diagnosing UC. LASSO was used the glmnet/R package to screen circRNAs with the optimal predictive features being observed in the circRNA-miRNA-mRNA. Such circRNAs, as the initial part of the ceRNA network, had features with nonzero coefficients in the LASSO regression model[24].
The nomogram was used to establish a model for the expression level of circRNAs and the risk of ulcerative colitis, and the corresponding value on each variable axis was determined according to the expression level of the feature. A vertical line can be formed to intersect the upper ruler at each point on the feature, where the intersection represents the score. The total score was calculated by adding the feature scores. The total score is found on the lower scale and by making a vertical line down that intersects with the risk of ulcerative colitis. Finally, the degree of correlation between circRNAs and the prevalence of ulcerative colitis were determined[25]. To evaluate the performance of the model, this study used the C-index, area under the receiver operating characteristic (ROC) curve, and decision curve analysis (DCA) to verify the diagnostic efficacy of the model. DCA is a decision curve analysis performed by quantifying the net benefit at different threshold probabilities to determine the clinical usefulness of a nomogram. The analytical model can predict the correlation between circRNA and UC and obtain the net benefit at each threshold probability, with P < 0.05 considered statistically significant[26].
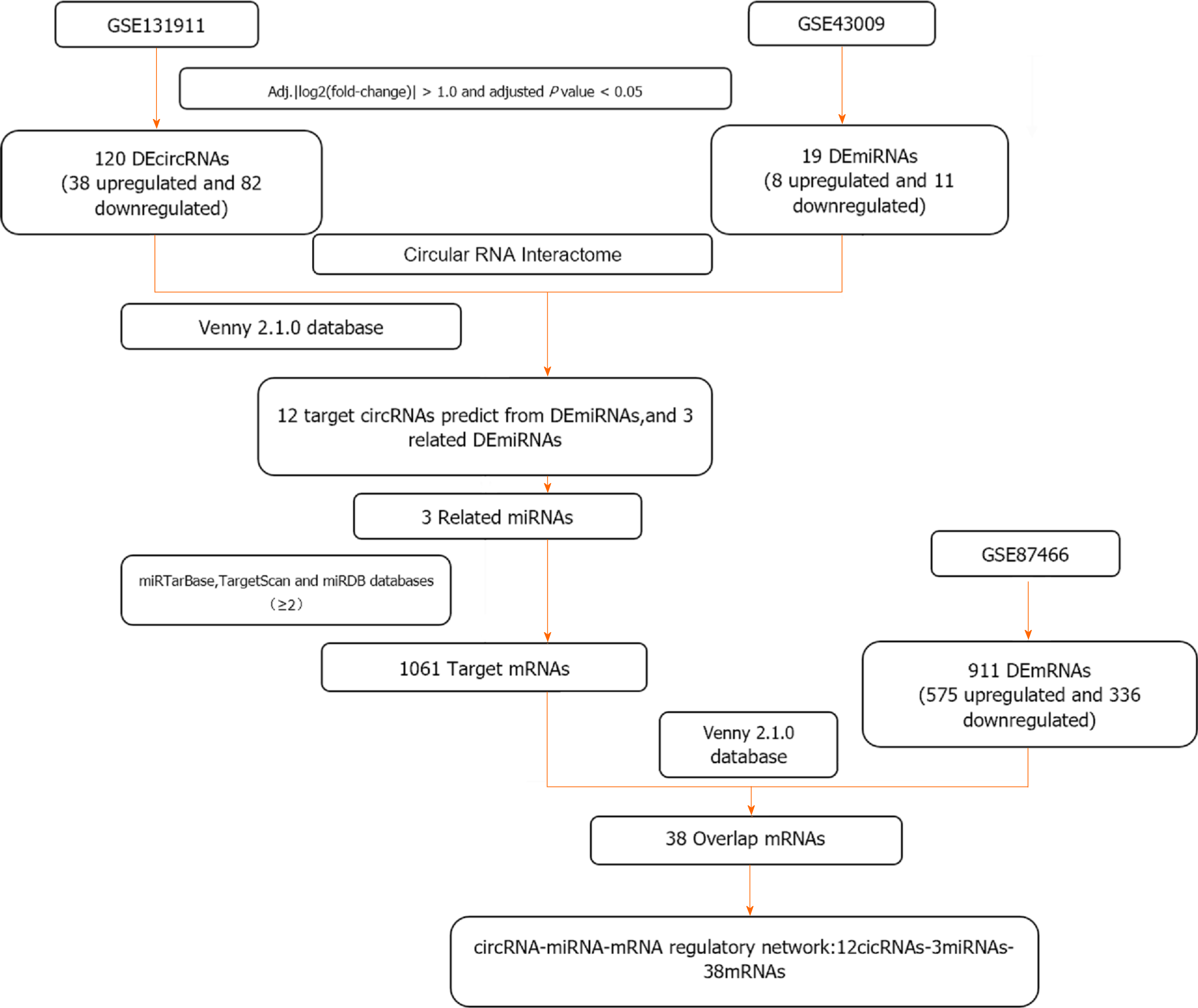
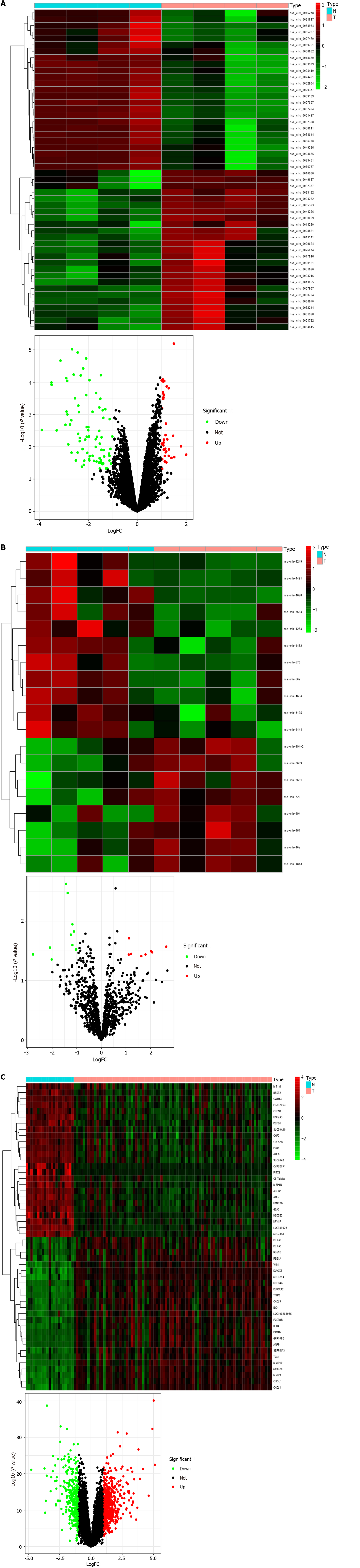
A flowchart illustrating the selection process is shown in Figure 1. The GSE131911 dataset identified 120 DEcircRNAs (38 upregulated and 82 downregulated). Similarly, 19 DEmiRNAs (8 upregulated and 11 downregulated) and 910 DEmRNAs (575 upregulated and 336 downregulated) were identified in GSE43009 and GSE87466, respectively. Subsequently, a heatmap and volcano plot of DEcircRNAs (Figure 2A), DEmiRNAs, and DEmRNAs (Figure 2B and C) were constructed.
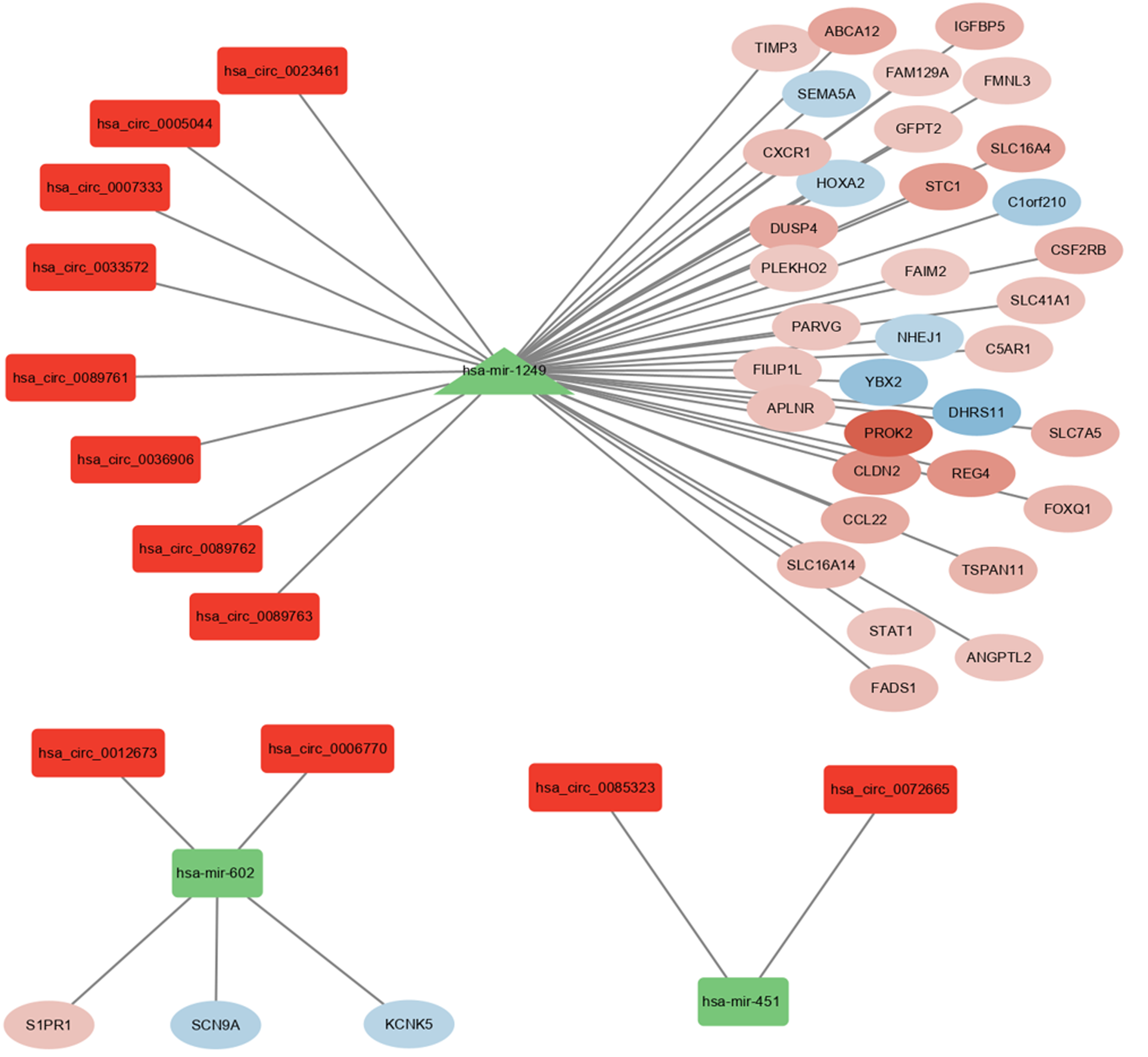
First, the target miRNAs predicted by DEcircRNAs and DEmiRNAs were intersected, after which 12 circRNAs and 3 miRNAs were identified. Second, we predicted target mRNAs from three miRNA databases (≥ 2) and considered their intersection with DEmRNAs. Finally, by integrating potential target mRNAs, related miRNAs, and related circRNAs, 12 overlapping circRNAs (hsa_circ_0023461, hsa_circ_0005044, hsa_circ_0007333, hsa_circ_0033572, hsa_circ_0089761, hsa_circ_0036906, hsa_circ_0089762, hsa_circ_0089763, hsa_circ_0012673, hsa_circ_0006770, hsa_circ_0085323, and hsa_circ_0072665), 3 miRNAs, and 38 target mRNAs were allowed to enter the circRNA-miRNA-mRNA network for further study (Figure 3).
GO (Figure 4A) and KEGG (Figure 4B) enrichment analyses were performed on the 38 DEmRNAs derived from the previous process. Biological process (BP) analysis revealed that DEmRNAs were predominantly enriched in cellular divalent inorganic cation homeostasis (GO:0072503) and divalent inorganic cation homeostasis (GO:0072507). To the best of our knowledge, the present study is the first to suggest that the occurrence of colitis may be related to homeostasis of divalent inorganic cations. When considering cellular component (CC), basolateral plasma membrane (GO:0016323), basal plasma membrane (GO:0009925), and basal part of the cell (GO:0045178) were the top three enriched terms. Imbalances in epithelium-matrix interactions have been discussed as a pathomechanism in UC, resulting in dysfunction of the mucous barrier of the colon[27]. Active transmembrane transporter activity (GO:0022804) and secondary active transmembrane transporter activity (GO:0015291) are related to molecular function. Transporters can influence the disposition of chemicals within the intestine by participating in their absorption, distribution, and elimination[28]. KEGG pathway analysis showed that the mRNAs were also considerably concentrated in the T-cell receptor signalling pathway, which confirmed that UC is an autoimmune disease (hsa:04660).
From the 12 differentially expressed circRNAs and 8 samples, LASSO was used to screen the circRNAs with optimal predictive features. Features with nonzero coefficients were selected in the LASSO regression model. LASSO regression selection revealed that hsa_circ_0085323 and hsa_circ_0036906 were important diagnostic circRNAs (Figures 5A and B).
The optimal model predictors (hsa_circ_0085323 and hsa_circ_0036906) screened from the differential circRNA data in the ceRNA network were used to establish a nomogram to predict the risk of UC (Figure 6A). The Harrell C-index of the model was analysed to quantify the discriminative performance of the nomogram, and the model discrimination performance was 1. The ROC curve, which represents the measurement of the diagnostic accuracy of the predictive model, was 1 (Figure 6B). The decision curve integrates the above models, and the maximum area under the curve is the maximum benefit (Figure 6C). Because few models can achieve perfect prediction performance, the overfitting results presented by this verification method may be due to the small amount of relevant data. The data amount will be further expanded and related research will be conducted in the future.
UC is one of the most serious intestinal diseases and has a poor prognosis. The elevated recurrence rate and therapy resistance of ulcerative colitis after treatment have inspired researchers to explore further the molecular mechanisms and new therapeutic targets and diagnostic markers. Recent progress in circRNA research has revealed the core aspects of circRNA biogenesis and biology. In particular, circRNA is closely related to the pathogenesis of UC. CircRNAs have a relatively stable structure owing to their circular structure. Knockout or overexpression of circRNAs can regulate downstream miRNA-mRNAs. Research on the circRNA-miRNA-mRNA regulatory network has been deeply involved in various tumours, making it vital for the diagnosis, prognosis, and treatment of UC[29].
Following the development of high-throughput genome sequencing technologies and the availability of diverse open databases, computer analysis enables us to build a circRNA-miRNA-mRNA network to help us better understand the cross-talk between RNAs and elucidate the occurrence and development of UC from complex genetic interactions[30]. Numerous studies have demonstrated that circRNAs can modulate intestinal immune imbalances, such as through the autophagy pathway, and can even be used as biomarkers for the diagnosis of IBD[11].
Our study compared UC and normal intestinal mucosa with datasets retrieved from the GEO database to identify DEcircRNAs, DEmiRNAs, and DEmRNAs. CircRNA-miRNA and miRNA-mRNA interactions were subsequently predicted, and 12 circRNAs, 3 miRNAs, and 38 mRNAs were selected to construct the ceRNA network. GO and KEGG enrichment analyses of mRNA in this ceRNA network were also performed. Furthermore, this circRNA-miRNA-mRNA network enhances the precision of potential candidate biomarkers for disease diagnosis and treatment, as it narrows the scope of research.
CircRNAs are naturally occurring RNAs highly expressed in the eukaryotic transcriptome[31]. CircRNAs can target miRNAs with high intensity, reducing their ability to target mRNAs, and are thus optimal biomarkers for UC. We identified two circRNAs (hsa_circ_0085323 and hsa_circ_0036906) using LASSO regression analysis. In addition, we constructed and internally validated a new model to calculate UC risk based on hsa_circ_0085323 and hsa_circ_0036906. Our model used the nomogram because it has been used extensively as a prognostic tool in oncology and medicine[32]. To our knowledge, our study is the first to apply a nomogram to UC. In addition, molecular and subsequent molecular mechanisms were identified in this study, which is an innovative combination.
We performed correlation analyses between the two predicted circRNAs and downstream miRNAs in the ceRNA network. Interestingly, studies have found that hsa-miR-451 is downregulated in colorectal cancer (CRC), which can play a tumour suppressor role by targeting the macrophage migration inhibitory factor and can also inhibit the growth of CRC cells by downregulating the PI3K/AKT pathway[33,34]. Differential analysis showed that hsa_circ_0085323 was upregulated in UC, upstream of hsa-miR-451, confirming that hsa_circ_0085323 increased the risk of UC if it scored higher in the nomogram. Although there are few studies on hsa_circ_0085323, additional studies on hsa-miR-451 can provide important evidence. Similarly, miR-1249 can directly target heterogeneous nuclear ribonucleoprotein K to accelerate the malignant phenotype of hepatocellular carcinoma[35]. Prokineticin 2 (PROK2) is an inflammatory cytokine-like molecule mainly expressed by macrophages and neutrophils infiltrating tissue injury sites and is upregulated in biopsy samples from patients with UC[36]. PROK2 is a target gene in the ceRNA network of UC, upstream of which is miR-1249. P53-induced miR-1249 may suppress CRC growth, metastasis, and angiogenesis by targeting vascular endothelial growth factor and high mobility group A2 HMGA2[37]. Hsa_circ_0036906 can act as a sponge for hsa-miR-1249, but the mechanism underlying its regulation of hsa-miR-1249 has not yet been investigated. In the prediction model, the lower the expression of hsa_circ_0036906, the higher the score on the nomogram, which is inconsistent with the circRNA-miRNA-mRNA regulatory network. The specific mechanisms of action of hsa_circ_0036906 and hsa-miR-1249 require further experimental verification. Furthermore, recent studies have explored ceRNA networks in UC, revealing certain distinctions from our own research. For instance, Xu et al[38] performed a comprehensive analysis that revealed four central genes regulated by NF-κB, ultimately constructing an lncRNA-miRNA-transcription factor (NF-κB) interaction network. Another study aimed to demonstrate the high diagnostic accuracy of two mRNAs (CTLA4 and STAT1) within the ceRNA network in UC[39]. In contrast, we focused on circRNA as a potential clinical diagnostic marker, deviating from conventional approaches, with circRNA possibly offering improved specificity. Over the past two years, only one article has analysed three mRNAs (FGG, KRT10, and TLR4), six miRNAs (hsa-miR-6875-5p, hsa-miR-1908-5p, hsa-miR-186-5p, hsa-miR-4436a, hsa-miR-520a-5p, and hsa-miR-6763-5p), two lncRNAs (XIST and NEAT1), and two chromosomal regions (NM_001039703 and NM_006267) that produce the most effective circRNAs, participating in the non-coding RNA-associated ceRNA network related to IBD. These elements may serve as potential therapeutic targets. Further research into ceRNA networks in IBD is essential for future investigations[40].
In summary, we successfully constructed a gut mucosa-derived ceRNA regulatory network associated with circRNAs and provided insights into the interactions between various RNA transcripts in UC. We established a promising nomogram model for UC disease risk diagnosis based on circRNA characteristic parameters, indicating biomarkers and subsequent molecular mechanisms. This model and ceRNA network can help predict disease occurrence and treatment.
Our study had certain limitations: (1) The sample size for the dataset obtained from the available public database was too small because the human sample sequencing data related to UC are few, thereby limiting the construction of the ceRNA network only from these three datasets; (2) The clinical sample data were too small, and predicting the risk of all Chinese UC patients was impossible. The prevalence of UC tends to be individualised. We searched the GEO data set again, looking for a similar data set for sample merging and de-batching. However, we only managed to find the same GSE142106. However, this data set is a mouse sample and did not match the type of sample we had studied before; therefore, we ended up having to use that unique data set. We hope to test our hypothesis by expanding human samples in clinical trials in the future; (3) Risk factor analysis did not include all potential factors affecting changes in circRNA expression; (4) Although the robustness of our nomograms was extensively checked by internal validation using bootstrapping tests, no external validation was performed. Experimental techniques, such as qPCR, should be used to detect circRNA expression to validate the accuracy of our model. At a deeper level, we also look forward to the results of experimental validation in clinical samples. We wish to validate the results of this paper; however, adverse effects may be inevitable. Under the premise that there is no logical problem, both negative and positive results are worth thinking about; and (5) We found that hsa_circ_0085323 and hsa_circ_0036906 have no corresponding homologous genes in mice and rats[41]. Therefore, the next step of qPCR validation must be achieved in humans. As a result of temporal limitations and various additional considerations, the incorporation of ethical authorization, along with adherence to the Helsinki Declaration and acquisition of patient consent, has yet to be actualized and will be duly acknowledged in a forthcoming publication. After passing ethics, we made every effort to collect blood samples from patients with UC for experimental validation.
This study for the first time presents the potential role of the circRNA-miRNA-mRNA regulatory network in UC in a relatively intuitive way and provides a novel idea for constructing predictive nomograms from DEcircRNAs in the ceRNA network. Diagnostic nomograms based on DEcircRNA signatures may provide a more intuitive method for UC disease risk prediction and facilitate more optimized treatment strategies.
Ulcerative colitis (UC) is an inflammatory bowel disease that affects the mucosal and submucosal layers of the colon and rectum. With advancements in the diagnosis and treatment of UC, the prospect of rapidly increasing the number of drugs with new targets is anticipated. However, despite progress in the biological understanding, information regarding the pathogenesis of UC remains limited. Therefore, exploring the molecular mechanisms of UC is of paramount importance for formulating appropriate therapeutic strategies and diagnosing the disease.
The primary focus of the research is to construct a competing endogenous RNA (ceRNA) network in UC and elucidate the mechanistic role of this network in the pathogenesis of UC. Additionally, using circular RNA (circRNA) as characteristic parameters, a diagnostic model for UC has been developed. Future endeavors involve expanding clinical UC sample data, applying the research methodology and approach outlined in this study, and conducting experimental validation. The goal is to further identify a more comprehensive ceRNA network and circRNAs with enhanced clinical diagnostic value.
The primary objective of this study is to construct a ceRNA network in UC and identify circRNAs serving as diagnostic biomarkers for UC. Ultimately, we successfully built a circRNA-miRNA-mRNA network in UC, identifying two circRNAs with clinical diagnostic value. Further exploration in this direction is anticipated to contribute to the innovation of diagnostic biomarkers for UC and provide additional insights into the crucial significance of non-coding RNA networks in UC.
Three GSE datasets related to UC were obtained from the Gene Expression Omnibus database. Difference analysis was performed on the three GSE data sets, and differences in circRNA, miRNA and mRNA were identified. Difference analysis was performed on the three GSE data sets, and differences in circRNA, miRNA and mRNA were identified. A circRNA-miRNA-mRNA regulatory network was constructed, and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were performed on the differential mRNAs. A circRNA-miRNA-mRNA regulatory network was constructed, and GO and KEGG analyses were performed on the differential mRNAs. circRNA predictors in UC models were screened and a model was established and validated.
We have successfully constructed a ceRNA regulatory network originating from intestinal mucosa associated with circRNAs, providing insights into the interactions among various RNA transcripts in UC. Utilizing circRNA characteristic parameters, we developed a promising nomogram model for UC disease risk diagnosis, highlighting the involvement of biomarkers and subsequent molecular mechanisms. This model and the ceRNA network can aid in predicting the onset and treatment of the disease. Further experimental verification in clinical UC samples is needed in the future.
This study introduces the theoretical framework of circRNA as a diagnostic biomarker for clinical identification of UC. The novel approach proposed involves the circRNA-miRNA-mRNA network as a mechanistic pathway for subsequent therapeutic interventions in UC.
The future research direction will focus on observing the significance of circRNAs as diagnostic biomarkers for UC within a clinical context.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Immunology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sitkin S, Russia S-Editor: Gong ZM L-Editor: A P-Editor: Cai YX
| 1. | Al-Horani R, Spanudakis E, Hamad B. The market for ulcerative colitis. Nat Rev Drug Discov. 2022;21:15-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 2. | Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389:1756-1770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2199] [Cited by in RCA: 2484] [Article Influence: 310.5] [Reference Citation Analysis (2)] |
| 3. | Hirten RP, Sands BE. New Therapeutics for Ulcerative Colitis. Annu Rev Med. 2021;72:199-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 100] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 4. | Mohanapriya R, Akshaya RL, Selvamurugan N. A regulatory role of circRNA-miRNA-mRNA network in osteoblast differentiation. Biochimie. 2022;193:137-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 5. | Xiang S, Wu Y, Shi H, Xue L, Luo K, Ding Y. Circular RNA hsa_circ_0001445 in plasma as a novel biomarker for osteoporosis in postmenopausal women. Biomark Med. 2020;14:1599-1607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 6. | Chen G, Tang W, Wang S, Long C, He X, Yang D, Peng S. Promising diagnostic and therapeutic circRNAs for skeletal and chondral disorders. Int J Biol Sci. 2021;17:1428-1439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 7. | Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 389] [Cited by in RCA: 798] [Article Influence: 159.6] [Reference Citation Analysis (0)] |
| 8. | Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1186] [Cited by in RCA: 1501] [Article Influence: 136.5] [Reference Citation Analysis (0)] |
| 9. | Liu L, Wang H, Yu S, Gao X, Liu G, Sun D, Jiang X. An Update on the Roles of circRNA-ZFR in Human Malignant Tumors. Front Cell Dev Biol. 2021;9:806181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Zhu P, Zhu X, Wu J, He L, Lu T, Wang Y, Liu B, Ye B, Sun L, Fan D, Wang J, Yang L, Qin X, Du Y, Li C, Ren W, Wu X, Tian Y, Fan Z. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat Immunol. 2019;20:183-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 171] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 11. | Lin L, Zhou G, Chen P, Wang Y, Han J, Chen M, He Y, Zhang S. Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease? Cell Death Dis. 2020;11:456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 12. | Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 801] [Article Influence: 114.4] [Reference Citation Analysis (0)] |
| 13. | Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol. 2016;937:3-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 625] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 14. | Li B, Li Y, Li L, Yu Y, Gu X, Liu C, Long X, Zuo X. Hsa_circ_0001021 regulates intestinal epithelial barrier function via sponging miR-224-5p in ulcerative colitis. Epigenomics. 2021;13:1385-1401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Zhou P, Zheng G, Li Y, Wu D, Chen Y. Construction of a circRNA-miRNA-mRNA Network Related to Macrophage Infiltration in Hepatocellular Carcinoma. Front Genet. 2020;11:1026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | He Y, Chen Y, Tong Y, Long W, Liu Q. Identification of a circRNA-miRNA-mRNA regulatory network for exploring novel therapeutic options for glioma. PeerJ. 2021;9:e11894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Hu YA, Zhu Y, Liu G, Yao X, Yan X, Yang Y, Wang W, Zou X, Li X. Expression profiles of circular RNAs in colon biopsies from Crohn's disease patients by microarray analysis. J Clin Lab Anal. 2021;35:e23788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Li K, Strauss R, Ouahed J, Chan D, Telesco SE, Shouval DS, Canavan JB, Brodmerkel C, Snapper SB, Friedman JR. Molecular Comparison of Adult and Pediatric Ulcerative Colitis Indicates Broad Similarity of Molecular Pathways in Disease Tissue. J Pediatr Gastroenterol Nutr. 2018;67:45-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 19. | Yang L, Bian Y, Li Z, Yan Y, Li J, Li W, Zeng L. Identification of potential biomarkers and pathways in ulcerative colitis with combined public mRNA and miRNA expression microarray data analysis. J Gastrointest Oncol. 2019;10: 847-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 622] [Cited by in RCA: 911] [Article Influence: 101.2] [Reference Citation Analysis (0)] |
| 21. | Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, Lu R, Jurisica I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46:D360-D370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 409] [Article Influence: 68.2] [Reference Citation Analysis (0)] |
| 22. | Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127-D131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 855] [Cited by in RCA: 1997] [Article Influence: 399.4] [Reference Citation Analysis (0)] |
| 23. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 33583] [Article Influence: 1599.2] [Reference Citation Analysis (0)] |
| 24. | Wang H, Zhang L, Liu Z, Wang X, Geng S, Li J, Li T, Ye S. Predicting medication nonadherence risk in a Chinese inflammatory rheumatic disease population: development and assessment of a new predictive nomogram. Patient Prefer Adherence. 2018;12:1757-1765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 25. | Liu Y, Pang Z, Zhao X, Zeng Y, Shen H, Du J. Prognostic model of AU-rich genes predicting the prognosis of lung adenocarcinoma. PeerJ. 2021;9:e12275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 26. | Fan X, Zou X, Liu C, Peng S, Zhang S, Zhou X, Wang T, Zhu W. Global analysis of miRNA-mRNA regulation pair in bladder cancer. World J Surg Oncol. 2022;20:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 27. | Kuo WT, Shen L, Zuo L, Shashikanth N, Ong MLDM, Wu L, Zha J, Edelblum KL, Wang Y, Nilsen SP, Turner JR. Inflammation-induced Occludin Downregulation Limits Epithelial Apoptosis by Suppressing Caspase-3 Expression. Gastroenterology. 2019;157:1323-1337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 167] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 28. | Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev. 2010;62:1-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 660] [Cited by in RCA: 586] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 29. | Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3273] [Cited by in RCA: 3151] [Article Influence: 525.2] [Reference Citation Analysis (0)] |
| 30. | Wang Z, Ji X, Gao L, Guo X, Lian W, Deng K, Xing B. Comprehensive In Silico Analysis of a Novel Serum Exosome-Derived Competitive Endogenous RNA Network for Constructing a Prognostic Model for Glioblastoma. Front Oncol. 2021;11:553594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and Characterizing circRNA-Protein Interaction. Theranostics. 2017;7:4183-4191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 486] [Cited by in RCA: 485] [Article Influence: 60.6] [Reference Citation Analysis (0)] |
| 32. | Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26:1364-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1306] [Cited by in RCA: 2306] [Article Influence: 135.6] [Reference Citation Analysis (0)] |
| 33. | Bai H, Wu S. miR-451: A Novel Biomarker and Potential Therapeutic Target for Cancer. Onco Targets Ther. 2019;12:11069-11082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 34. | Li HY, Zhang Y, Cai JH, Bian HL. MicroRNA-451 inhibits growth of human colorectal carcinoma cells via downregulation of Pi3k/Akt pathway. Asian Pac J Cancer Prev. 2013;14:3631-3634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 35. | Shu H, Hu J, Deng H. miR-1249-3p accelerates the malignancy phenotype of hepatocellular carcinoma by directly targeting HNRNPK. Mol Genet Genomic Med. 2019;7:e00867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 36. | Watson RP, Lilley E, Panesar M, Bhalay G, Langridge S, Tian SS, McClenaghan C, Ropenga A, Zeng F, Nash MS. Increased prokineticin 2 expression in gut inflammation: role in visceral pain and intestinal ion transport. Neurogastroenterol Motil. 2012;24:65-75, e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 37. | Chen X, Zeng K, Xu M, Liu X, Hu X, Xu T, He B, Pan Y, Sun H, Wang S. P53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis. 2019;10:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 38. | Xu M, Kong Y, Chen N, Peng W, Zi R, Jiang M, Zhu J, Wang Y, Yue J, Lv J, Zeng Y, Chin YE. Identification of Immune-Related Gene Signature and Prediction of CeRNA Network in Active Ulcerative Colitis. Front Immunol. 2022;13:855645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 39. | Dong L, Zhang R, Huang Q, Shen Y, Li H, Yu S, Wu Q. Construction, bioinformatics analysis, and validation of competitive endogenous RNA networks in ulcerative colitis. Front Genet. 2022;13:951243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 40. | Elahimanesh M, Najafi M. Cross talk between bacterial and human gene networks enriched using ncRNAs in IBD disease. Sci Rep. 2023;13:7704. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 41. | Zhang Z, Qian H, Wang L, Tao Z, Cheng K, Wang K, Xie Y, Zhang L. Construction of a circRNA-miRNA-mRNA Regulatory Network for Coronary Artery Disease by Bioinformatics Analysis. Cardiol Res Pract. 2022;2022:4017082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |