Published online Mar 6, 2024. doi: 10.12998/wjcc.v12.i7.1215
Peer-review started: October 25, 2023
First decision: December 31, 2023
Revised: January 14, 2024
Accepted: February 6, 2024
Article in press: February 6, 2024
Published online: March 6, 2024
Processing time: 127 Days and 14.4 Hours
Although the etiology of nonalcoholic fatty liver disease (NAFLD) has not been thoroughly understood, the emerging roles of anthropometric indicators in assessing and predicting the risk of NAFLD have been highlighted by accumulating evidence.
To evaluate the causal relationships between five anthropometric indicators and NAFLD employing Mendelian randomization (MR) design.
The Anthropometric Consortium provided genetic exposure data for five anthropometric indicators, including hip circumference (HC), waist circumference (WC), waist-to-hip ratio (WHR), body mass index (BMI), and body fat percentage (BF). Genetic outcome data for NAFLD were obtained from the United Kingdom Biobank and FinnGen Consortium. Genome-wide significant single nucleotide polymorphisms were chosen as instrumental variables. Univariable MR (UVMR) and multivariable MR (MVMR) designs with analytical approaches, including inverse variance weighted (IVW), MR-Egger, weighted median (WM), and weighted mode methods, were used to assess the causal relationships between anthropometric indicators and NAFLD.
Causal relationships were revealed by UVMR, indicating that a higher risk of NAFLD was associated with a per-unit increase in WC [IVW: odds ratio (OR) = 2.67, 95%CI: 1.42-5.02, P = 2.25 × 10−3], and BF was causally associated with an increased risk of NAFLD (WM: OR = 2.23, 95%CI: 1.07-4.66, P = 0.033). The presence of causal effects of WC on the decreased risk of NAFLD was supported by MVMR after adjusting for BMI and smoking. However, no causal association between BF and NAFLD was observed. In addition, other causal relationships of HC, WHR (BMI adjusted), and BMI with the risk of NAFLD were not retained after FDR correction.
This study establishes a causal relationship, indicating that an increase in WC is associated with a higher risk of NAFLD. This demonstrates that a suitable decrease in WC is advantageous for preventing NAFLD.
Core Tip: Previous studies have demonstrated the potential significance of anthropometric indicators in the development of nonalcoholic fatty liver disease (NAFLD). Nevertheless, inconsistencies exist in the results of these studies, and the causal association remains unclear. Abdominal obesity, measured by waist circumference (WC), is a risk factor for NAFLD, as demonstrated by previous studies. Nevertheless, many of these studies were cross-sectional or considered only a single measurement, neglecting a comprehensive evaluation of changes in WC over time and the effect of long-term development and lifestyle changes. Consequently, establishing a causal relationship between anthropometric indicators and NAFLD requires further robust evidence.
- Citation: Xiao XP, Dai YJ, Zhang Y, Yang M, Xie J, Chen G, Yang ZJ. Investigating the causal associations between five anthropometric indicators and nonalcoholic fatty liver disease: Mendelian randomization study. World J Clin Cases 2024; 12(7): 1215-1226
- URL: https://www.wjgnet.com/2307-8960/full/v12/i7/1215.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i7.1215
Excessive fat deposition in liver cells characterizes nonalcoholic fatty liver disease (NAFLD). Currently, due to lifestyle changes and insufficient daily physical activity, NAFLD has emerged as the most severe chronic disease in society[1]. Reports indicate that NAFLD affects a quarter of the global population, with a prevalence rate of 24% and a tendency toward younger age groups[2,3]. Although the etiology of NAFLD has not been thoroughly understood, the emerging roles of anthropometric indicators in assessing and predicting the risk of NAFLD have been highlighted by accumulating evidence[4]. Numerous previous observational studies have reported relationships between the risk of NAFLD and noninvasive quantitative measurements of the body, such as anthropometric indicators, which comprise height, weight, hip circumference (HC), waist circumference (WC), waist-to-hip ratio (WHR), body mass index (BMI), and body fat percentage (BF)[4,5]. Contradictory findings have been obtained in some other studies, indicating no relationship between anthropometric indicators and the risk of NAFLD[6,7]. The causal relationships between anthropometric indicators and NAFLD risk remain undetermined, considering these inconsistent findings and the absence of randomized controlled studies.
Mendelian randomization (MR) is a novel epidemiological tool that employs genetic data to investigate the causal relationship between exposure and outcome[8]. Generally, genetic variants are independent of disease state and are randomly assigned to offspring through the maternal generation. Consequently, the limitations of conventional observational design can be overcome, and biases such as potential confounders and reverse causality can be minimized[9,10]. Previous studies have demonstrated the use of MR to investigate causal relationships between NAFLD and numerous diseases, including cardiovascular disease and psoriasis[11,12].
In this study, the causality of five anthropometric indicators, including HC, WC, WHR (BMI adjusted), BMI, and BF, with the risk of NAFLD, was investigated using genetic summary statistics with Univariable MR (UVMR) and multivariable MR (MVMR) frameworks to establish a foundation for the prevention and management of this high-burden disease.
In this MR analysis, the causal effects of various modifiable exposures on outcomes were estimated using single nucleotide polymorphisms (SNPs) as instrumental variables (IVs). However, three basic assumptions must be satisfied[13]. First, IVs must exhibit a high correlation with the exposure factor (correlation assumption) to minimize weak IV bias[14]. Second, the outcome should only be influenced by the identified IVs through exposure and not through other factors, expressed as "no horizontal pleiotropy" (exclusionary assumption)[15]. Third, to identify genuine causal associations, IVs should be independent of confounders in exposure-outcome associations (independence assumption)[16].
A two-stage design was used in this study, with the first phase employing bidirectional UVMR analyses to analyze the causal relationship between the five anthropometric measures and NAFLD. In the second phase, MVMR analyses were conducted to establish causality after adjusting for potential confounders, such as BMI and smoking. These findings were validated using databases from various sources. Figure 1 shows the study design.
Five anthropometric measures, including HC, WC, WHR (BMI adjusted), BMI, and BF, were chosen as exposure points. Shungin et al[17] reported a genome-wide association study (GWAS) that included up to 224459 individuals, from which summary statistics for HC, WC, and WHR (BMI adjusted) were extracted[17]. The summary data for BMI were obtained from another GWAS, which involved 700000 participants[18]. Genetic summary data for BF were obtained from GWAS data published by Neale Lab in 2017, which included 331117 participants (http://www.nealelab.is/uk-biobank). All exposure datasets were conducted in European ancestry.
The threshold was set at P < 5 × 10−08 to choose SNPs related to anthropometric metrics as IVs. Furthermore, SNPs with strong linkage disequilibrium (R2 < 0.001, window size = 10000 kb) were deleted to avoid possible bias. Significantly heterogeneous SNPs were excluded, and the remaining anthropometric-associated SNPs were selected as valid IVs using the heterogeneity test. In some of the exposure-outcome analyses, some SNPs were excluded due to the lack of available proxies, as they were not present in the outcome (Supplementary Table 1). Finally, a total of 807 anthropometric-associated SNPs were available, including 18 HC-associated SNPs, 46 WC-associated SNPs, 32 WHR (BMI adjusted)-associated SNPs, 485 BMI-associated SNPs, and 226 BF-associated SNPs (Supplementary Table 1).
Concerning the outcome datasets, genetic associations for NAFLD agent-related SNPs were extracted from the United Kingdom biobank, which included all European subjects, comprising 8434 cases and 770180 controls[19]. The GWAS findings were harmonized with those of GWAS exposed to the same effect allele. Effects allele frequencies (minor allele frequency ≤ 0.5) were employed to harmonize the palindromic SNPs. For IVs that could not be found in the results, attempts were made to include suitable proxy SNPs (r2 ≥ 0.8) by searching for them on the website (http://www.mulinlab.org/vportal/index.html).
This study was conducted using public open data from the MRC-IEU database and the FinnGen research project; thus, no ethical review was necessary.
UVMR analysis: Inverse variance weighted (IVW), weighted median (WM), weighted model, and MR Egger methods were used to evaluate the causal effects of HC, WC, WHR (BMI adjusted), and BF (per SD) on the risk of NAFLD. The IVW model, which was defined as the primary approach, can provide causal effect estimates with optimal precision when all IVs are valid and minimize the impact of heterogeneity. To assess weak IV bias, the variance (R2) and approximated F-statistic for per-exposure explained by IVs were calculated. This evaluation, which is based on the inherent flaws in IV selection, is generally considered to be sufficiently instrumental for F-statistics greater than 10[20,21]. The formula used is F = R2 × (N - 2)/(1 - R2), where R2 represents the variance of exposure explained by each IV[22]. In addition, FDR correction was implemented to control false positives that tend to occur due to multiple testing.
MVMR analyses: Smoking and BMI were the primary confounders associated with exposure (anthropometric indicators) and outcome (NAFLD). Therefore, to determine the independent effects of anthropometric indicators on the risk of NAFLD development, MVMR analyses were employed. In MVMR analyses, MR-PRESSO models were employed to correct for horizontal pleiotropy in causal effects, estimate heterogeneity, and exclude potential outliers. A significance level of P < 0.05 in the MVMR analysis was considered indicative of statistical significance.
Sensitivity analysis: To ensure the stability of the study's results, several sensitivity analyses were conducted. First, the nonexistence of horizontal pleiotropy, a fundamental premise for satisfying causal inference, was analyzed using the MR-PRESSO approach. Findings with a P value greater than 0.05 indicated the nonexistence of horizontal pleiotropy[23]. Second, Cochran's Q statistic was employed to eliminate heterogeneity of IVs, with P values over 0.05 indicating the absence of heterogeneity in the IVs[24]. Furthermore, leave-one-out (LOO) analyses were performed to exclude single SNPs exerting a substantial influence on the findings[25]. An online tool "mRnd" was used to calculate the statistical efficacy (power). R programming software (version 4.0.3) with the "MR," "TwoSampleMR," "MVMR," and "MRPRESSO" packages was used to perform all statistical analyses and data visualization.
Anthropometric indicators replication: Published anthropometric indicators GWAS data were extracted from other consortiums (P < 5 × 10-08, R2 < 0.001, window size = 10000 kb). The GWAS data for HC were obtained from Neale Lab's 2017 summary database, which included 336639 individuals in this study (http://www.nealelab.is/uk-biobank). Summary data for WC and BF were derived from the MRC-IEU database and exported from the GWAS pipeline using the United Kingdom Biobank's pheasant-derived variables. Furthermore, the Within Family GWAS consortium was the source of data for WHR (BMI adjusted). In this genetic epidemiological study, sample analyses of related individuals (such as siblings or parent-child triples) were performed (https://www.withinfamilyconsortium.com). The BMI data were obtained from the GWAS study conducted by Chittani et al[26], which included 236781 individuals[26]. All data were extracted from published studies, and consequently, no additional ethical review was necessary. Figure 1 shows the details of the exposure and outcome data.
NAFLD replication: Outcome data were obtained from FinnGen, a large cohort study comprising genomic and health data from 500000 Finland BioBank individuals (https://www.finngen.fi). Similar allele frequency distributions to other European populations were observed in Finland; however, significant strengths, including uncommon variants in intricate phenotypes and unique group genetic history, were noted[27].
A total of 807 anthropometric indicator-associated SNPs, including 18 HC-associated SNPs, 46 WC-associated SNPs, 32 WHR (BMI adjusted)-associated SNPs, 485 BMI-associated SNPs, and 226 BF-associated SNPs, were identified from three independent GWAS analyses (Supplementary Table 1). Among the 807 anthropometric indicator-associated SNPs, all F-statistics were greater than 10, indicating a low likelihood of weak IVs among the included SNPs. In addition, the selected IVs explained approximately 0.84% (HC), 1.13% (WC), 1.56% [WHR (BMI adjusted)], 4.85% (BMI), and 3.60% (BF) of the phenotype variances. Low degrees of sample overlap between exposure and outcome were indicated by the sample overlap results, with overlap rates within datasets being less than 15%.
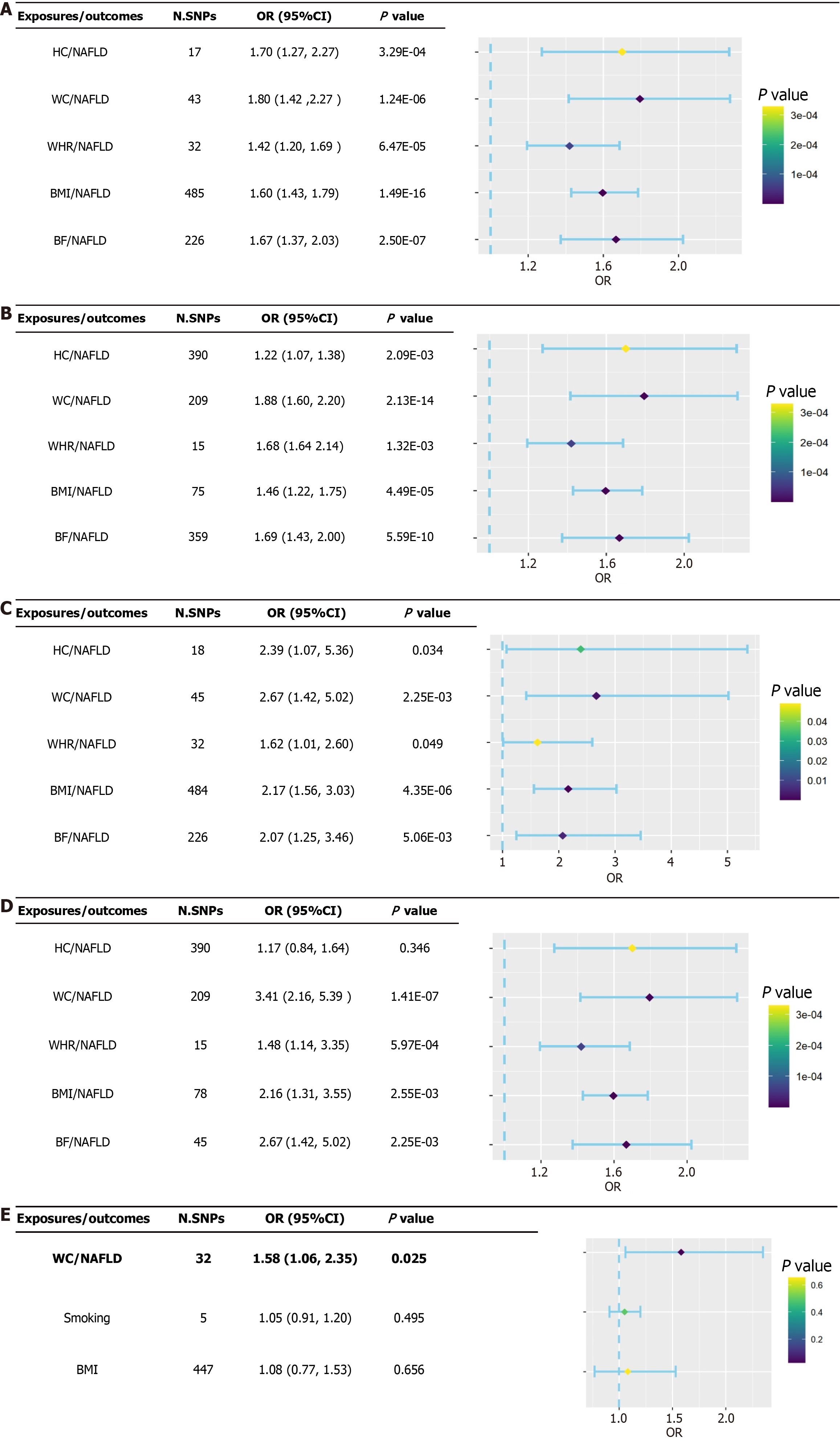
All findings were based on the IVW random-effects model because evidence of horizontal pleiotropy was not found (P > 0.05), but heterogeneity was present (P < 0.05). The findings of the IVW approach revealed causal relationships between five anthropometric indicators and NAFLD: HC (ORIVW = 1.70, 95%CI: 1.27-2.27, P = 3.29 × 10−04; ORWM = 1.76, 95%CI: 1.25-2.48, P = 1.20 × 10−03), WC (ORIVW = 1.80, 95%CI: 1.42-2.27, P = 1.24 × 10−06; ORWM = 1.88, 95%CI: 1.40-2.51, P = 2.42 × 10−05), WHR (BMI adjusted) (ORIVW = 1.42, 95%CI: 1.20-1.69, P = 6.47 × 10−05; ORWM = 1.35, 95%CI: 1.06-1.71, P = 1.40 × 10−03), BMI (ORIVW = 1.60, 95%CI: 1.43-1.79, P = 1.49 × 10−16; ORWM = 1.64, 95%CI: 1.38-1.94, P = 1.16 × 10−08), and BF (ORIVW = 1.67, 95%CI: 1.37-2.04, P = 2.50 × 10−07; ORWM = 1.91, 95%CI: 1.50-2.43, P = 1.36 × 10−07; Figure 2A and Supplementary Table 2). Furthermore, sensitivity analyses using the LOO approach indicated that no single SNPs drove these findings after the stepwise elimination of individual SNPs (all error lines were on either side of zero; Supplementary Figures 1-5). Considering the positive false due to multiple testing, significant causal relationships between HC, WC, WHR (BMI adjusted), BMI, BF, and NAFLD were still observed in the results after FDR correction (P < 0.01; Supplementary Figure 6).
No significant genetic predictive correlation was observed when NAFLD was tested as an exposure factor for inverse correlation, as indicated by estimates derived from the IVW method (P > 0.05). This further supports the notion that reversed causal relationships with NAFLD do not confound anthropometric indicators (Supplementary Table 2). In addition, the result of no significant genetic prediction of associations between NAFLD and anthropometric indicators was supported by sensitivity analysis.
With the above IVs selection standard, 1052 SNPs associated with anthropometric indicators, including 390 HC-associated SNPs, 209 WC-associated SNPs, 15 WHR- associated SNPs, 79 BMI-associated SNPs, and 359 BF-associated SNPs, were selected for causal estimation in the replicated exposures (Supplementary Table 1). For HC, WC, WHR (BMI adjusted), BMI, and BF, the F-statistics were all over 10, indicating that the chosen IVs were sufficiently robust and not susceptible to the influence of weak IVs. Furthermore, the statistical power of these replication exposures was similarly calculated, and the selected IVs were shown to explain approximately 5.50% (HC), 3.38% (WC), 0.99% [WHR (BMI adjusted)], 2.17% (BMI), and 4.60% (BF) of the phenotype variances.
Subsequently, the two sources of exposure (discovery and replication) were analyzed in a bidirectional MR analysis with outcomes from various GWAS (United Kingdom biobank and FinnGen). The causal relationship between an increased risk of NAFLD and higher WC, WHR (BMI adjusted), BMI, and BF was supported by our cross-validation comparison of four sets of MR findings; sensitivity analysis findings remained consistent. Although the existence of heterogeneity between SNPs was indicated by the evidence, the overall level of pleiotropy based on the MR-Egger intercept was not significant. In our inverse MR analysis of anthropometric indicators (discovery database) with NAFLD (FinnGen research project), the presence of NAFLD resulted in a decrease in BMI, indicating that the previously observed causal relationship between BMI and NAFLD was influenced by reverse causality bias. In addition, FDR correction was equally performed on the validated results, resulting in the loss of some weak causal relationships. The findings of the four MR analyses supported the fact that an increase in WC (ORIVW = 1.80, 95%CI: 1.42-2.27, P = 1.24 × 10-06; ORWM = 1.88, 95%CI: 1.40-2.51, P = 2.42 × 10-05) and BF (ORIVW = 1.67, 95%CI: 1.43-1.79, P = 2.50 × 10-07; ORWM = 1.91, 95%CI: 1.50-2.43, P = 1.36 × 10-07) would result in a higher risk of NAFLD after the exclusion of reverse causality (Figure 2B-D and Supplementary Tables 3-5). Supplementary Figure 6 shows the IVW results of the FDR correction.
After adjusting for BMI and smoking, the findings of the MVMR demonstrated that the relationship between the genetically determined increase in WC and an increase in the risk of NAFLD remained significant (OR = 1.58, 95%CI: 1.06-2.35, P = 0.025), indicating that WC may be a factor for increased risk of NAFLD. Nevertheless, a causal relationship between BF and the risk of NAFLD was not supported by the MVMR findings (ORIVW = 1.02, 95%CI: 0.60-1.73, P = 0.941; Figure 2E). Furthermore, causal relationships between HC, WHR (BMI adjusted), BMI, and the risk of NAFLD were not supported by the cross-validation findings.
The primary causes of the increased global burden of NAFLD are changes in lifestyle and dietary habits, and identifying risk factors is particularly crucial for disease prevention and control[28]. Previous studies have demonstrated that anthropometric indicators may primarily be risk factors for developing NAFLD[29]. Nevertheless, conflicting findings exist from various studies, and the causal relationships still need to be clarified[4,30]. Risky relationships between WC and BF for NAFLD were observed from repeated-validation TSMR. After excluding the confounding effects of BMI and smoking, the direct causal effect of BF on NAFLD could not be pursued in subsequent MVMR studies.
As a critical indicator of abdominal obesity (AO) patterns, a strong relationship exists between WC and the early risk of all-cause mortality. An increase of 10 cm in WC for an individual results in an increase of 11% points in the risk of all-cause mortality[31]. Furthermore, positive relationships with the risk of morbidity and mortality of major chronic diseases were exhibited by WC[32]. The importance of WC as an indicator of population health and long-term adverse outcomes was emphasized by these results. In addition, a relationship between WC and NAFLD was observed. A study in an adolescent population indicated that AO was a predictor of pulmonary fibrosis in children and adolescents with NAFLD, with WC as the primary measure[33]. Furthermore, a Korean study discovered a direct relationship between a larger WC and an increased risk of developing NAFLD[33]. This result was supported by a study of the Iranian population[34]. Although numerous studies have demonstrated that AO is a risk factor for NAFLD, with WC being the primary measure, most of them have been cross-sectional studies or single-measurement findings and the long-term dynamic changes in an individual's WC in response to changes in growth, development, and lifestyle, among other things, have not been sufficiently evaluated. From a genetic viewpoint, our results indicated a causal relationship between greater WC and higher NAFLD. The biological relationship can be explained by several possible mechanisms. On the one hand, AO reflects the excessive accumulation of visceral adiposity (VAT), which increases WC; on the other hand, an increase in WC results in a substantial collection of VAT, enabling the continuous secretion of pro-inflammatory cytokines, such as tumor necrosis factor-α. This activates inflammatory signaling pathways, thereby mediating metabolic homeostasis and insulin sensitivity of the organism[35-37].
BF measures the amount of fat in a person's body and has been identified as a crucial risk factor for cardiometabolic[38]. BF is strongly related to the risk of NAFLD, as indicated by the Rotterdam study, and this relationship is more visible in the female population. Furthermore, in a longitudinal relationship investigating the effect of BF changes on NAFLD incidence and remission, Kim et al[39] demonstrated that increased BF was longitudinally associated with increased risk of NAFLD and negatively associated with NAFLD regression[39]. From the two replication datasets, our combined TSMR findings indicated that increased BF substantially increased the risk of NAFLD. After excluding the effects of BMI and smoking, MVMR rejected a direct causal impact of BF on NAFLD.
Furthermore, the relationships of HC, WHR (BMI adjusted), and BMI with NAFLD have been analyzed in numerous studies. A cross-sectional study of the prevalence of NAFLD in pathologically overweight women in South India revealed that BMI, BF, and BWP are three indicators that can be used to a large degree as indicators of the development of NAFLD[40]. Improved cardiovascular metabolism is associated with HC. The development of NAFLD may be triggered by excessive HC, as it has been closely associated with muscle mass, and improving muscle mass will reduce insulin resistance and decrease the likelihood of NAFLD[41,42]. Indicators commonly employed to evaluate nutritional conditions and obesity, such as BMI and WHR (BMI adjusted), primarily reflect the whole-body fat problem rather than VAT. WC is more reflective of the volume of VAT than BMI[43]. Consequently, some studies support visceral obesity with WC and BF as primary measures of a crucial risk factor for NAFLD[44]. Based on European GWAS pooled data, a previous study reported partially different findings compared with our findings, indicating that WHR may be a potential risk factor for NAFLD[45]. Several factors could explain this difference. First, the WHR (BMI adjusted) data used in the GWAS were corrected for BMI, indicating that changes in WHR independent of BMI might not be fully captured by the selection of WHR (BMI adjusted) SNPs. Second, the causal inferences included fewer SNPs, resulting in limited phenotypic differences and reduced statistical power to detect a true causal association between WHR and NAFLD risk. Furthermore, differences in study populations and sample sizes could have contributed to the inconsistent findings. To address these challenges, cross-repeated validation was performed using two independent GWAS datasets to eliminate bias resulting from data selection and enhance the credibility of our findings.
Several limitations require attention. First, the exposure and outcome datasets were obtained from European ancestry; therefore, these findings may not be generalizable to other populations with different genetic backgrounds. Further studies are required to validate these results in other ethnic populations. Second, although the F-statistic can be used to evaluate the first hypothesis, verifying the second and third hypotheses is generally challenging and may lead to potential bias. Third, the causal relationship between anthropometric indicators and NAFLD in different gender/age groups cannot be investigated due to the lack of personal demographic information on anthropometric indicators.
Although previous studies have extensively investigated the relationship between AO and NAFLD, there are limited studies on the association between other anthropometric measures[45,46]. This study expands and validates these results by incorporating a wider range of anthropometric measures, offering additional supporting evidence for a causal association in early-stage NAFLD. Compared with previous studies, this study has numerous notable advantages. Although the association between WC, BMI, and BMI-adjusted WHR and NAFLD has been partially examined in previous studies, evidence for a causal association between other anthropometric indicators (such as HC and BF) and NAFLD remains limited. To the best of our knowledge, the causal association between the five major anthropometric indicators and NAFLD was comprehensively assessed for the first time in this study using UVMR and MVMR approaches. This study provides valuable causal evidence and directionality for the early prediction and diagnosis of NAFLD. The results of this study were replicated using GWAS data from a European population to enhance the reliability of the findings and ensure robust conclusions. Data from two distinct sources were examined in this repli
This study demonstrates that genetically determined increased WC maintains a positive and causal association with NAFLD, even in the presence of confounders, including BMI and smoking. This underscores the potential of WC as a reliable indicator for the early identification and diagnosis of NAFLD.
This study offers the first comprehensive assessment of causal associations between five anthropometric measures and nonalcoholic fatty liver disease (NAFLD) by using both Univariate Mendelian randomization (MR) and Multivariable MR methods. Considering the possibility of potential chance in the results, we additionally selected another exposure and outcome Genome-wide association study data from European population for replication, including a cross-analysis of two different sources of data.
Although the etiology of NAFLD has not been thoroughly understood, the emerging roles of anthropometric indicators in assessing and predicting the risk of NAFLD have been highlighted by accumulating evidence. Numerous previous observational studies have reported relationships between the risk of NAFLD and noninvasive quantitative measurements of the body, such as anthropometric indicators, which comprise height, weight, hip circumference, waist circumference (WC), waist-to-hip ratio, body mass index (BMI), and body fat percentage. Contradictory findings have been obtained in some other studies, indicating no relationship between anthropometric indicators and the risk of NAFLD. The causal relationships between anthropometric indicators and NAFLD risk remain undetermined, considering these inconsistent findings and the absence of randomized controlled studies.
This study demonstrates that genetically determined increased WC maintains a positive and causal association with NAFLD, even in the presence of confounders, including BMI and smoking. This underscores the potential of WC as a reliable indicator for the early identification and diagnosis of NAFLD.
MR is a novel epidemiological tool that employs genetic data to investigate the causal relationship between exposure and outcome. Generally, genetic variants are independent of disease state and are randomly assigned to offspring through the maternal generation. Consequently, the limitations of conventional observational design can be overcome, and biases such as potential confounders and reverse causality can be minimized. Previous studies have demonstrated the use of MR to investigate causal relationships between NAFLD and numerous diseases, including cardiovascular disease and psoriasis.
Genetically determined increased WC maintains a positive and causal association with NAFLD, even in the presence of confounders, including BMI and smoking. Several limitations require attention. First, the exposure and outcome datasets were obtained from European ancestry; therefore, these findings may not be generalizable to other populations with different genetic backgrounds. Further studies are required to validate these results in other ethnic populations. Second, although the F-statistic can be used to evaluate the first hypothesis, verifying the second and third hypotheses is generally challenging and may lead to potential bias. Third, the causal relationship between anthropometric indicators and NAFLD in different sex/age groups cannot be investigated due to the lack of personal demographic information on anthropometric indicators.
This study demonstrates that genetically determined increased WC maintains a positive and causal association with NAFLD, even in the presence of confounders, including BMI and smoking. This underscores the potential of WC as a reliable indicator for the early identification and diagnosis of NAFLD.
Future studies should consider using WC as an auxiliary measurement for identifying NAFLD.
The authors express their gratitude to the Figdraw website and the CHiPlot website for their provision of scientific drawing tools.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Ulasoglu C, Turkey S-Editor: Li L L-Editor: A P-Editor: Zhao S
| 1. | Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62:S47-S64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1516] [Cited by in RCA: 2140] [Article Influence: 214.0] [Reference Citation Analysis (0)] |
| 2. | Nobili V, Alisi A, Valenti L, Miele L, Feldstein AE, Alkhouri N. NAFLD in children: new genes, new diagnostic modalities and new drugs. Nat Rev Gastroenterol Hepatol. 2019;16:517-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 218] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 3. | Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4054] [Cited by in RCA: 3786] [Article Influence: 540.9] [Reference Citation Analysis (2)] |
| 4. | Almeda-Valdes P, Aguilar-Salinas CA, Uribe M, Canizales-Quinteros S, Méndez-Sánchez N. Impact of anthropometric cut-off values in determining the prevalence of metabolic alterations. Eur J Clin Invest. 2016;46:940-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Agbim U, Carr RM, Pickett-Blakely O, Dagogo-Jack S. Ethnic Disparities in Adiposity: Focus on Non-alcoholic Fatty Liver Disease, Visceral, and Generalized Obesity. Curr Obes Rep. 2019;8:243-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 6. | Ahadi M, Molooghi K, Masoudifar N, Namdar AB, Vossoughinia H, Farzanehfar M. A review of non-alcoholic fatty liver disease in non-obese and lean individuals. J Gastroenterol Hepatol. 2021;36:1497-1507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 7. | Tobari M, Hashimoto E, Taniai M, Ikarashi Y, Kodama K, Kogiso T, Tokushige K, Takayoshi N, Hashimoto N. Characteristics of non-alcoholic steatohepatitis among lean patients in Japan: Not uncommon and not always benign. J Gastroenterol Hepatol. 2019;34:1404-1410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 8. | Au Yeung SL, Gill D. Standardizing the reporting of Mendelian randomization studies. BMC Med. 2023;21:187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 32] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 9. | Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: A review. Res Synth Methods. 2019;10:486-496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 461] [Cited by in RCA: 997] [Article Influence: 166.2] [Reference Citation Analysis (0)] |
| 10. | O'Donnell JA, Zheng T, Meric G, Marques FZ. The gut microbiome and hypertension. Nat Rev Nephrol. 2023;19:153-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 130] [Reference Citation Analysis (0)] |
| 11. | Näslund-Koch C, Bojesen SE, Gluud LL, Skov L, Vedel-Krogh S. Non-alcoholic fatty liver disease is not a causal risk factor for psoriasis: A Mendelian randomization study of 108,835 individuals. Front Immunol. 2022;13:1022460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 12. | Au Yeung SL, Borges MC, Wong THT, Lawlor DA, Schooling CM. Evaluating the role of non-alcoholic fatty liver disease in cardiovascular diseases and type 2 diabetes: a Mendelian randomization study in Europeans and East Asians. Int J Epidemiol. 2023;52:921-931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 13. | Birney E. Mendelian Randomization. Cold Spring Harb Perspect Med. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 352] [Article Influence: 117.3] [Reference Citation Analysis (0)] |
| 14. | Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42:1497-1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 586] [Cited by in RCA: 1270] [Article Influence: 141.1] [Reference Citation Analysis (0)] |
| 15. | Ding P, VanderWeele TJ, Robins JM. Instrumental variables as bias amplifiers with general outcome and confounding. Biometrika. 2017;104:291-302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 16. | Border R, O'Rourke S, de Candia T, Goddard ME, Visscher PM, Yengo L, Jones M, Keller MC. Assortative mating biases marker-based heritability estimators. Nat Commun. 2022;13:660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 40] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 17. | Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, Workalemahu T, Wu JMW, Buchkovich ML, Heard-Costa NL, Roman TS, Drong AW, Song C, Gustafsson S, Day FR, Esko T, Fall T, Kutalik Z, Luan J, Randall JC, Scherag A, Vedantam S, Wood AR, Chen J, Fehrmann R, Karjalainen J, Kahali B, Liu CT, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bragg-Gresham JL, Buyske S, Demirkan A, Ehret GB, Feitosa MF, Goel A, Jackson AU, Johnson T, Kleber ME, Kristiansson K, Mangino M, Leach IM, Medina-Gomez C, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Stančáková A, Sung YJ, Tanaka T, Teumer A, Van Vliet-Ostaptchouk JV, Yengo L, Zhang W, Albrecht E, Ärnlöv J, Arscott GM, Bandinelli S, Barrett A, Bellis C, Bennett AJ, Berne C, Blüher M, Böhringer S, Bonnet F, Böttcher Y, Bruinenberg M, Carba DB, Caspersen IH, Clarke R, Daw EW, Deelen J, Deelman E, Delgado G, Doney AS, Eklund N, Erdos MR, Estrada K, Eury E, Friedrich N, Garcia ME, Giedraitis V, Gigante B, Go AS, Golay A, Grallert H, Grammer TB, Gräßler J, Grewal J, Groves CJ, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heikkilä K, Herzig KH, Helmer Q, Hillege HL, Holmen O, Hunt SC, Isaacs A, Ittermann T, James AL, Johansson I, Juliusdottir T, Kalafati IP, Kinnunen L, Koenig W, Kooner IK, Kratzer W, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindström J, Lobbens S, Lorentzon M, Mach F, Magnusson PK, Mahajan A, McArdle WL, Menni C, Merger S, Mihailov E, Milani L, Mills R, Moayyeri A, Monda KL, Mooijaart SP, Mühleisen TW, Mulas A, Müller G, Müller-Nurasyid M, Nagaraja R, Nalls MA, Narisu N, Glorioso N, Nolte IM, Olden M, Rayner NW, Renstrom F, Ried JS, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Sennblad B, Seufferlein T, Sitlani CM, Smith AV, Stirrups K, Stringham HM, Sundström J, Swertz MA, Swift AJ, Syvänen AC, Tayo BO, Thorand B, Thorleifsson G, Tomaschitz A, Troffa C, van Oort FV, Verweij N, Vonk JM, Waite LL, Wennauer R, Wilsgaard T, Wojczynski MK, Wong A, Zhang Q, Zhao JH, Brennan EP, Choi M, Eriksson P, Folkersen L, Franco-Cereceda A, Gharavi AG, Hedman ÅK, Hivert MF, Huang J, Kanoni S, Karpe F, Keildson S, Kiryluk K, Liang L, Lifton RP, Ma B, McKnight AJ, McPherson R, Metspalu A, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Olsson C, Perry JR, Reinmaa E, Salem RM, Sandholm N, Schadt EE, Scott RA, Stolk L, Vallejo EE, Westra HJ, Zondervan KT; ADIPOGen Consortium; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GEFOS Consortium; GENIE Consortium; GLGC; ICBP; International Endogene Consortium; LifeLines Cohort Study; MAGIC Investigators; MuTHER Consortium; PAGE Consortium; ReproGen Consortium, Amouyel P, Arveiler D, Bakker SJ, Beilby J, Bergman RN, Blangero J, Brown MJ, Burnier M, Campbell H, Chakravarti A, Chines PS, Claudi-Boehm S, Collins FS, Crawford DC, Danesh J, de Faire U, de Geus EJ, Dörr M, Erbel R, Eriksson JG, Farrall M, Ferrannini E, Ferrières J, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gieger C, Gudnason V, Haiman CA, Harris TB, Hattersley AT, Heliövaara M, Hicks AA, Hingorani AD, Hoffmann W, Hofman A, Homuth G, Humphries SE, Hyppönen E, Illig T, Jarvelin MR, Johansen B, Jousilahti P, Jula AM, Kaprio J, Kee F, Keinanen-Kiukaanniemi SM, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuulasmaa K, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimäki T, Lyssenko V, Männistö S, Marette A, Matise TC, McKenzie CA, McKnight B, Musk AW, Möhlenkamp S, Morris AD, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Palmer LJ, Penninx BW, Peters A, Pramstaller PP, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schwarz PE, Shuldiner AR, Staessen JA, Steinthorsdottir V, Stolk RP, Strauch K, Tönjes A, Tremblay A, Tremoli E, Vohl MC, Völker U, Vollenweider P, Wilson JF, Witteman JC, Adair LS, Bochud M, Boehm BO, Bornstein SR, Bouchard C, Cauchi S, Caulfield MJ, Chambers JC, Chasman DI, Cooper RS, Dedoussis G, Ferrucci L, Froguel P, Grabe HJ, Hamsten A, Hui J, Hveem K, Jöckel KH, Kivimaki M, Kuh D, Laakso M, Liu Y, März W, Munroe PB, Njølstad I, Oostra BA, Palmer CN, Pedersen NL, Perola M, Pérusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sinisalo J, Slagboom PE, Snieder H, Spector TD, Stefansson K, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Veronesi G, Walker M, Wareham NJ, Watkins H, Wichmann HE, Abecasis GR, Assimes TL, Berndt SI, Boehnke M, Borecki IB, Deloukas P, Franke L, Frayling TM, Groop LC, Hunter DJ, Kaplan RC, O'Connell JR, Qi L, Schlessinger D, Strachan DP, Thorsteinsdottir U, van Duijn CM, Willer CJ, Visscher PM, Yang J, Hirschhorn JN, Zillikens MC, McCarthy MI, Speliotes EK, North KE, Fox CS, Barroso I, Franks PW, Ingelsson E, Heid IM, Loos RJ, Cupples LA, Morris AP, Lindgren CM, Mohlke KL. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187-196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1401] [Cited by in RCA: 1179] [Article Influence: 117.9] [Reference Citation Analysis (0)] |
| 18. | Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM; GIANT Consortium. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641-3649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 936] [Cited by in RCA: 1480] [Article Influence: 246.7] [Reference Citation Analysis (0)] |
| 19. | Ghodsian N, Abner E, Emdin CA, Gobeil É, Taba N, Haas ME, Perrot N, Manikpurage HD, Gagnon É, Bourgault J, St-Amand A, Couture C, Mitchell PL, Bossé Y, Mathieu P, Vohl MC, Tchernof A, Thériault S, Khera AV, Esko T, Arsenault BJ. Electronic health record-based genome-wide meta-analysis provides insights on the genetic architecture of non-alcoholic fatty liver disease. Cell Rep Med. 2021;2:100437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 104] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 20. | Yarmolinsky J, Bonilla C, Haycock PC, Langdon RJQ, Lotta LA, Langenberg C, Relton CL, Lewis SJ, Evans DM; PRACTICAL Consortium, Davey Smith G, Martin RM. Circulating Selenium and Prostate Cancer Risk: A Mendelian Randomization Analysis. J Natl Cancer Inst. 2018;110:1035-1038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 21. | Au Yeung SL, Zhao JV, Schooling CM. Evaluation of glycemic traits in susceptibility to COVID-19 risk: a Mendelian randomization study. BMC Med. 2021;19:72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Burgess S, Thompson SG; CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40:755-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2753] [Cited by in RCA: 2330] [Article Influence: 166.4] [Reference Citation Analysis (0)] |
| 23. | Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017;14:577-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 487] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 24. | Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology. 2017;28:30-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 373] [Cited by in RCA: 1284] [Article Influence: 183.4] [Reference Citation Analysis (0)] |
| 25. | Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27:R195-R208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 794] [Cited by in RCA: 983] [Article Influence: 140.4] [Reference Citation Analysis (0)] |
| 26. | Chittani M, Zaninello R, Lanzani C, Frau F, Ortu MF, Salvi E, Fresu G, Citterio L, Braga D, Piras DA, Carpini SD, Velayutham D, Simonini M, Argiolas G, Pozzoli S, Troffa C, Glorioso V, Kontula KK, Hiltunen TP, Donner KM, Turner ST, Boerwinkle E, Chapman AB, Padmanabhan S, Dominiczak AF, Melander O, Johnson JA, Cooper-Dehoff RM, Gong Y, Rivera NV, Condorelli G, Trimarco B, Manunta P, Cusi D, Glorioso N, Barlassina C. TET2 and CSMD1 genes affect SBP response to hydrochlorothiazide in never-treated essential hypertensives. J Hypertens. 2015;33:1301-1309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 27. | Lim ET, Würtz P, Havulinna AS, Palta P, Tukiainen T, Rehnström K, Esko T, Mägi R, Inouye M, Lappalainen T, Chan Y, Salem RM, Lek M, Flannick J, Sim X, Manning A, Ladenvall C, Bumpstead S, Hämäläinen E, Aalto K, Maksimow M, Salmi M, Blankenberg S, Ardissino D, Shah S, Horne B, McPherson R, Hovingh GK, Reilly MP, Watkins H, Goel A, Farrall M, Girelli D, Reiner AP, Stitziel NO, Kathiresan S, Gabriel S, Barrett JC, Lehtimäki T, Laakso M, Groop L, Kaprio J, Perola M, McCarthy MI, Boehnke M, Altshuler DM, Lindgren CM, Hirschhorn JN, Metspalu A, Freimer NB, Zeller T, Jalkanen S, Koskinen S, Raitakari O, Durbin R, MacArthur DG, Salomaa V, Ripatti S, Daly MJ, Palotie A; Sequencing Initiative Suomi (SISu) Project. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 255] [Cited by in RCA: 302] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 28. | Satapathy SK, Sanyal AJ. Epidemiology and Natural History of Nonalcoholic Fatty Liver Disease. Semin Liver Dis. 2015;35:221-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 247] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 29. | Kuang M, Sheng G, Hu C, Lu S, Peng N, Zou Y. The value of combining the simple anthropometric obesity parameters, Body Mass Index (BMI) and a Body Shape Index (ABSI), to assess the risk of non-alcoholic fatty liver disease. Lipids Health Dis. 2022;21:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 30. | Keating SE, Parker HM, Hickman IJ, Gomersall SR, Wallen MP, Coombes JS, Macdonald GA, George J, Johnson NA. NAFLD in clinical practice: Can simple blood and anthropometric markers be used to detect change in liver fat measured by (1) H-MRS? Liver Int. 2017;37:1907-1915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 31. | Mehran L, Amouzegar A, Fanaei SM, Masoumi S, Azizi F. Anthropometric measures and risk of all-cause and cardiovascular mortality: An 18 years follow-up. Obes Res Clin Pract. 2022;16:63-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 32. | Dong J, Ni YQ, Chu X, Liu YQ, Liu GX, Zhao J, Yang YB, Yan YX. Association between the abdominal obesity anthropometric indicators and metabolic disorders in a Chinese population. Public Health. 2016;131:3-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Manco M, Bedogni G, Marcellini M, Devito R, Ciampalini P, Sartorelli MR, Comparcola D, Piemonte F, Nobili V. Waist circumference correlates with liver fibrosis in children with non-alcoholic steatohepatitis. Gut. 2008;57:1283-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 34. | Motamed N, Sohrabi M, Ajdarkosh H, Hemmasi G, Maadi M, Sayeedian FS, Pirzad R, Abedi K, Aghapour S, Fallahnezhad M, Zamani F. Fatty liver index vs waist circumference for predicting non-alcoholic fatty liver disease. World J Gastroenterol. 2016;22:3023-3030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 64] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 35. | Ross R, Neeland IJ, Yamashita S, Shai I, Seidell J, Magni P, Santos RD, Arsenault B, Cuevas A, Hu FB, Griffin BA, Zambon A, Barter P, Fruchart JC, Eckel RH, Matsuzawa Y, Després JP. Waist circumference as a vital sign in clinical practice: a Consensus Statement from the IAS and ICCR Working Group on Visceral Obesity. Nat Rev Endocrinol. 2020;16:177-189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 483] [Cited by in RCA: 1025] [Article Influence: 205.0] [Reference Citation Analysis (0)] |
| 36. | Gastaldelli A, Cusi K, Pettiti M, Hardies J, Miyazaki Y, Berria R, Buzzigoli E, Sironi AM, Cersosimo E, Ferrannini E, Defronzo RA. Relationship between hepatic/visceral fat and hepatic insulin resistance in nondiabetic and type 2 diabetic subjects. Gastroenterology. 2007;133:496-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 413] [Cited by in RCA: 452] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 37. | Kawano Y, Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol. 2013;48:434-441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 545] [Cited by in RCA: 700] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 38. | Alferink LJM, Trajanoska K, Erler NS, Schoufour JD, de Knegt RJ, Ikram MA, Janssen HLA, Franco OH, Metselaar HJ, Rivadeneira F, Darwish Murad S. Nonalcoholic Fatty Liver Disease in The Rotterdam Study: About Muscle Mass, Sarcopenia, Fat Mass, and Fat Distribution. J Bone Miner Res. 2019;34:1254-1263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 39. | Kim D, Chung GE, Kwak MS, Kim YJ, Yoon JH. Effect of longitudinal changes of body fat on the incidence and regression of nonalcoholic fatty liver disease. Dig Liver Dis. 2018;50:389-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 40. | Atri A, Jiwanmall SA, Nandyal MB, Kattula D, Paravathareddy S, Paul TV, Thomas N, Kapoor N. The Prevalence and Predictors of Non-alcoholic Fatty Liver Disease in Morbidly Obese Women - A Cross-sectional Study from Southern India. Eur Endocrinol. 2020;16:152-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 41. | Després JP. Body fat distribution and risk of cardiovascular disease: an update. Circulation. 2012;126:1301-1313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 760] [Cited by in RCA: 953] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 42. | Younossi ZM. Non-alcoholic fatty liver disease - A global public health perspective. J Hepatol. 2019;70:531-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 943] [Cited by in RCA: 1455] [Article Influence: 242.5] [Reference Citation Analysis (1)] |
| 43. | Romero-Corral A, Somers VK, Sierra-Johnson J, Korenfeld Y, Boarin S, Korinek J, Jensen MD, Parati G, Lopez-Jimenez F. Normal weight obesity: a risk factor for cardiometabolic dysregulation and cardiovascular mortality. Eur Heart J. 2010;31:737-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 438] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 44. | Lee S, Kim KW, Lee J, Park T, Khang S, Jeong H, Song GW, Lee SG. Visceral adiposity as a risk factor for lean non-alcoholic fatty liver disease in potential living liver donors. J Gastroenterol Hepatol. 2021;36:3212-3218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 45. | Ning L, Sun J. Associations between body circumference and testosterone levels and risk of metabolic dysfunction-associated fatty liver disease: a mendelian randomization study. BMC Public Health. 2023;23:602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 46. | Gagnon E, Pelletier W, Gobeil É, Bourgault J, Manikpurage HD, Maltais-Payette I, Abner E, Taba N, Esko T, Mitchell PL, Ghodsian N, Després JP, Vohl MC, Tchernof A, Thériault S, Arsenault BJ. Mendelian randomization prioritizes abdominal adiposity as an independent causal factor for liver fat accumulation and cardiometabolic diseases. Commun Med (Lond). 2022;2:130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |