Published online Mar 16, 2024. doi: 10.12998/wjcc.v12.i8.1395
Peer-review started: December 1, 2023
First decision: December 7, 2023
Revised: January 8, 2024
Accepted: February 18, 2024
Article in press: February 18, 2024
Published online: March 16, 2024
Processing time: 101 Days and 15.9 Hours
As a well-known fact to the public, gestational diabetes mellitus (GDM) could bring serious risks for both pregnant women and infants. During this important investigation into the linkage between GDM patients and their altered expression in the serum, proteomics techniques were deployed to detect the differentially expressed proteins (DEPs) of in the serum of GDM patients to further explore its pathogenesis, and find out possible biomarkers to forecast GDM occurrence.
To investigation serum proteins differentially expressed in GDM were assessed using isobaric tag for relative and absolute quantitation (iTRAQ) proteomics and bioinformatics analyses.
Subjects were divided into GDM and normal control groups according to the IADPSG diagnostic criteria. Serum samples were randomly selected from four cases in each group at 24-28 wk of gestation, and the blood samples were identified by applying iTRAQ technology combined with liquid chromatography-tandem mass spectrometry. Key proteins and signaling pathways associated with GDM were identified by bioinformatics analysis, and the expression of key proteins in serum from 12 wk to 16 wk of gestation was further verified using enzyme-linked immunosorbent assay (ELISA).
Forty-seven proteins were significantly differentially expressed by analyzing the serum samples between the GDM gravidas as well as the healthy ones. Among them, 31 proteins were found to be upregulated notably and the rest 16 proteins were downregulated remarkably. Bioinformatic data report revealed abnormal expression of proteins associated with lipid metabolism, coagulation cascade activation, complement system and inflammatory response in the GDM group. ELISA results showed that the contents of RBP4, as well as ANGPTL8, increased in the serum of GDM gravidas compared with the healthy ones, and this change was found to initiate from 12 wk to 16 wk of gestation.
GDM symptoms may involve abnormalities in lipid metabolism, coagulation cascade activation, complement system and inflammatory response. RBP4 and ANGPTL8 are expected to be early predictors of GDM.
Core Tip: Gestational diabetes mellitus (GDM) poses a great risk to mothers and infants. During this research, proteomics techniques were applied to investigate the differentially expressed proteins in the serum of the GDM patients to further explore its pathogenesis, and to search for potential biomarkers for disease forecast. Subjects were divided into GDM and normal control groups according to the IADPSG diagnostic criteria. Key proteins and signaling pathways associated with GDM were identified by bioinformatics analysis, and the expression of key proteins in serum from 12 wk to 16 wk of gestation was further verified using enzyme-linked immunosorbent assay.
- Citation: Cao WL, Yu CP, Zhang LL. Serum proteins differentially expressed in gestational diabetes mellitus assessed using isobaric tag for relative and absolute quantitation proteomics. World J Clin Cases 2024; 12(8): 1395-1405
- URL: https://www.wjgnet.com/2307-8960/full/v12/i8/1395.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i8.1395
The prevalence rate of gestational diabetes mellitus (GDM) is high during the whole pregnancy process, and could also cause serious harm to both mother and fetus[1]. There are two main types of diabetes in pregnancy, one is pre-existing diabetes before gestation, called pre-pregnancy DM, and the other is diabetes that first occurs after pregnancy, called GDM[2]. According to the related reports, among the gravidas with diabetes mellitus, 90% belong to the GDM category. If the condition of GDM is severe or the blood sugar is not well controlled, it will have a greater impact on both the pregnant woman and the fetus, while high blood sugar can cause abnormal development of the embryo and even intrauterine death[3]. Till now, the pathologic development and leading causes of GDM remain to be further discovered, but the known factors include genetics, involvement of inflammatory factors, impaired lipid metabolism, and abnormal expression of estrogen and progesterone receptors[4-6]. Therefore, it is important to understand the pathogenesis of GDM for early prevention and treatment and to reduce maternal and fetal mortality.
Since the placenta is directly related to maternal circulation, potential changes in protein expression in the placenta should also lead to the alterations of the that in serum. Therefore, one interesting assumption could be raised about the differentially Expressed Proteins (DEPs): The alterations (changes in categories and quantity for example) of DEPs could be developed as a tool to discover serum markers that predict GDM specificity and will also help to further investigate the pathogenesis of GDM. But this should only work under the condition that the above DEPs changes could be detected from the GDM patient’s serum.
Isobaric tag for relative and absolute quantitation (iTRAQ) technology is a tandem mass spectrometry-based protein quantification technique that allows absolute or relative quantification of proteins[7]. This technique possesses the following merits: Improved sensitivity, superior resolution, and enhanced reproducibility when contrasted with the alternative ones. The iTRAQ technique combined with (LC-MS/MS) can screen a large number of DEPs, and the study of these DEPs provides a strong theoretical basis for identifying biomarkers of GDM in the future. However, few studies have been reported on the application of iTRAQ technology to identify GDM[8]. In this study, we propose to apply iTRAQ experiments and LC-MS/MS (liquid chromatography-tandem mass spectrometry) analysis to detect DEPs in the serum of GDM patients and perform bioinformatics analysis, which will help to understand the pathogenic mechanism of GDM to find its potential early serum markers.
Blood samples were collected from pregnant women at 12 wk to 16 wk and 24 wk to 28 wk without any treatment from October 2020 to April 2021 at the Maternal and Child Health Hospital of Hubei Province. The pregnancy outcomes of the pregnant women were followed, and the samples were divided into GDM and control groups according to the IADPSG diagnostic criteria, and finally 4 samples from each of the GDM and control groups were selected for protein quantification.
The aging condition of the GDM patients was calculated as 29.8 years ± 2.7 years (mean ± SD), with a gestation of 26.1 wk ± 1.0 wk. In the control group, this statistic was counted as 30.1 years ± 1.4 years, with a gestation of 26.4 wk ± 1.2 wk. There was no statistically significant difference in age (P = 0.74) and gestational week (P = 0.81) between the two groups. The experiment was approved by the Medical Ethics Committee of the Maternal and Child Health Hospital of Hubei Province, and all participating subjects signed the consent form.
For more precise experimental data, high-abundance proteins were eliminated via PierceTM Top 12 Abundant Protein Depletion Spin Columns (Thermo Fisher). Protein concentration was assessed using the Bradford method, and the quality of samples was evaluated by running through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Followed by protein quantification, a centrifugal tube containing 60 μg protein solutions was combined with 5 μL dithiothreitol at 37 °C for 1 h. Subsequently, 20 μL iodoacetamide was added, and the reaction lasted for 1 h at room temperature. All samples were pipetted into the ultrafiltration tube, and the filtrate was discarded post-centrifugation. UA buffer (100 μL) was added and centrifuged under 14000 g for 10 min (twice repeats). Fifty mmol/L NH4HCO3 (50 μL) was then added to the filtrate after centrifugation (thrice repeats). Trypsin buffer (40 μL) was added and shaken under 600 rpm for 1 min. The sample then underwent enzymatic hydrolysis at 37 °C for 12 h to 18 h, and a 75 μg sample was extracted. Following labeling, equal volumes of each sample were mixed and desalted using a C18 cartridge.
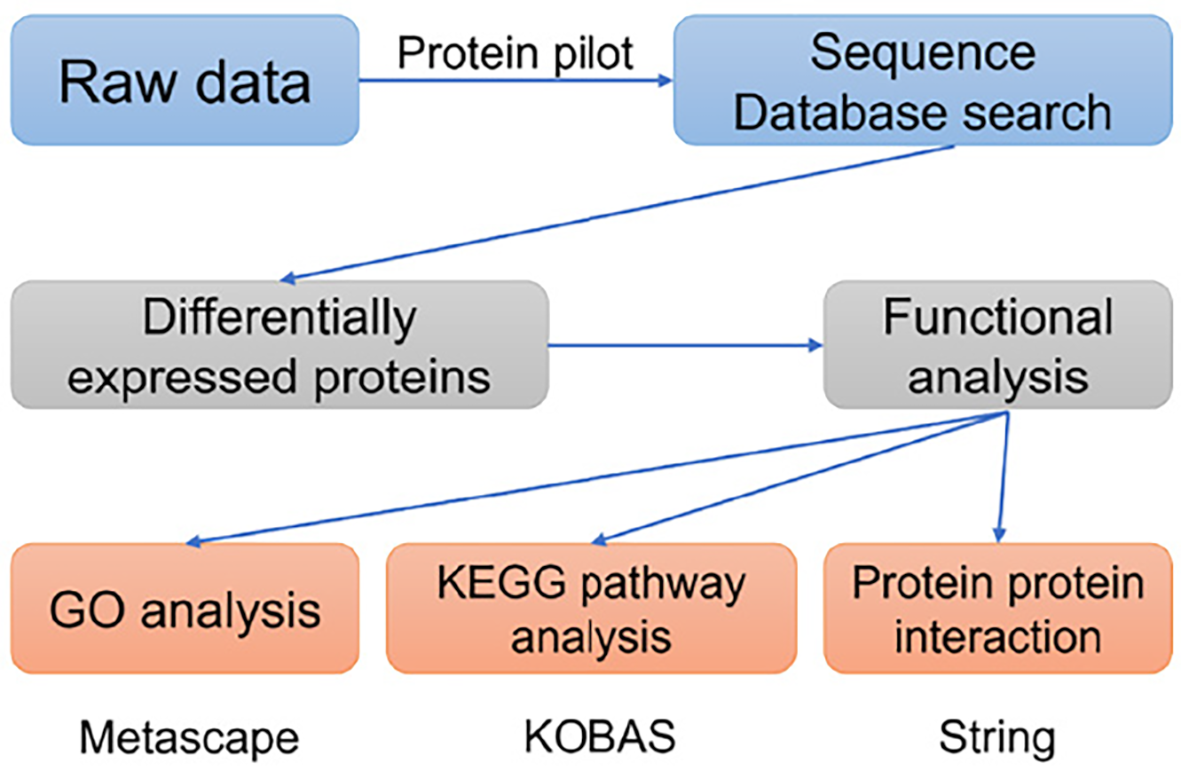
The labeled samples were redissolved in 40 μL 0.1% formic acid aqueous solution. The peptides were loaded onto a C18-reversed phase column (3 μm-C18 resin, 75 μm × 15 cm). The mobile phase is 2% methyl cyanide/0.1% formic acid/98% water and 80% methyl cyanide/0.08% formic acid/20% water. The gradient: 0 to 68 min, the B phase rises from 7% to 36% linearly; 68 min to 75 min, rises from 36% to 100% linearly. Each sample was separated by capillary high-performance liquid chromatography and analyzed by Orbitrap Fusion Lumos mass spectrometer (Thermo Science). After the data were collected, the data were processed according to the flowchart shown in Figure 1. ProteinPilot Software 4.2 (Inc., La Jolla) was used to search the database uniprot_human, FASTA to obtain different results. The protein fold changes of at least 1.2 (in both replicates) were obtained and were significantly different between the groups at P < 0.05 as a significant difference.
Metascape serves as an online tool for gene and protein annotation, visualization, and integration discovery (http://metascape.org). Therefore, this tool was deployed to conduct Gene Ontology (GO) analyses. For Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, the KOBAS online analysis database (http://kobas.cbi.pku.edu.cn/) was employed. Meanwhile, the protein-protein interaction (PPI) network among the DEPs were constructed using the online String tool (https://cn.string-db.org/). For the comparisons between groups, statistical significance was determined with a two-sided P value < 0.05.
RBP4 and ANGPTL8 proteins were tested to be upregulated in the GDM patients after running the enzyme-linked immunosorbent assay (ELISA) experiment using the kit (specification and catalog number: 96T, OM626395). The sample collection and quantified evaluation were conducted under the producer’s usage manual. The hematology specimens from each of the GDM patients and healthy gravidas were selected for ELISA, all of which were obtained from 12 wk to 16 wk of gestation.
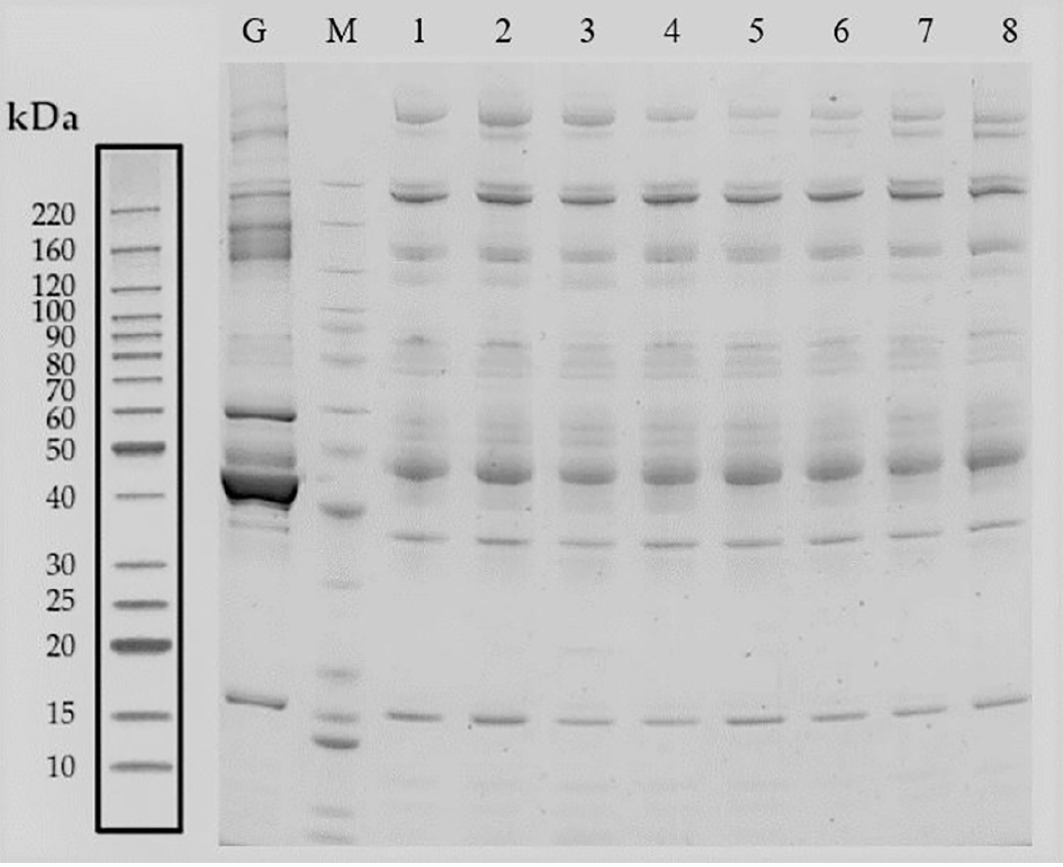
The hematology specimens from both groups of GDM and control were collected, followed by separation through SDS-PAGE electrophoresis. The total proteins from 8 samples were successfully separated within the molecular weight range of 10-220 kDa, ensuring no protein was degraded as well as efficient removal of high-abundance proteins. After imaging the proteins on the film, a similar band exists in all of the 8 individuals, suggesting that the proteins extracted could meet the requirements of subsequent assays (Figure 2).
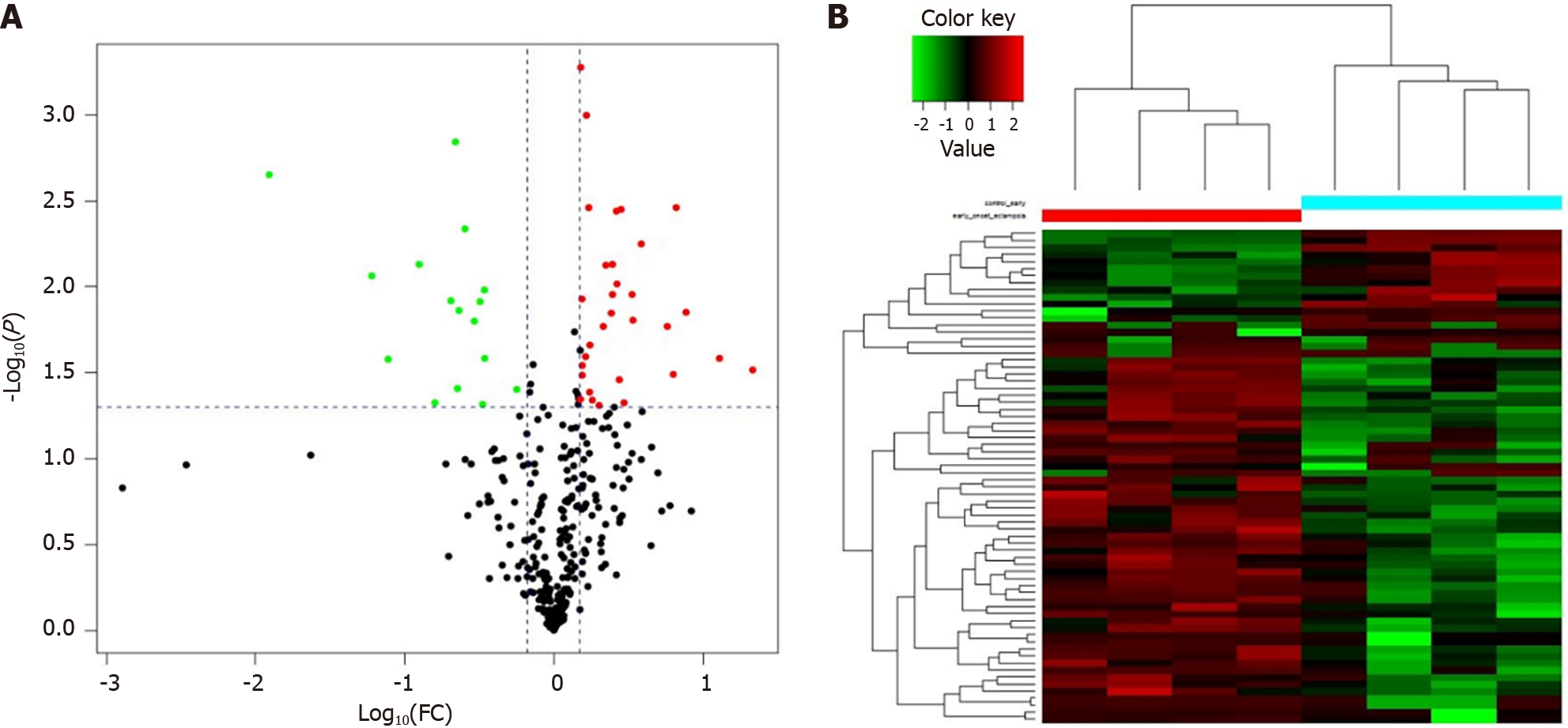
LC-MS/MS technique is popular and important for analyzing proteins in serum specimens. Two hundred and fifteen DEPs were discovered in the GDM patients and gravidas, of which 47 proteins were identified with iTRAQ ratios > 1.50 or < 0.67, with statistically significant differences (P < 0.05). The data reveals that 31 proteins were detected with an expression elevation and the rest 16 proteins were detected with an expression inhibition in the GDM gravidas compared with the healthy women (Table 1). The protein content alterations were observed between the two groups, and these changes were then formed a cluster diagram (Figure 3).
| Regulated type | Protein accession | Gene name | Protein description | Fold change |
| Up | Q6UXH0 | ANGPTL8 | Angiopoietin like 8 | 5.86 |
| Up | P02753 | RBP4 | Retinol binding protein 4 | 4.81 |
| Up | P02649 | APOE | Apolipoprotein E | 3.86 |
| Up | P08505 | IL-6 | Interleukin 6 | 3.40 |
| Up | P05121 | PIA-1 | Plasminogen activator inhibitor-1 | 3.17 |
| Up | P02671 | FGA | Fibrinogen alpha chain | 2.82 |
| Up | P06396 | GSN | Gelsolin | 2.74 |
| Up | O88947 | F10 | Coagulation factor X | 2.65 |
| Up | P05164 | MPO | Myeloperoxidase | 2.56 |
| Up | Q6Q788 | F9 | Apolipoprotein A5 | 2.51 |
| Up | J3KPA1 | CRISP3 | Cysteine-rich secretory protein 3 | 2.50 |
| Up | P00748 | F12 | Coagulation factor XII | 2.47 |
| Up | P27169 | PON1 | EGF containing fibulin extracellular matrix protein 1 | 2.31 |
| Up | P49913 | CAMP | Cathelicidin antimicrobial peptide | 2.24 |
| Up | P13671 | C6 | Complement C6 | 2.23 |
| Up | P16070 | CD44 | CD44 antigen | 2.13 |
| Up | Q76LX8 | ADAMTS13 | Von willebrand factor-cleaving protease | 2.01 |
| Up | P00451 | F8 | Coagulation factor VIII | 1.96 |
| Up | P02743 | APCS | Serum amyloid P-component | 1.95 |
| Up | Q16853 | AOC3 | Membrane primary amine oxidase | 1.95 |
| Up | Q9NZP8 | C8B | Complement C1r subcomponent-like protein | 1.92 |
| Up | A0A024R035 | C9 | Complement C9 | 1.86 |
| Up | P09871 | C1S | Complement C1s subcomponent | 1.83 |
| Up | P49908 | SELENOP | Selenoprotein P | 1.81 |
| Up | Q14624 | ITIH4 | Inter-alpha-trypsin inhibitor heavy chain 4 | 1.75 |
| Up | B4DPQ0 | C1R | Complement C1r subcomponent | 1.73 |
| Up | P19827 | ITIH1 | Inter-alpha-trypsin inhibitor heavy chain 1 | 1.71 |
| Up | P08709 | F7 | Coagulation factor VII | 1.71 |
| Up | P04275 | VWF | von Willebrand factor | 1.65 |
| Up | C0JYY2 | APOB | Apolipoprotein B | 1.55 |
| Up | P00740 | F9 | Coagulation factor IX | 1.52 |
| Down | P05543 | SERPINA7 | Thyroxine-binding globulin | 0.56 |
| Down | P01024 | C3 | Complement C3 | 0.43 |
| Down | P01008 | SERPINC1 | Serpin family C member 1 | 0.34 |
| Down | P07333 | CSF1R | Macrophage colony-stimulating factor 1 receptor | 0.34 |
| Down | Q12860 | CNTN1 | Contactin 1 | 0.33 |
| Down | P04070 | PROC | Protein C, inactivator of coagulation factors Va and VIIIa | 0.29 |
| Down | P01871 | IGHM | Immunoglobulin heavy constant mu | 0.25 |
| Down | Q15485 | FCN2 | Ficolin 2 | 0.23 |
| Down | Q15848 | ADPN | Adiponectin | 0.23 |
| Down | Q8WWZ8 | OIT3 | Oncoprotein-induced transcript 3 protein | 0.22 |
| Down | Q9UNU2 | C4B | Complement C4-B | 0.20 |
| Down | O95445 | APOM | Apolipoprotein M | 0.16 |
| Down | P04278 | SHBG | Sex hormone-binding globulin | 0.15 |
| Down | Q14213 | EBI3 | Interleukin-27 subunit beta | 0.13 |
| Down | O43866 | CD5L | CD5 antigen-like | 0.06 |
| Down | P06733 | ENO1 | Alpha-enolase | 0.02 |
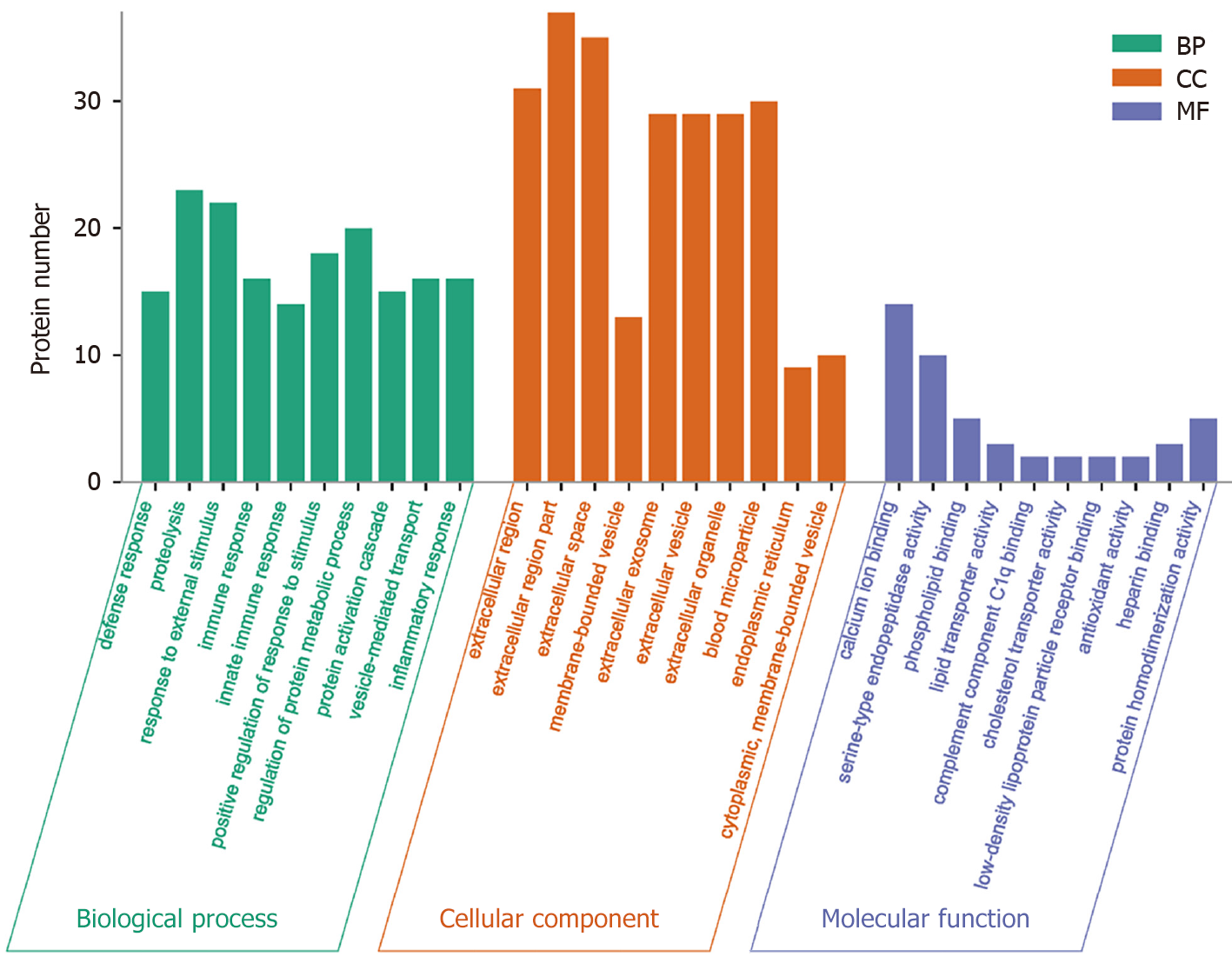
GO analysis, a widely employed and powerful analyzing tool in bioinformatics, encompasses three ontologies: Cell composition (CC), molecular function (MF), and biological process (BP). The outcomes of the GO functional annotation analysis reveal the DEPs quantity under either column among the mentioned ontologies. GO functional enrichment analysis reveals noteworthy functional elements among the DEPs, which will help researchers find out the related affected biomolecular functions by analyzing the notable changes. As a well-known fact, proteins are usually found to collaborate into a unit to manifest their biological activities in organisms, and based on the merits introduced for analyzing the protein expression difference, pathway-related analysis will become easier and deepen the comprehension of the relationship between alterations and functions, for instance, the key biochemical metabolic and signaling transduction pathways linked to DEPs.
The analyzing report of the GO function annotation has been depicted in Figure 4. Under the BP category, most proteins were closely related to the function “proteolysis” (n = 23 proteins), for instance, the top three up-regulated proteins RBP4, FGA, and PON1. Under the CC category, a significant proportion of proteins were closely related to the function “extracellular region part” (n = 37 proteins), for instance, the top three up-regulated proteins ANGPTL8, RBP4, and GSN. Regarding the MF category, the majority of proteins were closely related to the function “calcium ion binding” (n = 14 proteins), for instance, the top three up-regulated proteins ANGPTL8, FGA, and ADAMTS13.
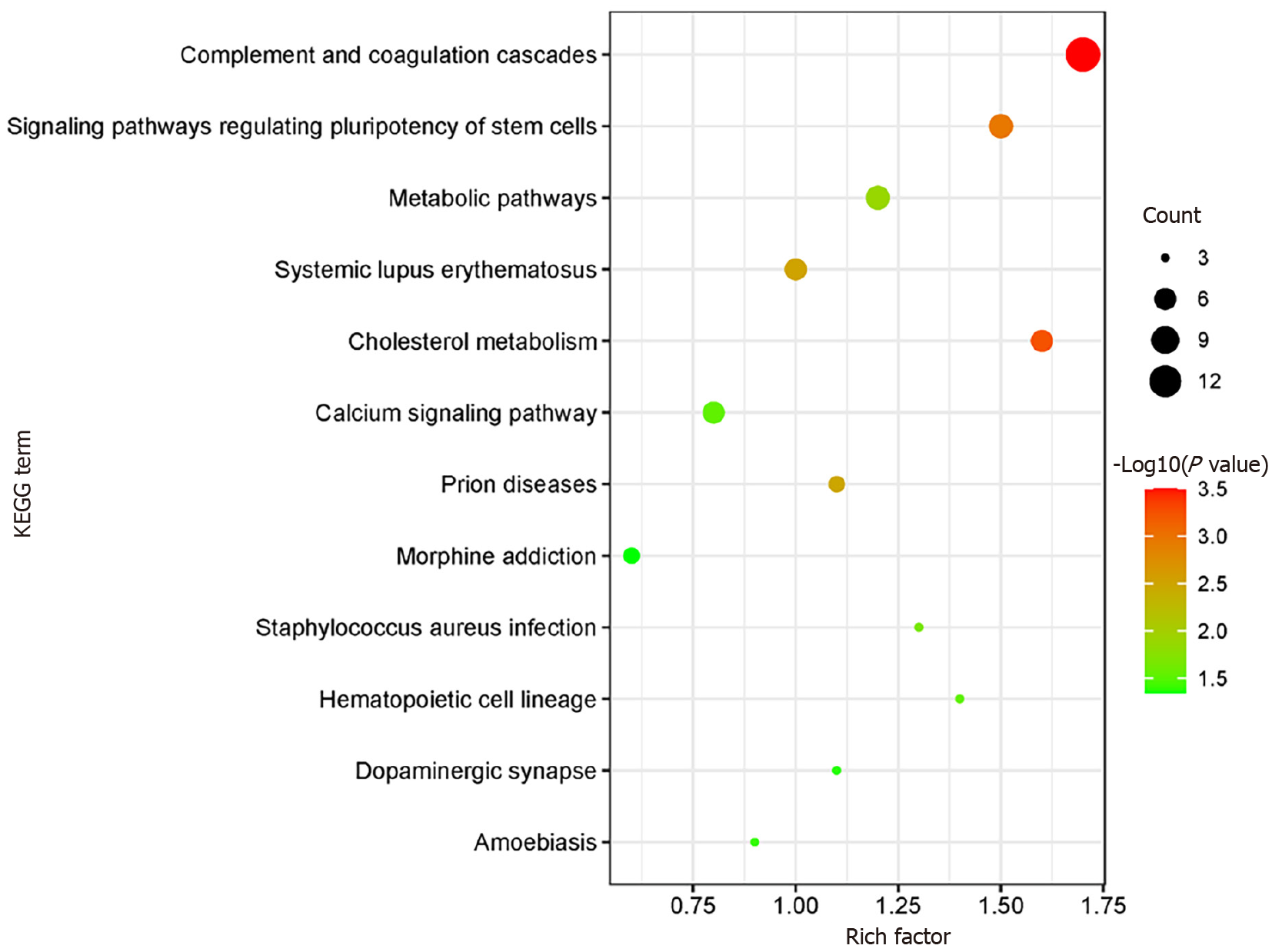
KOBAS online analysis tool was used to identify functions associated with DEPs and KEGG signaling pathways. Results of KEGG pathway enrichment show that complement and coagulation cascades, signaling pathways regulating pluripotency of stem cells metabolic pathways, and cholesterol metabolism are the main signaling pathways in the GDM group (Figure 5).
The KOBAS online analysis tool was employed to discern functions linked to DEPs and KEGG signaling pathways. The analysis report of KEGG pathway enrichment indicates that the primary signaling pathways in the GDM group involve complement and coagulation cascades, signaling pathways regulating pluripotency of stem cells, metabolic pathways, and cholesterol metabolism (Figure 5).
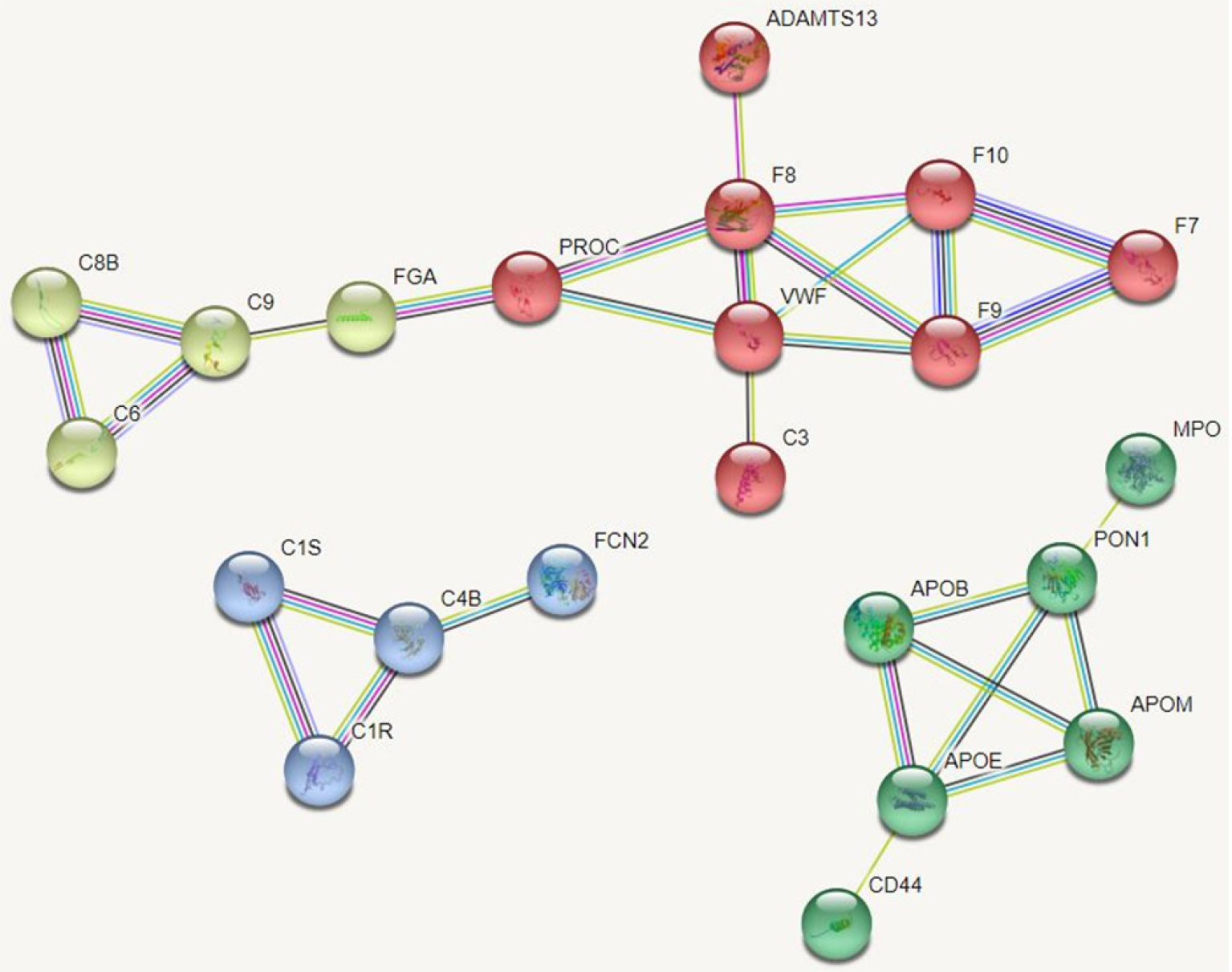
Protein-protein interaction networks (PPI networks) are composed of individual proteins that interact with each other to participate in various aspects of life processes such as biological signaling, regulation of gene expression, energy and material metabolism, and cell cycle regulation. Systematic analysis of the interaction relationships of a large number of proteins in biological systems is important for understanding how proteins work in biological systems and for understanding the functional connections between proteins. Studying the interaction network between proteins also helps to uncover the core regulatory genes. A PPI network of DEPs was constructed and the core proteins were clustered with different colors clustering was performed. The results showed that core proteins are mainly involved in the coagulation cascade, lipid metabolism, and inflammatory activation (Figure 6).
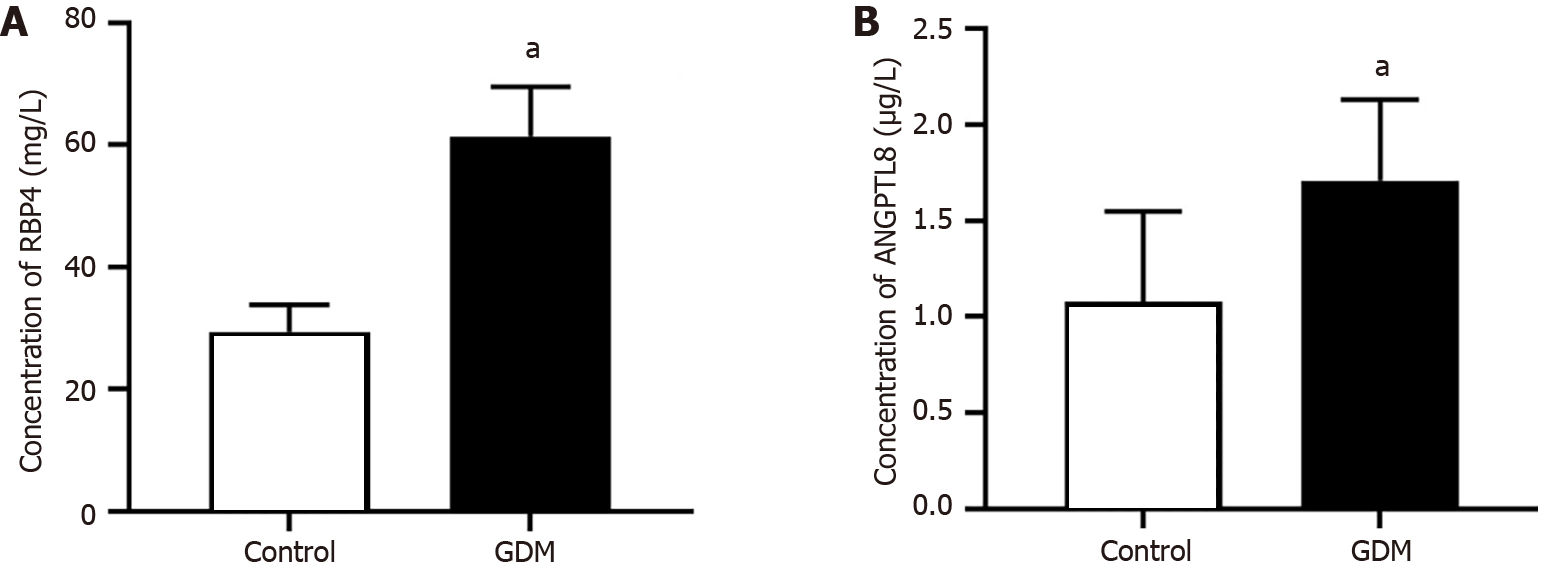
Serum levels of RBP4 and ANGPTL8 were measured by ELISA. The serum concentrations of RBP4 and ANGPTL8 were 61.39 mg/L ± 8.26 mg/L and 1.71 μg/L ± 0.42 μg/L in the GDM group and 29.37 mg/L ± 4.32 mg/L and 1.08 μg/L ± 0.47 μg/L in the control group. The data showed that the above two protein concentrations were significantly higher in the GDM group than in the control group (Figure 7).
The etiology of GDM is not fully understood, and current research suggests that it is a multifactorial disease specific to pregnancy[9]. Biomarkers have great potential in predicting GDM, and several biomarkers have been identified that can be involved in early prediction but are not widely used in clinical practice[10-12]. An in-depth understanding of the pathogenesis of GDM can guide its early intervention and help improve pregnancy outcomes and fetal prognosis. At present, proteomics technology has achieved some results in the study of GDM[13].
In this study, the analysis of serum protein profiles of pregnant women from 11 wk to 16 wk of gestation revealed that 47 DEPs were identified in the GDM group, of which 31 were up-regulated and 16 were down-regulated. Bioinformatics analysis revealed that the BPs of these proteins in vivo mainly include coagulation, lipid metabolism, complement activation, and acute inflammatory response. These BPs have been found by investigators to be involved in the pathophysiological processes of GDM. Interestingly, these DEPs are also present in preeclampsia, type 1 diabetes, type 2 diabetes, recurrent abortion, and other diseases. Since differential expression of these proteins has been identified in early or mid-gestation, these proteins may be potential biomarkers for the early diagnosis of GDM.
In normal pregnancy, the coagulation and fibrinolytic systems of pregnant women are altered relative to those of non-pregnant women to help cope with labor and reduce postpartum hemorrhage. The expression levels of coagulation factors VIII, IX, X, XII, FGA, and vWF were reported to be significantly increased in pregnant women with GDM relative to healthy pregnant women, while coagulation factor V was significantly decreased, which is also consistent with our results[14-16]. This result suggests that a hypercoagulable blood state in early pregnancy predicts a greater likelihood of GDM in later gestation. In addition, we found that SERPINC1 and PROC expression were downregulated in the GDM group. SERPINC1 has the effect of inhibiting thrombin and coagulation factors IX and X, thus suppressing the anticoagulant state, which makes pregnant women with GDM more inclined to a hypercoagulable state in early pregnancy[17].
Insulin resistance in peripheral tissues and increased inflammatory response are two characteristics of normal pregnant women, which are more prominent in pregnant women with GDM[18-20]. We identified several complement proteins and inflammatory response-related proteins, including C3, C4a, C6, C9, C8B, C1S, and C1R. A recent study showed that maternal blood levels of C3a and C4a were significantly downregulated in GDM at the time of delivery compared to controls[21]. The actual mechanism by which plasma complement proteins are reduced in pregnant women with GDM is not known. The actual mechanism of reduced plasma complement protein in gravidas with GDM, and their low complement protein levels may lead to defective virus neutralization or cell lysis and increase the risk of infection in women with GDM. It may also lead to defective clearance of immune complexes and cause an inflammatory response.
In addition, four proteins associated with inflammation were identified, including GSN, ITIH1, ITIH4, and IL-6. In previous studies, these four proteins have been associated with inflammatory processes, including GDM-related inflammatory responses[22-24]. GSN is a promising therapeutic candidate as it has the advantage of modulating several inflammatory pathways. ITIH1 is a serine protease inhibitor that carries hyaluronic acid in the plasma and plays a role in the process of inflammation and carcinogenesis. ITIH4 is an IL-6-dependent acute phase protein that may act as an anti-inflammatory protein. In animal experiments, ITIH4 may help predict acute rejection after subclinical pancreas transplantation[25].
According to the recent studies of GDM, it is widely believed to play an important role in insulin resistance, therefore two proteins related to insulin resistance that are highly expressed in the GDM group are of interest to us, and they are also the most notable ones among the DEPs in the GDM gravidas. Both RBP4 and ANGPTL8 are a protein synthesized by the liver and adipose tissue, and they are both associated with insulin resistance. Studies have also reported significant differences in their expression in pregnant women with GDM, as well as a close relationship to induce GDM occurrence[26,27]. We confirmed the serum RBP4 and ANGPTL8 expression levels of pregnant women with GDM by ELISA, and the results were consistent with our mass spectrometry results. Therefore, RBP4 and ANGPTL8 are expected to be potential biomarkers for early prediction of GDM. However, this study has some shortcomings, such as a small sample size, a single study subject, and geographical restrictions on the source of cases. Therefore, it is necessary to increase the sample size, include multi-center study subjects, and expand the source range of cases in future studies to confirm the accuracy of this study.
In summary, iTRAQ combined with LC-MS/MS was effective in screening DEPs in the serum of GDM patients. We used bioinformatics to analyze DEPs with significantly altered expression levels to provide potential indicators for the prediction of GDM and to provide ideas for studying the pathogenesis of GDM. The pathogenesis of GDM appears to be related to dysfunction of lipid metabolism, activation of the coagulation cascade, the complement system, and the inflammatory response. This study provides several proteins in serum that may be of potential value in the clinical diagnosis of GDM, contributing to future studies on the prediction, prevention, and pathogenesis of GDM. However, the pathogenesis of GDM still requires a more in-depth study of these proteins, which is a shortcoming of this study.
The development of gestational diabetes mellitus is closely correlated with the differential expression of serum proteins.
The researchers found significant changes in the expression levels of some proteins in the serum of patients with gestational diabetes.
With the intensive investigation of these differentially expressed proteins, we can expect to make more breakthroughs in the field of gestational diabetes.
By using isobaric tag for relative and absolute quantitation (iTRAQ) proteomic and bioinformatics analysis methods, the investigators successfully revealed the differences in serum protein expression in gestational diabetes patients.
The enzyme-linked immunosorbent assay results showed that the serum RBP 4 and ANGPTL8 expression were higher in the GDM group between 12 wk and 16 wk of gestation than in the control group.
GDM development may be related to abnormal lipid metabolism, activation of coagulation cascade, complement system and inflammatory response. RBP4 and ANGPTL8 may be the early predictors of GDM.
RBP4 and ANGPTL8 are expected to be early predictors of GDM.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Obstetrics and gynecology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ahadi A, Iran S-Editor: Chen YL L-Editor: A P-Editor: Yu HG
| 1. | Bidhendi Yarandi R, Vaismoradi M, Panahi MH, Gåre Kymre I, Behboudi-Gandevani S. Mild Gestational Diabetes and Adverse Pregnancy Outcome: A Systemic Review and Meta-Analysis. Front Med (Lausanne). 2021;8:699412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 2. | Berger H, Melamed N, Murray-Davis B, Hasan H, Mawjee K, Barrett J, McDonald SD, Geary M, Ray JG; Diabetes, Obesity and Hypertension in Pregnancy Research Network (DOH-NET) and the Southern Ontario Obstetrical Network (SOON) Investigators. Prevalence of Pre-Pregnancy Diabetes, Obesity, and Hypertension in Canada. J Obstet Gynaecol Can. 2019;41:1579-1588.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 3. | Kouhkan A, Najafi L, Malek M, Baradaran HR, Hosseini R, Khajavi A, Khamseh ME. Gestational diabetes mellitus: Major risk factors and pregnancy-related outcomes: A cohort study. Int J Reprod Biomed. 2021;19:827-836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 4. | Pantham P, Aye IL, Powell TL. Inflammation in maternal obesity and gestational diabetes mellitus. Placenta. 2015;36:709-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 424] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 5. | Fatima SS, Jamil Z, Alam F, Malik HZ, Madhani SI, Ahmad MS, Shabbir T, Rehmani MN, Rabbani A. Polymorphism of the renalase gene in gestational diabetes mellitus. Endocrine. 2017;55:124-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | Miao Z, Wu H, Ren L, Bu N, Jiang L, Yang H, Zhang J, Guo X. Long-Term Postpartum Outcomes of Insulin Resistance and β-cell Function in Women with Previous Gestational Diabetes Mellitus. Int J Endocrinol. 2020;2020:7417356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 7. | Moulder R, Bhosale SD, Goodlett DR, Lahesmaa R. Analysis of the plasma proteome using iTRAQ and TMT-based Isobaric labeling. Mass Spectrom Rev. 2018;37:583-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 8. | Zhao D, Shen L, Wei Y, Xie J, Chen S, Liang Y, Chen Y, Wu H. Identification of candidate biomarkers for the prediction of gestational diabetes mellitus in the early stages of pregnancy using iTRAQ quantitative proteomics. Proteomics Clin Appl. 2017;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | Johns EC, Denison FC, Norman JE, Reynolds RM. Gestational Diabetes Mellitus: Mechanisms, Treatment, and Complications. Trends Endocrinol Metab. 2018;29:743-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 519] [Article Influence: 74.1] [Reference Citation Analysis (0)] |
| 10. | Omazić J, Viljetić B, Ivić V, Kadivnik M, Zibar L, Müller A, Wagner J. Early markers of gestational diabetes mellitus: what we know and which way forward? Biochem Med (Zagreb). 2021;31:030502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 11. | Di Filippo D, Wanniarachchi T, Wei D, Yang JJ, Mc Sweeney A, Havard A, Henry A, Welsh A. The diagnostic indicators of gestational diabetes mellitus from second trimester to birth: a systematic review. Clin Diabetes Endocrinol. 2021;7:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Çetin C, Güngör ND, Yavuz M. First trimester glycosylated hemoglobin for gestational diabetes mellitus screening. Taiwan J Obstet Gynecol. 2021;60:899-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 13. | Shen L, Zhao D, Chen Y, Zhang K, Chen X, Lin J, Li C, Iqbal J, Zhao Y, Liang Y, Wei Y, Feng C. Comparative Proteomics Analysis of Serum Proteins in Gestational Diabetes during Early and Middle Stages of Pregnancy. Proteomics Clin Appl. 2019;13:e1800060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 14. | Liu F, Zhao C, Liu L, Ding H, Huo R, Shi Z. Peptidome profiling of umbilical cord plasma associated with gestational diabetes-induced fetal macrosomia. J Proteomics. 2016;139:38-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 15. | Liu X, Sun J, Wen X, Duan J, Xue D, Pan Y, Zhang W, Cheng X, Wang C. Proteome profiling of gestational diabetes mellitus at 16-18 weeks revealed by LC-MS/MS. J Clin Lab Anal. 2020;34:e23424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Abdel Gader AG, Khashoggi TY, Habib F, Awadallah SB. Haemostatic and cytokine changes in gestational diabetes mellitus. Gynecol Endocrinol. 2011;27:356-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Kovac M, Mitic G, Mikovic Z, Mandic V, Miljic P, Mitrovic M, Tomic B, Bereczky Z. The influence of specific mutations in the AT gene (SERPINC1) on the type of pregnancy related complications. Thromb Res. 2019;173:12-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Zheng T, Chen H. Resveratrol ameliorates the glucose uptake and lipid metabolism in gestational diabetes mellitus mice and insulin-resistant adipocytes via miR-23a-3p/NOV axis. Mol Immunol. 2021;137:163-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 19. | Garamvölgyi Z, Prohászka Z, Rigó J Jr, Kecskeméti A, Molvarec A. Increased circulating heat shock protein 70 (HSPA1A) levels in gestational diabetes mellitus: a pilot study. Cell Stress Chaperones. 2015;20:575-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Alamolhoda SH, Yazdkhasti M, Namdari M, Zakariayi SJ, Mirabi P. Association between C-reactive protein and gestational diabetes: a prospective study. J Obstet Gynaecol. 2020;40:349-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Lappas M. Lower circulating levels of complement split proteins C3a and C4a in maternal plasma of women with gestational diabetes mellitus. Diabet Med. 2011;28:906-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Amirian A, Mahani MB, Abdi F. Role of interleukin-6 (IL-6) in predicting gestational diabetes mellitus. Obstet Gynecol Sci. 2020;63:407-416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 23. | Teng Y, Xuan S, Jiang M, Tian L, Tian J, Chang Q. Expression of H(2)S in Gestational Diabetes Mellitus and Correlation Analysis with Inflammatory Markers IL-6 and TNF-α. J Diabetes Res. 2020;2020:3085840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Gao J, Qin Z, Guan X, Guo J, Wang H, Liu S. Overexpression of GSN could decrease inflammation and apoptosis in EAE and may enhance vitamin D therapy on EAE/MS. Sci Rep. 2017;7:604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 25. | García-Gil FA, Lampreave F, Fuentes-Broto L, Carpintero R, Gonzalvo E, García JJ, Alvarez-Alegret R, Alfaro J, Orden I, Roda L. Inter-alpha-trypsin inhibitor heavy chain 4 as a marker of acute rejection in pancreas allotransplantation in pigs. Transplant Proc. 2010;42:3063-3069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 26. | Chen Y, Lv P, Du M, Liang Z, Zhou M, Chen D. Increased retinol-free RBP4 contributes to insulin resistance in gestational diabetes mellitus. Arch Gynecol Obstet. 2017;296:53-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Abdeltawab A, Zaki ME, Abdeldayem Y, Mohamed AA, Zaied SM. Circulating micro RNA-223 and angiopoietin-like protein 8 as biomarkers of gestational diabetes mellitus. Br J Biomed Sci. 2021;78:12-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |