Published online Mar 16, 2024. doi: 10.12998/wjcc.v12.i8.1454
Peer-review started: November 13, 2023
First decision: December 26, 2023
Revised: December 29, 2023
Accepted: January 30, 2024
Article in press: January 30, 2024
Published online: March 16, 2024
Processing time: 119 Days and 20.9 Hours
A rare autosomal recessive genetic disorder, 3M syndrome, is characterized by severe intrauterine and postnatal growth retardation. Children with 3M syndrome typically exhibit short stature, facial deformities, long tubular bones, and high vertebral bodies but generally lack mental abnormalities or other organ damage. Pathogenic genes associated with 3M syndrome include CUL7, OBSL1 and CCDC8. The clinical and molecular characteristics of patient with 3M syn
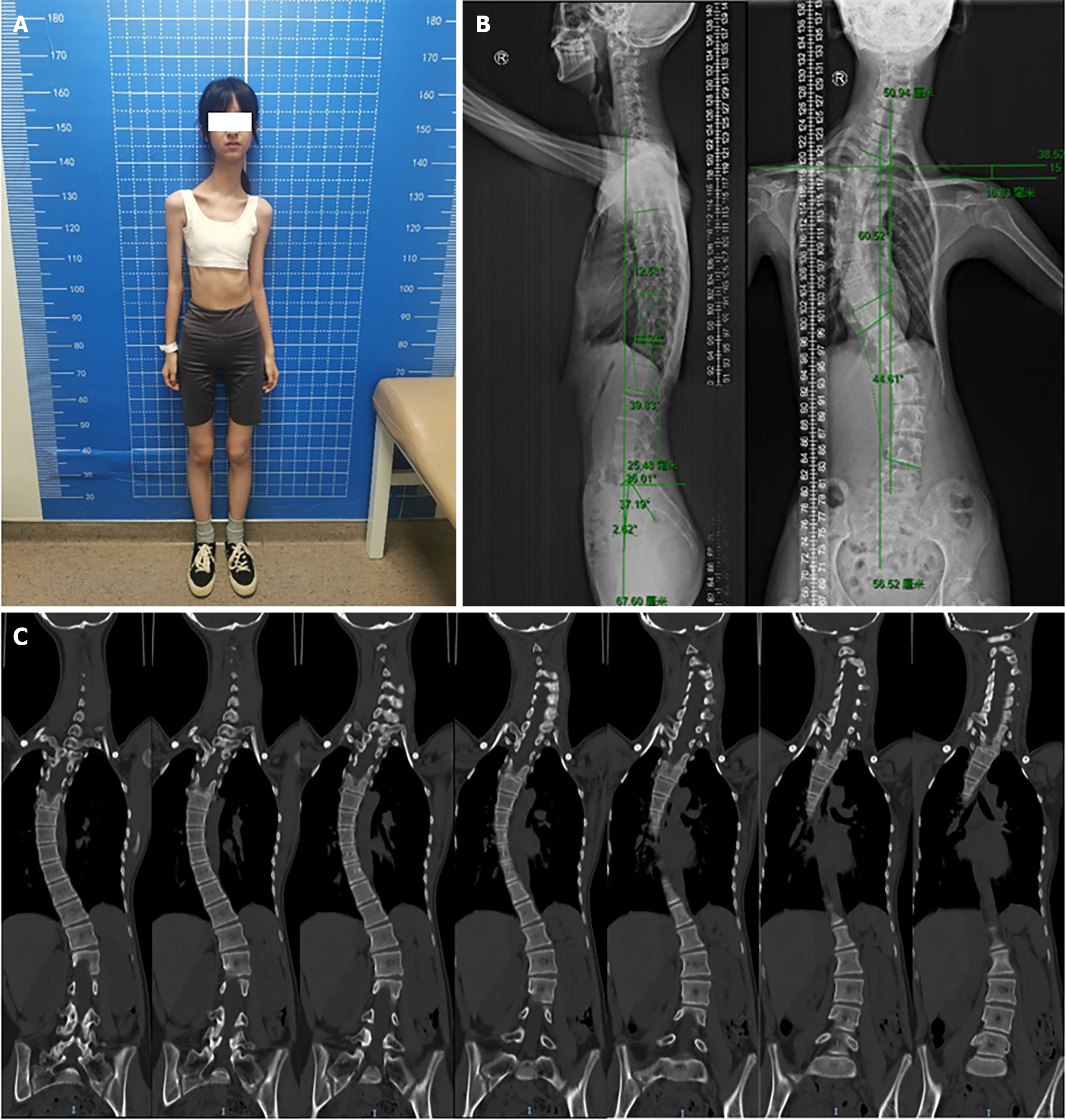
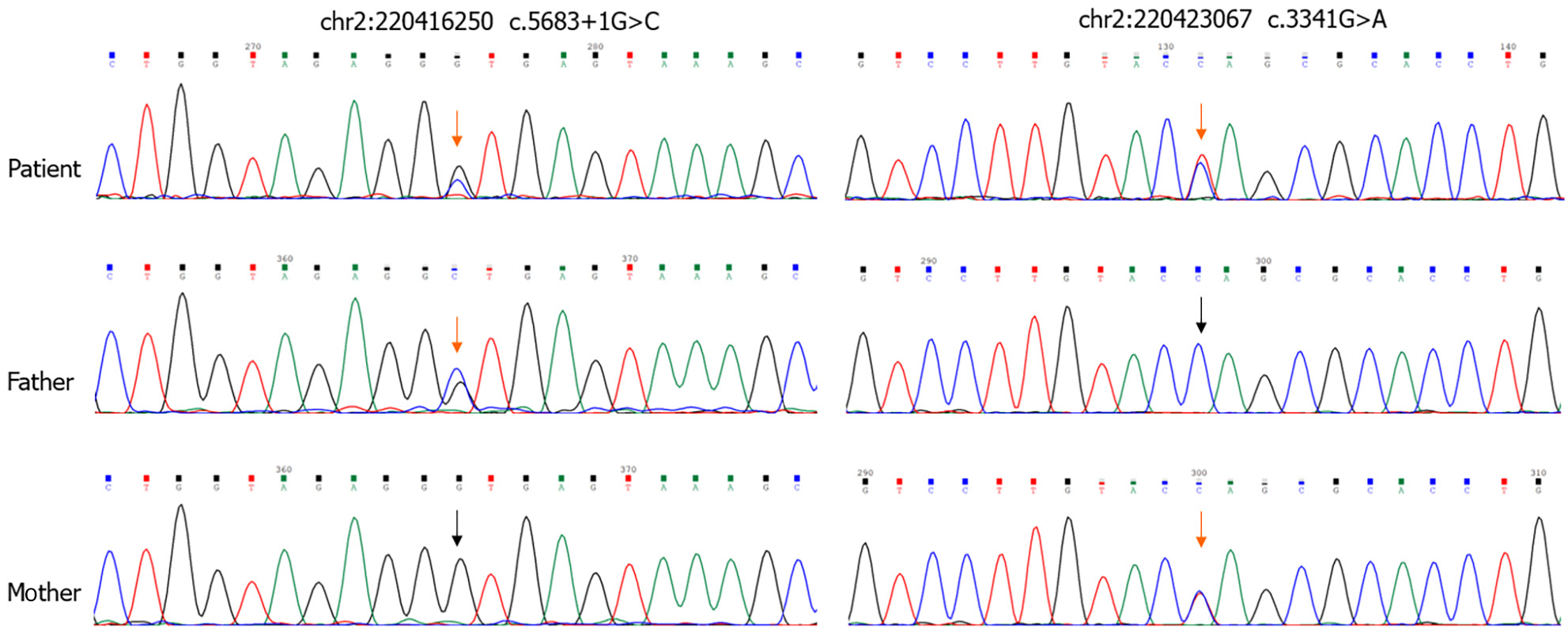
In this case, the patient displayed square shoulders, scoliosis, long slender tubular bones, and normal neurological development. Notably, the patient did not exhibit the typical dysmorphic facial features, relative macrocephaly, or growth retardation commonly observed in individuals with 3M syndrome. Whole exon sequencing revealed a novel heterozygous c.56681+1G>C (Splice-3) variant and a previously reported nonsense heterozygous c.3341G>A (p.Trp1114Ter) variant of OBSL1. Therefore, it is important to note that the clinical features of 3M syndrome may not always be observable, and genetic confirmation is often required. Additionally, the identification of the c.5683+1G>C variant in OBSL1 is notewor
Our study identified a new variant (c.5683+1G>C) of OBSL1 that contributes to expanding the molecular profile of 3M syndrome.
Core Tip: 3M syndrome, a rare autosomal recessive genetic disease that is characterized by severe intrauterine and postnatal growth retardation. Children with 3M syndrome typically exhibit short stature, facial deformities, long tubular bones, and high vertebral bodies, while lacking mental abnormalities or other organ damage. The pathogenic genes associated with 3M syndrome include CUL7, OBSL1 and CCDC8. When encountering patients with short stature and dysmorphic features alongside normal intelligence, it is essential to consider 3M syndrome as a differential diagnosis. Our study has identified a new variant in the OBSL1 gene, contributing to the expanding molecular profile of 3M syndrome.
- Citation: Luo MR, Dai SM, Li Y, Wang Q, Liu H, Gao P, Liu JY, Chen J, Zhao SJ, Yin GY. 3M syndrome patient with a novel mutation: A case report. World J Clin Cases 2024; 12(8): 1454-1460
- URL: https://www.wjgnet.com/2307-8960/full/v12/i8/1454.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i8.1454
3M syndrome (OMIM#273750), a rare autosomal recessive disease, was first discovered in 1975 by Miller et al[1]. Since then, approximately 200 cases have been reported worldwide, including eight in China.
Notably, the prevalence of 3M syndrome may be underestimated due to the possibility of normal mental development; therefore, the actual number of cases could be higher than that previously reported[2].
Typically, 3M syndrome is associated with distinctive facial deformities such as a triangular face, protruding forehead, flat nose, upturned nostrils, full lips, and a wide jaw. However, in the present case, the facial features appeared normal. Additionally, skeletal abnormalities, including hyperlordosis, slender long bones, prominent fleshy heels, joint hypermobility, and congenital hip dislocation, are often observed in individuals with 3M syndrome[3]. Other physical characteristics that may be present include a short neck, square shoulders, sternal abnormalities, narrow thorax, winged scapulae, lordosis, joint laxity, brachydactyly of the fifth finger, and rocker-bottom feet[3]. It is important to note that the intellectual and endocrine functions were unaffected[4].
Diagnosis of 3M syndrome is primarily based on clinical and radiological findings, which are further confirmed through genetic analysis. Three pathogenic genes associated with 3MS have been identified: CUL7 on chromosome 6p21.1, OBSL1 on chromosome 2q35-36.1, and CCDC8 on chromosome 19q13.2-q13.32[5-7]. Because of the different mutated genes, 3M syndrome is categorized into three distinct types: type 1, type 2 and type 3, with incidence rates of 77.5%, 16.3% and 6%, respectively[8].
CUL7 is responsible for encoding the cullin 7 (CUL7) protein, which serves as a scaffold protein and is a vital component of an E3 ubiquitin ligase enzyme. Nonsense or missense mutations in the CUL7 gene prevent the substrate from ubiquitinating, degrading, and accumulating in the body. OBSL1 encodes a cytoskeletal adaptor protein that is primarily localized within the prenuclear region. The exact function of obscurin-like 1 (OBSL1) is still under investigation; however, recent studies have highlighted its interaction with the protein encoded by CCDC8. This interaction is essential for p54-mediated apoptosis in cells. The function of coiled-coil domain-containing protein 8 (CCDC8) remains largely unknown. Unraveling the precise mechanisms governing this intricate relationship is crucial for understanding the physiological and pathological implications associated with these proteins[9].
In this report, we present a comprehensive assessment of the clinical and molecular manifestations in a 3M syndrome patient. We performed whole-exon sequencing to confirm the diagnosis and identified a novel variant of OBSL1, thereby expanding our understanding of the molecular spectrum of this syndrome in the Chinese population.
A 15-year-old woman was referred to the orthopedic clinic for a lateral curvature.
The patient was found to have high and low shoulders, spinal deformity, and low back pain after walking 2 years ago. Notably, the patient did not have any relevant past interventions with outcomes for the final diagnosis.
The patient was generally in good health. No medical history or close contact history of diabetes, hypertension, coronary heart disease, hepatitis, tuberculosis or other infectious diseases, no surgical history, no trauma history, no blood product infusion history, no food or drug allergy history, vaccination history as planned.
Sequencing analysis revealed that the patient's parents were heterozygous carriers, with the father carrying the c.5683+1G>C variant and the mother carrying the c.3341G>A variant (Figure 1).
She was 166 cm tall and weighed only 30 kg, with a body mass index of 10.89. Further examination revealed several dysmorphic features, including square shoulders, slender tubular bones, scoliosis, and a small pelvis (Figure 2). The endocrine function and intelligence of the patient were normal (Table 1). The patient presented with tachycardia, anterior mitral valve prolapse, and mild tricuspid valve insufficiency.
| Age | 15 yr |
| Gender | Female |
| Weight | 30 kg |
| Height | 166 cm |
| BMI | 10.89 |
| Facial features | |
| Relative macrocephaly | - |
| Dolichocephaly | - |
| Triangular face | - |
| Hypoplastic midface | - |
| Fleshy nasal tip | - |
| Long philtrum | - |
| Full lips | - |
| Pointed chin | - |
| Musculoskeletal features | |
| Short broad neck | - |
| Square shoulders | + |
| Short thorax | - |
| Sternal anomaly | - |
| Hyperlordosis | - |
| Generalized or isolated joint hypermobility | - |
| Prominent heels, and pes planus | - |
| Genitourinary anomalies | - |
| Intelligence | Normal |
| Radiographic features | |
| Long slender tubular bones | + |
| Tall vertebra | - |
| Wedging of the thoracic vertebral bodies | - |
| Thoracic kyphoscoliosis | + |
| Small pelvis | + |
| Molecular findings | |
| Karyotype | 46, XX |
| Gene | OBSL1(NM_015311) |
| Variation (DNA) | c.5683+1G>C and c.3341G>A |
| Variation (protein) | Splice-3 and p.Trp1114Ter |
| Novelty | Novel and Known |
| ACMG classification | Pathogenic |
Lung function tests indicated restrictive ventilation dysfunction, with maximum voluntary ventilation of > 70%. EDTA-anticoagulated blood samples were collected from the patients after obtaining informed consent from their guardians. Genomic DNA was extracted from whole blood using standard procedures. In this study, we used whole-exon sequencing to sequence CUL7 (NM_001168370), OBSL1 (NM_015311) and CCDC8 (NM_032040) genes[10]. In this case, the heterozygous variants in OBSL1 (NM_015311) were c.5683+1G>C (Splice-3) and c.3341G>A (p.Trp1114Ter). OBSL1 consists of a protein with 4 tandem N-terminal immunoglobulin (Ig)-like domains, a central fibronectin domain, and 13 C-terminal Ig domains. The nonsense mutation c.3341G>A (p.Trp1114Ter) is located in exon10, the IGc2 domain of the protein (amino acid positions 1095–1160). The mutation prematurely terminates the translation of the protein and prevents subsequent amino acid synthesis, which includes several domains. The IG domain may be involved in a variety of functions in proteins, such as intercellular recognition, cell surface receptors, muscle structure, and the immune system, so the mutation may affect the normal physiological function of the protein. The c.5683+1G>C mutation is located on the intron, which is located at the classical splicing site and is predicted by the software to potentially affects mRNA splicing, which leads to decreased OBSL1 protein expression and loss of protein function, contributing to disease occurrence. Notably, the c.5683+1G>C variant has not been previously reported. Sequencing analysis revealed that the patient’s parents were heterozygous carriers, with the father carrying the c.5683+1G>C variant and the mother carrying the c.3341G>A variant (Figure 1).
3M syndrome with scoliosis.
Spinal orthopedic surgery.
The patient was still alive.
The case from a Chinese family with imaging characteristics and dysmorphological findings were partially consistent with those of 3M syndrome. The patient was evaluated by a proficient clinical orthopedist. Demographic information, family medical history, clinical manifestations, and radiological findings were extracted from hospital records.
Clinical records were retrospectively reviewed to extract the epidemiological data (sex, age, consanguinity, geographical origin, and personal history); clinical features (facial dysmorphisms, psychomotor and intellectual development, thoracic deformities, spinal anomalies, and limb abnormalities); and radiological findings (radiography and CT)[2]. All patient-specific information was properly deidentified to ensure privacy.
3M syndrome is a rare autosomal recessive disease. This syndrome causes severe intrauterine and postnatal growth retardation, facial deformities and skeletal malformations; however, intelligence and endocrine function remain unaffected. Its diagnosis is primarily based on gene sequencing; however, the prognosis remains unclear.
In previous studies, the disease locus gene was mapped to chromosome 6p21.1 using a homozygosity mapping strategy, leading to the identification of mutations in the CUL7 gene. CUL7 (6p21.1) is the major gene associated with 3M syndrome, accounting for 77.5% of cases[5]. Other mutations involve OBSL1 (2q35) in 16.3% of cases and CCDC8 (19q13.33) in 6% of cases[11].
CUL7, OBSL1 and CCDC8 physically interact with each other to form the 3M complex, which plays a crucial role in maintaining microtubule and genome integrity[12]. Disruption of this complex results in microtubule damage, abnormal chromosomal separation and cell death. However, the exact mechanism underlying 3M syndrome development remains unclear. It is evident that there is still much to be discovered about the intricate functions and interactions of CUL7, OBSL1 and CCDC8. Moreover, the detailed mechanisms responsible for the growth impairments observed in the 3M syndrome remain largely unclear.
According to the human gene mutation database (http://www.hgmd.cf.ac.uk), 88 published variants of CUL7 and 36 of OBSL1 lead to 3M syndrome. The most common variants include frameshift, nonsense and missense mutations. In this case, one variant was a nonsense mutation, whereas the other was affected by splicing. It is rare to have two mutations simultaneously, which may also lead to more severe developmental abnormalities; however, the patient’s intellectual development remains unaffected. Similarly, determining whether a direct relationship exists between scoliosis and genetic mutations in patients requires additional case data.
It is noteworthy that CUL7, OBSL1 and CCDC8 are responsible for 98% of 3MS cases without a consistent genotype–phenotype correlation[8]. For orthopedists, it is important to provide symptomatic treatment and follow-up care, considering the high likelihood of encountering patients with this syndrome. In this case, growth hormone (GH) therapy remains controversial owing to individual differences and the treatment effectiveness[11]. Although the patient had normal GH levels, GH therapy should be considered to address the severe short stature commonly associated with 3M syndrome despite the absence of growth restriction in this patient. The duration of treatment should be determined based on the height gain and growth rate.
While the prenatal diagnosis of 3M syndrome is a topic of debate due to normal intelligence, preimplantation genetic diagnosis is of great importance for families aiming to have a healthy child.
In conclusion, when encountering patients with short stature and dysmorphic features along with normal intelligence, it is essential to consider 3M syndrome as a differential diagnosis. Genetic sequencing of CUL7, OBSL1 and CCDC8 is necessary to confirm the clinical diagnosis and provide appropriate genetic counseling.
Our study identified a new variant (c.5683+1G>C) of OBSL1 that contributes to expanding the molecular profile of 3M syndrome.
We would like to thank the patient and her family for supporting this work.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: Foreign member of the Russian Academy of Sciences; Chairman of Orthopedic Branch of Jiangsu Medical Association; Chairman designate of Orthopaedic Branch of Jiangsu Medical Association.
Specialty type: Orthopedics
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Sánchez-Puig N, Mexico; Zha L, China S-Editor: Liu JH L-Editor: Kerr C P-Editor: Yu HG
| 1. | Miller JD, McKusick VA, Malvaux P, Temtamy S, Salinas C. The 3-M syndrome: a heritable low birthweight dwarfism. Birth Defects Orig Artic Ser. 1975;11:39-47. [PubMed] |
| 2. | Isik E, Arican D, Atik T, Ooi JE, Darcan S, Ozen S, Simsek Kiper PO, Utine E, Cogulu O, Ozkinay F. A rare cause of syndromic short stature: 3M syndrome in three families. Am J Med Genet A. 2021;185:461-468. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 3. | Simsek-Kiper PO, Taskiran E, Kosukcu C, Arslan UE, Cormier-Daire V, Gonc N, Ozon A, Alikasifoglu A, Kandemir N, Utine GE, Alanay Y, Alikasifoglu M, Boduroglu K. Further expanding the mutational spectrum and investigation of genotype-phenotype correlation in 3M syndrome. Am J Med Genet A. 2019;179:1157-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Lugli L, Bertucci E, Mazza V, Elmakky A, Ferrari F, Neuhaus C, Percesepe A. Pre- and post-natal growth in two sisters with 3-M syndrome. Eur J Med Genet. 2016;59:232-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Huber C, Dias-Santagata D, Glaser A, O'Sullivan J, Brauner R, Wu K, Xu X, Pearce K, Wang R, Uzielli ML, Dagoneau N, Chemaitilly W, Superti-Furga A, Dos Santos H, Mégarbané A, Morin G, Gillessen-Kaesbach G, Hennekam R, Van der Burgt I, Black GC, Clayton PE, Read A, Le Merrer M, Scambler PJ, Munnich A, Pan ZQ, Winter R, Cormier-Daire V. Identification of mutations in CUL7 in 3-M syndrome. Nat Genet. 2005;37:1119-1124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 138] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 6. | Hanson D, Murray PG, Sud A, Temtamy SA, Aglan M, Superti-Furga A, Holder SE, Urquhart J, Hilton E, Manson FD, Scambler P, Black GC, Clayton PE. The primordial growth disorder 3-M syndrome connects ubiquitination to the cytoskeletal adaptor OBSL1. Am J Hum Genet. 2009;84:801-806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Hanson D, Murray PG, O'Sullivan J, Urquhart J, Daly S, Bhaskar SS, Biesecker LG, Skae M, Smith C, Cole T, Kirk J, Chandler K, Kingston H, Donnai D, Clayton PE, Black GC. Exome sequencing identifies CCDC8 mutations in 3-M syndrome, suggesting that CCDC8 contributes in a pathway with CUL7 and OBSL1 to control human growth. Am J Hum Genet. 2011;89:148-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | Huber C, Munnich A, Cormier-Daire V. The 3M syndrome. Best Pract Res Clin Endocrinol Metab. 2011;25:143-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 9. | Khachnaoui-Zaafrane K, Ouertani I, Zanati A, Kandara H, Maazoul F, Mrad R. 3M syndrome: A Tunisian seven-cases series. Eur J Med Genet. 2022;65:104448. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 10. | Clayton PE, Hanson D, Magee L, Murray PG, Saunders E, Abu-Amero SN, Moore GE, Black GC. Exploring the spectrum of 3-M syndrome, a primordial short stature disorder of disrupted ubiquitination. Clin Endocrinol (Oxf). 2012;77:335-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Yan J, Yan F, Li Z, Sinnott B, Cappell KM, Yu Y, Mo J, Duncan JA, Chen X, Cormier-Daire V, Whitehurst AW, Xiong Y. The 3M complex maintains microtubule and genome integrity. Mol Cell. 2014;54:791-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 12. | Tüysüz B, Alp Ünkar Z, Turan H, Gezdirici A, Uludağ Alkaya D, Kasap B, Yeşil G, Vural M, Ercan O. Natural history of facial and skeletal features from neonatal period to adulthood in a 3M syndrome cohort with biallelic CUL7 or OBSL1 variants. Eur J Med Genet. 2021;64:104346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |