Published online Mar 16, 2024. doi: 10.12998/wjcc.v12.i8.1517
Peer-review started: December 13, 2023
First decision: December 21, 2023
Revised: January 3, 2024
Accepted: February 25, 2024
Article in press: February 25, 2024
Published online: March 16, 2024
Processing time: 89 Days and 10.6 Hours
Nonallelic homologous recombination (NAHR) of segmental duplications or low copy repeats (LCRs) result in DNA gain/loss and play an important role in the origin of genomic disorders.
A 3-year- old boy was referred for genetic analysis. Comparative genomic hybridization array analysis revealed a loss of 3776 kb in the 4p16.3 chromosomal region and a gain of 3201 kb in the 11p15.5p15.4 chromosomal region.
Genomic imbalances caused by NAHR in LCRs result in deletion and duplication syndromes.
Core Tip: Chromosome analysis of a young child with developmental delay, cleft palate, hearing loss, and mental retardation indicated 46, XY. Further genetic analysis with comparative genomic hybridization array revealed a deletion in the short arm of chromosome 4 and a gain in the short arm of chromosome 11. The patient’s phenotypic findings were associated with the genes affected by the genomic loss and gain.
- Citation: Kaya I. Detection of 4p16.3 deletion and 11p15.5p15.4 gain in a boy by comparative genomic hybridization array: A case report. World J Clin Cases 2024; 12(8): 1517-1522
- URL: https://www.wjgnet.com/2307-8960/full/v12/i8/1517.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i8.1517
One of the most common chromosomal abnormalities in humans is reciprocal translocation. Balanced reciprocal translocations occur in approximately 1 in 600 people[1]. Most carriers of balanced reciprocal translocations have normal phenotypes. However, molecular analyses have shown that in patients with abnormal phenotypes, seemingly balanced reciprocal chromosome translocations are accompanied by a chromosomal imbalance at the translocation break points or elsewhere in the genome[2].
Segmental duplications (SDs) or low copy repeats (LCRs) comprise 6.6% of the human genome and play an important role in the origin of genomic disorders. LCRs mainly cluster at pericentromeric, subtelomeric, and interstitial loci throughout the genome. LCRs are DNA blocks at least 1 kb in length. Recurrent genomic disorders result from meiotic misalignment of these blocks, which have very high sequence identity (> 90%). This meiotic misalignment rearranges the segment through nonallelic homologous recombination (NAHR), resulting in one chromatid with a deletion and another chromatid with reciprocal duplication. The consequent duplication syndromes usually have a milder phenotype than the deletion syndromes. Gene losses generally result in more severe phenotypic findings[3].
Wolf-Hirschhorn syndrome (WHS; OMIM 194190) is a microdeletion syndrome characterized by abnormal craniofacial features, prenatal and postnatal growth retardation, microcephaly, mental delay, seizures, and congenital heart malformations[4].
According to the literature, there is also considerable variability in the WHS phenotype, mostly due to differences in the underlying genomic defect. However, there is a core phenotype defined by short stature, low body weight, intellectual disability, “Greek warrior helmet” facies, seizures, and severe growth retardation[5]. Some of the structural defects, such as cleft lip and palate, occur more frequently in individuals with deletions larger than 3 megabases (Mb)[6].
WHS is caused by deletions in WHS critical regions 1 and 2 (WHSCR-1/2). WHSCR-1 is located approximately 2 Mb from the end of the chromosomal 4p16.3 region and encompasses the very closely situated WHS candidate 1 and 2 genes (WHSC1/WHSC2). WHSCR-2 is located at a distance of approximately 1.4-1.9 Mb from the end of the chromosomal 4p16.3 region and includes important genes such as FGFR3, LETM1, MSX, and WHSC1[7].
Silver-Russell syndrome (SRS; OMIM 180860) and Beckwith-Wiedemann syndrome (BWS; OMIM 130650) are congenital imprinting disorders that affect growth in clinically opposite ways. The expression of both syndromes usually depends on parental origin of the chromosome with the imprinted genes[8]. Paternal replication of the 11p15 chromosome region may result in BWS, while maternal replication of the same region can result in SRS. These two syndromes have numerous opposing phenotypes, particularly with respect to growth parameters. Differences in phenotype have been attributed to altered dosage of the imprinted genes that control growth within this region of 11p15[9].
A child with findings of developmental delay, cleft palate, hearing loss, and mental retardation was referred for genetic analysis.
Cleft palate was present from birth. Other findings emerged during postnatal development.
Unremarkable.
Unremarkable.
Cleft palate was present.
Informed consent was obtained from the patient’s family. Karyotype analysis was first performed from a peripheral blood sample according to the conventional cytogenetic analysis procedure. Analysis results were evaluated according to the 2013 International System for Human Cytogenetic Nomenclature. As the second step, genomic DNA was obtained from the peripheral blood sample using the QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol, for use in comparative genomic hybridization (CGH) array analysis. Enzyme cleavage, labeling, purification, hybridization, and post-hybridization washing were performed according to the working protocol from Agilent Technologies, Inc. The Agilent Oligonucleotide Microarray (8x60K microarray system) platform was used and the results were analyzed using Agilent CytoGenomic Edition 2.5.8.1/GRCh 37/hg19) software.
There is not imaging examination in the case.
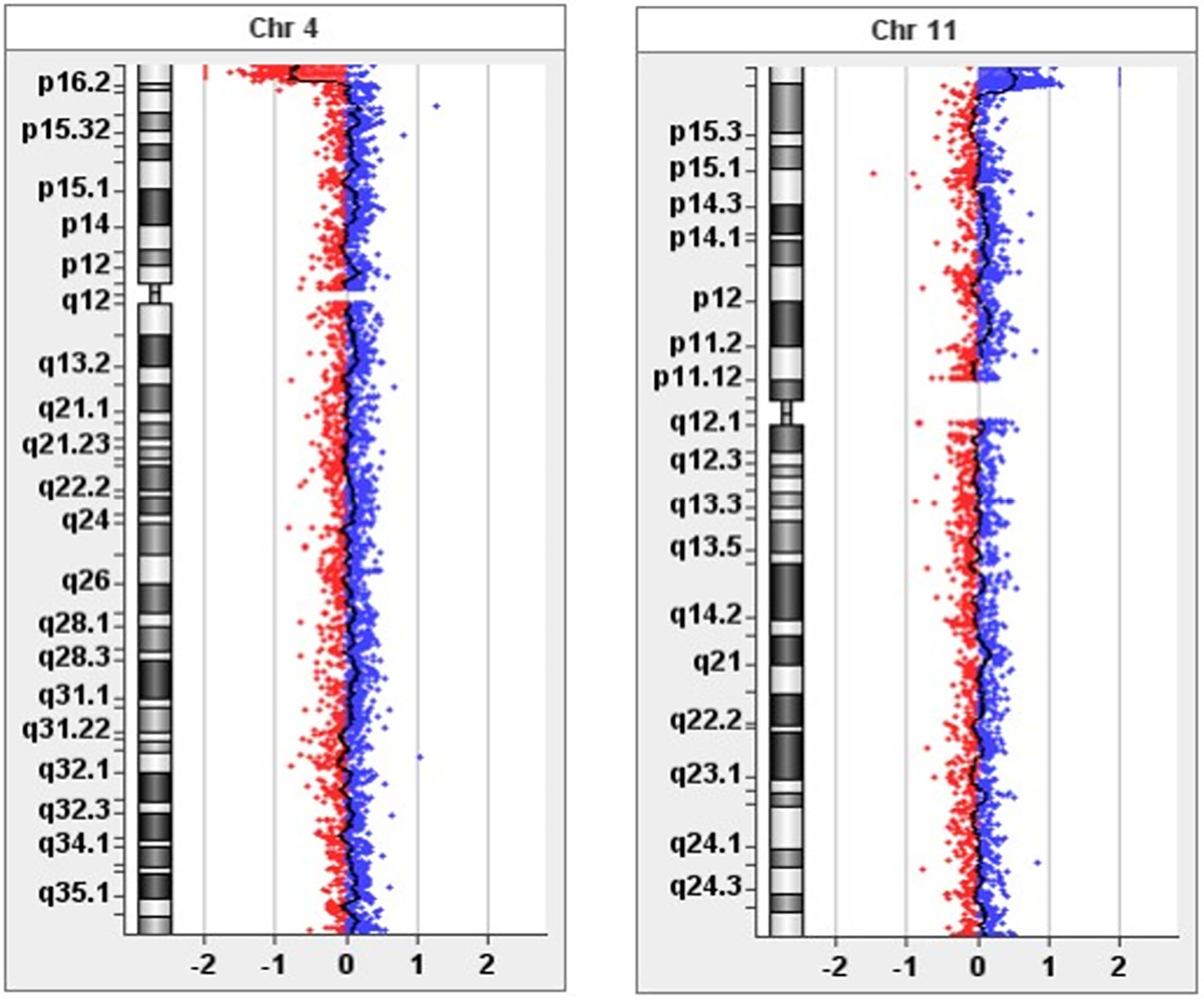
Karyotype analysis yielded normal results. CGH array analysis revealed a loss of 3,776 kb in the 4p16.3 region and gain of 3,201 kb in the 11p15.5p15.4 region (Figure 1). The genes in the relevant regions are shown in Table 1. This case report aimed to present the genotype-phenotype relationship in light of the literature.
| Chromosomal | Genes |
| 4p16.3 | ZNF141, PDE6B, MYL5, CPLX1, GAK, DGKQ, IDUA, FGFRL1, RNF212, SPON2, CTBP1, MAEA, SLBP, TACC3, FGFR3, LETM1, WHSC1, WHSC2, POLN, RNF4, TNIP2, SH3BP2, ADD1, NOP14, HTT, HGFAC, DOK7, LRPAP1, ADRA2C, ZNF595, ZNF718, ZNF876P, ZNF732, ABCA11P, ZNF721, PIGG, ATP5I, MFSD7, PCGF3, LOC100129917, TMEM175, SLC26A1, TMED11P, LOC100130872, C4orf42, KIAA1530, CRIPAK, FAM53A, TMEM129, SCARNA22, MIR943, C4orf48, NAT8L, HAUS3, MXD4, ZFYVE28, LOC402160, FAM193A, MFSD10, C4orf10, GRK4, C4orf44, RGS12, LOC100133461 |
| 11p15.5p15.4 | ODF3, SIRT3, IFITM2, IFITM1, IFITM3, PKP3, SIGIRR, PTDSS2, HRAS, MIR210, SCT, DRD4, TALDO1, SLC25A22, PNPLA2, CD151, POLR2L, TSPAN4, AP2A2, MUC6, MUC2, MUC5B, TOLLIP, DUSP8, KRTAP5-1, CTSD, TNNI2, LSP1, TNNT3, MRPL23, H19, IGF2, INS, TH, ASCL2, C11orf21, TSPAN32, CD81, TSSC4, TRPM5, KCNQ1, KCNQ1DN, CDKN1C, PHLDA2, NAP1L4, CARS, OSBPL5, MRGPRG, MRGPRE, ZNF195, BET1L, RIC8A, PSMD13, NLRP6, ATHL1, IFITM5, B4GALNT4, ANO9, RNH1, LRRC56, C11orf35, RASSF7, LOC143666, PHRF1, IRF7, CDHR5, DEAF1, TMEM80, EPS8L2, PDDC1, CEND1, LRDD, RPLP2, SNORA52, EFCAB4A, CHID1, LOC255512, BRSK2, MOB2, LOC338651, KRTAP5-2, KRTAP5-3, KRTAP5-4, KRTAP5-5, FAM99A, FAM99B, KRTAP5-6, LOC402778, SYT8, MIR4298, LOC100133545, MIR675, INS-IGF2, MIR483, IGF2AS, KCNQ1OT1, SLC22A18AS, SLC22A18, SNORA54, C11orf36 |
The case did not receive any treatment.
Genetic counseling was given to the family.
The loss in the 4p16.3 region in our patient is consistent with WHS (OMIM 194190). In this case, cleft palate was present as a craniofacial feature. Such structural defects in WHS are more common in individuals with deletions of more than 3 Mb[6], as in our case. The delays in growth and cognitive development observed in our patient were also consistent with WHS[10]. Definitive clinical signs are mapped to a 1.9-Mb region of 4p16.3 identified as a critical region (WHSCR-2). Typical WHS is a contiguous gene deletion disorder because evidence indicates that the core phenotype is caused by haploinsufficiency of numerous genes in the deleted critical region[5,10].
Wolf-Hirschhorn syndrome candidate 1 (WHSC1), also known as NSD2 and MMSET, is a histone-modifying enzyme related to both the characteristic facies and growth delay[11]. WHSC1 is a neurodevelopmental gene associated with growth delay, intellectual disability, and facial dysmorphism[12]. WHSC1 deletion is associated with many characteristic WHS features, including the distinct facial appearance[10]. WHSC1-mutant mice exhibited characteristics similar to some phenotypic features of WHS in humans, including developmental delay, growth retardation, heart defects, and midline and craniofacial defects. While heterozygous mice survived with varying degrees of the WHS phenotype, homozygotes showed more severe phenotypes and died shortly after birth[11].
Leucine Zipper/EF-hand-containing transmembrane protein 1 deletion has been associated with seizures[13], while oligodontia has been associated with MSX1 loss of function[14,15]. No causal gene (or genes) for hearing loss have yet been identified in WHS. Ahmed et al[7] showed that WHSC1 loss of function leads to failure of hair cell organization and stereocilia formation and innervation defects in the cochlea.
The overgrowth disorder BWS and growth restriction disorder SRS have been associated with various epigenetic and genetic defects affecting a set of imprint genes on chromosome 11p15.5[16]. Two imprinting control regions (ICR1 and ICR2) located in chromosomal region 11p15 control fetal and postnatal growth. ICR1 contains the maternally expressed H19 gene and paternally expressed IGF2 gene, while ICR2 contains the maternally expressed KCNQ1 and CDKN1C genes and the paternally expressed KCNQ1OT1 gene. Normally, genes expressed in one allele are imprinted (methylated) and silenced in the other allele. Imprinting disorders lead to abnormal expression of these genes and the clinical phenotypes of SRS or BWS[17]. In mouse models, knockouts lacking both the IGF2 and KCNQ1OT1 genes show fetal growth restriction, while ICR1 defects leading to biallicular IGF2 expression or transactivation of the IGF2 gene lead to fetal overgrowth. BWS is characterized by prenatal and postnatal overgrowth, organomegaly, abdominal wall defects, frequent hemihyperplasia, and an increased risk of childhood tumors. This syndrome is caused by genetic or epigenetic changes in the imprinted 11p15 region, leading to downregulation of maternally expressed genes and upregulation of paternally expressed genes, including IGF2. In contrast, chromosome 11p15 duplications of maternal origin have been identified in patients with fetal growth retardation and SRS (OMIM 180860). SRS is a clinically heterogeneous syndrome with prenatal and postnatal growth retardation, first described by Silver and Russell. It may present with short stature, prominent forehead, triangular face, low-set ears, clinodactyly of the fifth fingers, cafe-au-lait spots, genital abnormalities, blue sclera, syndactyly of the toes, and severe feeding difficulties[18].
The role of imprinting errors in SRS was first suggested because of maternal uniparental disomy 7 (UPD7) detected in 5%-10% of cases. More recently, loss of paternal methylation of ICR1 or maternal duplication of 11p15 has been demonstrated in one-third of SRS cases. In contrast to BWS, these defects result in IGF2 silencing and CDKN1C activation, respectively[16]. In summary, the etiology of SRS involves maternal UPD7, ICR1 hypomethylation, ICR2 hypome
Genetic analysis of the patient’s parents could not be performed because they did not return to the center. This is a limitation of our case report. From a genomic point of view, because of the patient’s 4p16.3 deletion and 11p15.5p15.4 duplication, we predict that a 4;11 translocation occurred in the family at least one generation earlier.
No phenotypic anomaly or mental retardation was present in any other family members. The phenotypical problems emerged after unbalanced transmission of the translocation from one of the parents to their offspring. As a result of balanced translocation segregation anomalies such as 4p monosomy and 11p15.5p15.4 region duplication, unbalanced transmission from parent to child is highly possible due to the segregation anomaly. It has also been demonstrated that recurrent translocation between chromosomes 4 and 11 occurs via NAHR mediated by interchromosomal paralog LCRs[19].
Growth retardation is a prominent phenotypic feature of both WHS and SRS. The developmental anomaly in this case was growth retardation. Therefore, the patient’s 3,201-kb gain in the 11p15.5p15.4 region does not support BWS, which is associated with overgrowth. The growth retardation in this case in consistent with the SRS phenotype. Due to the finding of growth retardation, the 11p15.5p15.4 duplication was thought to be of maternal origin. Large duplications affecting both ICRs at 11p15 are known to be associated with growth retardation/SRS or overgrowth/BWS, depending on the parental origin of the imbalance[20]. In our case, the 11p15.5p15.4 duplication was 3,201 kb in size and encompassed both the ICR1 and ICR2 regions. Therefore, there were two active copies of CDKN1C and H19, consistent with maternal duplication of the 11p15.5p15.4 region. Thus, the SRS phenotype reflects the overexpression of maternal CDKN1C[21]. The effect on growth-related genes is based on a gene dosage effect. Phenotypically, developmental delay, cleft palate, and mental retardation are consistent with WHS. In our case, a 3776-kb deletion was detected in 4p16.3. Regarding the patient’s hearing loss, stereocilia formation defect and innervation defect in the cochlea have been described in WHS[7]. Growth retardation is a finding observed in both WHS and SRS.
Genomic disorders are caused by meiotic misalignment of DNA paralogs with over 90% sequence identity. In LCRs, the segment affected by NAHR is rearranged to form one chromatid with a deletion with another with reciprocal duplication[3]. The condition seen in our case, which combined some phenotypic features of both WHS and SRS, is the result of an unbalanced genomic structure caused by the translocation segregation anomaly arising in one of the patient’s parents or previous generations.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Genetics and heredity
Country/Territory of origin: Turkey
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: De Nova JM, Spain S-Editor: Wang JL L-Editor: A P-Editor: Yu HG
| 1. | Van Dyke DL, Weiss L, Roberson JR, Babu VR. The frequency and mutation rate of balanced autosomal rearrangements in man estimated from prenatal genetic studies for advanced maternal age. Am J Hum Genet. 1983;35:301-308. [PubMed] |
| 2. | Sismani C, Kitsiou-Tzeli S, Ioannides M, Christodoulou C, Anastasiadou V, Stylianidou G, Papadopoulou E, Kanavakis E, Kosmaidou-Aravidou Z, Patsalis PC. Cryptic genomic imbalances in patients with de novo or familial apparently balanced translocations and abnormal phenotype. Mol Cytogenet. 2008;1:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 3. | Vervoort L, Vermeesch JR. Low copy repeats in the genome: from neglected to respected. Explor Med. 2023;4:166-175. [DOI] [Full Text] |
| 4. | Van Buggenhout G, Melotte C, Dutta B, Froyen G, Van Hummelen P, Marynen P, Matthijs G, de Ravel T, Devriendt K, Fryns JP, Vermeesch JR. Mild Wolf-Hirschhorn syndrome: micro-array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype-phenotype map. J Med Genet. 2004;41:691-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 91] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 5. | Zollino M, Orteschi D, Ruiter M, Pfundt R, Steindl K, Cafiero C, Ricciardi S, Contaldo I, Chieffo D, Ranalli D, Acquafondata C, Murdolo M, Marangi G, Asaro A, Battaglia D. Unusual 4p16.3 deletions suggest an additional chromosome region for the Wolf-Hirschhorn syndrome-associated seizures disorder. Epilepsia. 2014;55:849-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 6. | Rezai S, Wilansky J, Gottimukkala S, Chadee A, Rajegowda BH, Henderson CE. Wolf-hirschhorn syndrome (WHS), a case report and review of literature. J Pediatr Neonatal Care. 2016;5:00170. [DOI] [Full Text] |
| 7. | Ahmed M, Ura K, Streit A. Auditory hair cell defects as potential cause for sensorineural deafness in Wolf-Hirschhorn syndrome. Dis Model Mech. 2015;8:1027-1035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Õunap K. Silver-Russell Syndrome and Beckwith-Wiedemann Syndrome: Opposite Phenotypes with Heterogeneous Molecular Etiology. Mol Syndromol. 2016;7:110-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 9. | South ST, Whitby H, Maxwell T, Aston E, Brothman AR, Carey JC. Co-occurrence of 4p16.3 deletions with both paternal and maternal duplications of 11p15: modification of the Wolf-Hirschhorn syndrome phenotype by genetic alterations predicted to result in either a Beckwith-Wiedemann or Russell-Silver phenotype. Am J Med Genet A. 2008;146A:2691-2697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 10. | Zollino M, Murdolo M, Marangi G, Pecile V, Galasso C, Mazzanti L, Neri G. On the nosology and pathogenesis of Wolf-Hirschhorn syndrome: genotype-phenotype correlation analysis of 80 patients and literature review. Am J Med Genet C Semin Med Genet. 2008;148C:257-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 110] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 11. | Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 Lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 304] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 12. | Zollino M, Doronzio PN. Dissecting the Wolf-Hirschhorn syndrome phenotype: WHSC1 is a neurodevelopmental gene contributing to growth delay, intellectual disability, and to the facial dysmorphism. J Hum Genet. 2018;63:859-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Schlickum S, Moghekar A, Simpson JC, Steglich C, O'Brien RJ, Winterpacht A, Endele SU. LETM1, a gene deleted in Wolf-Hirschhorn syndrome, encodes an evolutionarily conserved mitochondrial protein. Genomics. 2004;83:254-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 71] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Nieminen P, Kotilainen J, Aalto Y, Knuutila S, Pirinen S, Thesleff I. MSX1 gene is deleted in Wolf-Hirschhorn syndrome patients with oligodontia. J Dent Res. 2003;82:1013-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Limeres J, Serrano C, De Nova JM, Silvestre-Rangil J, Machuca G, Maura I, Cruz Ruiz-Villandiego J, Diz P, Blanco-Lago R, Nevado J, Diniz-Freitas M. Oral Manifestations of Wolf-Hirschhorn Syndrome: Genotype-Phenotype Correlation Analysis. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Cardarelli L, Sparago A, De Crescenzo A, Nalesso E, Zavan B, Cubellis MV, Selicorni A, Cavicchioli P, Pozzan GB, Petrella M, Riccio A. Silver-Russell syndrome and Beckwith-Wiedemann syndrome phenotypes associated with 11p duplication in a single family. Pediatr Dev Pathol. 2010;13:326-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 17. | Vals MA, Kahre T, Mee P, Muru K, Kallas E, Žilina O, Tillmann V, Õunap K. Familial 1.3-Mb 11p15.5p15.4 Duplication in Three Generations Causing Silver-Russell and Beckwith-Wiedemann Syndromes. Mol Syndromol. 2015;6:147-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Netchine I, Rossignol S, Dufourg MN, Azzi S, Rousseau A, Perin L, Houang M, Steunou V, Esteva B, Thibaud N, Demay MC, Danton F, Petriczko E, Bertrand AM, Heinrichs C, Carel JC, Loeuille GA, Pinto G, Jacquemont ML, Gicquel C, Cabrol S, Le Bouc Y. 11p15 imprinting center region 1 loss of methylation is a common and specific cause of typical Russell-Silver syndrome: clinical scoring system and epigenetic-phenotypic correlations. J Clin Endocrinol Metab. 2007;92:3148-3154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 189] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 19. | Ou Z, Stankiewicz P, Xia Z, Breman AM, Dawson B, Wiszniewska J, Szafranski P, Cooper ML, Rao M, Shao L, South ST, Coleman K, Fernhoff PM, Deray MJ, Rosengren S, Roeder ER, Enciso VB, Chinault AC, Patel A, Kang SH, Shaw CA, Lupski JR, Cheung SW. Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome Res. 2011;21:33-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 20. | Begemann M, Spengler S, Gogiel M, Grasshoff U, Bonin M, Betz RC, Dufke A, Spier I, Eggermann T. Clinical significance of copy number variations in the 11p15.5 imprinting control regions: new cases and review of the literature. J Med Genet. 2012;49:547-553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 21. | Chiesa N, De Crescenzo A, Mishra K, Perone L, Carella M, Palumbo O, Mussa A, Sparago A, Cerrato F, Russo S, Lapi E, Cubellis MV, Kanduri C, Cirillo Silengo M, Riccio A, Ferrero GB. The KCNQ1OT1 imprinting control region and non-coding RNA: new properties derived from the study of Beckwith-Wiedemann syndrome and Silver-Russell syndrome cases. Hum Mol Genet. 2012;21:10-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |