Published online Feb 16, 2024. doi: 10.12998/wjcc.v12.i5.891
Peer-review started: December 9, 2023
First decision: December 14, 2023
Revised: January 1, 2024
Accepted: January 23, 2024
Article in press: January 23, 2024
Published online: February 16, 2024
Processing time: 53 Days and 2.4 Hours
Previous studies have indicated bidirectional associations between urate levels and inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD). However, it remains unclear whether the observations are causal because of confounding factors.
To investigate the causal associations between urate levels and IBD using bidirectional Mendelian randomization (MR).
Independent genetic variants for urate levels and IBD were selected as instrumental variables from published genome-wide association studies (GWASs). Summary statistics for instrument-outcome associations were retrieved from three separate databases for IBD (the UK Biobank, the FinnGen database and a large GWAS meta-analysis) and one for urate levels (a large GWAS meta-analysis). MR analyses included the inverse-variance-weighted method, weighted-median estimator, MR-Egger and sensitivity analyses (MR-PRESSO). A meta-analysis was also conducted to merge the data from separate outcome databases using a fixed-effects model.
Genetically higher serum urate levels were strongly associated with an increased risk of UC [odds ratio (OR): 1.95, 95% confidence interval (CI): 1.86-2.05] after outlier correction, and the ORs (95%CIs) for IBD and CD were 0.94 (95%CI: 0.86-1.03) and 0.91 (95%CI: 0.80-1.04), respectively. Animal studies have confirmed the positive association between urate levels and UC. Moreover, genetically predicted IBD was inversely related to urate levels (OR: 0.97, 95%CI: 0.94-0.99). However, no association was observed between genetically influenced UC or CD and urate levels.
Urate levels might be risk factors for UC, whereas genetically predicted IBD was inversely associated with urate levels. These findings provide essential new insight for treating and preventing IBD.
Core Tip: Previous observational studies have indicated the association between urate levels and inflammatory bowel disease (IBD) (including ulcerative colitis (UC) and Crohn’s disease). To overcome the limitations of conventional observational studies and investigate the causal association between urate levels and IBD, we conducted a bidirectional Mendelian randomization (MR) study. MR analysis revealed that higher urate levels may be risk factors for UC, and genetically predicted IBD was inversely associated with urate levels.
- Citation: Zhang S, Fang X, Kang L, Sui XY, Liu M, Luo YJ, Fu S, Li ZS, Zhao SB, Bai Y. Serum urate is associated with an increased risk of inflammatory bowel disease: A bidirectional Mendelian randomization study. World J Clin Cases 2024; 12(5): 891-902
- URL: https://www.wjgnet.com/2307-8960/full/v12/i5/891.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i5.891
Inflammatory bowel disease (IBD), comprising ulcerative colitis (UC) and Crohn’s disease (CD), is characterized by chronic inflammation and a prolonged duration in the gastrointestinal tract[1]. Epidemiological studies have confirmed that the incidence of IBD in developing countries has exceeded 0.3% with the rapid adoption of the Western lifestyle[1,2]. Specifically, there are 322 and 214 cases per 100000 for CD and 505 and 214 cases per 100000 for UC in Europe and the United States, respectively. The long course of IBD lasts throughout the patient’s life, and the risk of colorectal cancer is much greater than that in the general population[3,4]. The pathogenesis of IBD involves interplay between environmental risk factors (not limited to smoking, unfavorable lifestyles and diets) and genetic variants, resulting in inadequate intestinal immune activation and dysbiosis of the gut microbiota[5,6]. Previous studies demonstrated that depleted mucosal antioxidant defense was common in IBD and thus may impede mucosal repair and compromise the inflamed mucosa[7]. Over the past decade, the association between antioxidants and IBD has attracted considerable interest[7-10] in light of the strong association between antioxidant capacity and the severity and disease activity of IBD.
Urate is vital as an antioxidant for neutralizing hydroxyl, superoxide and peroxynitrite radicals, which can decrease oxidative stress in vivo[11,12]. Previous studies have indicated that the serum uric acid-to-creatinine ratio is positively correlated with disease activity in CD patients[13]. Increased urate levels were positively correlated with an increased risk of UC[14]. Moreover, the use of a clinical drug (allopurinol) improved the severity of colitis by reducing urate levels[15]. An animal study by Rahimian et al[9] further demonstrated that uric acid mediated the protective effects of inosine against colitis. Overall, the relationship between urate levels and IBD (including UC and CD) has not been well established. A recent Mendelian randomization (MR) study by Chen et al[16] did not support the causal effect of serum urate levels on UC or CD incidence. However, the causal effect of these polymorphisms remains elusive because of the limited number of single-nucleotide polymorphisms (SNPs) used as instrumental variables (IVs). However, the causal effect of IBD (including UC and CD) on urate levels remains unclear.
Using genetic variants identified through genome-wide association studies (GWAS), MR is a popular approach for investigating the causal relationship between exposures and outcomes[17]. Therefore, to overcome the limitations of conventional observational studies, we aimed to examine the potential bidirectional relationship between IBD (including UC and CD) and serum urate levels in the present MR study. In addition, we conducted in vivo animal studies to verify the association between urate levels and IBD. This study provides reliable insight into the causal associations between urate levels and IBD.
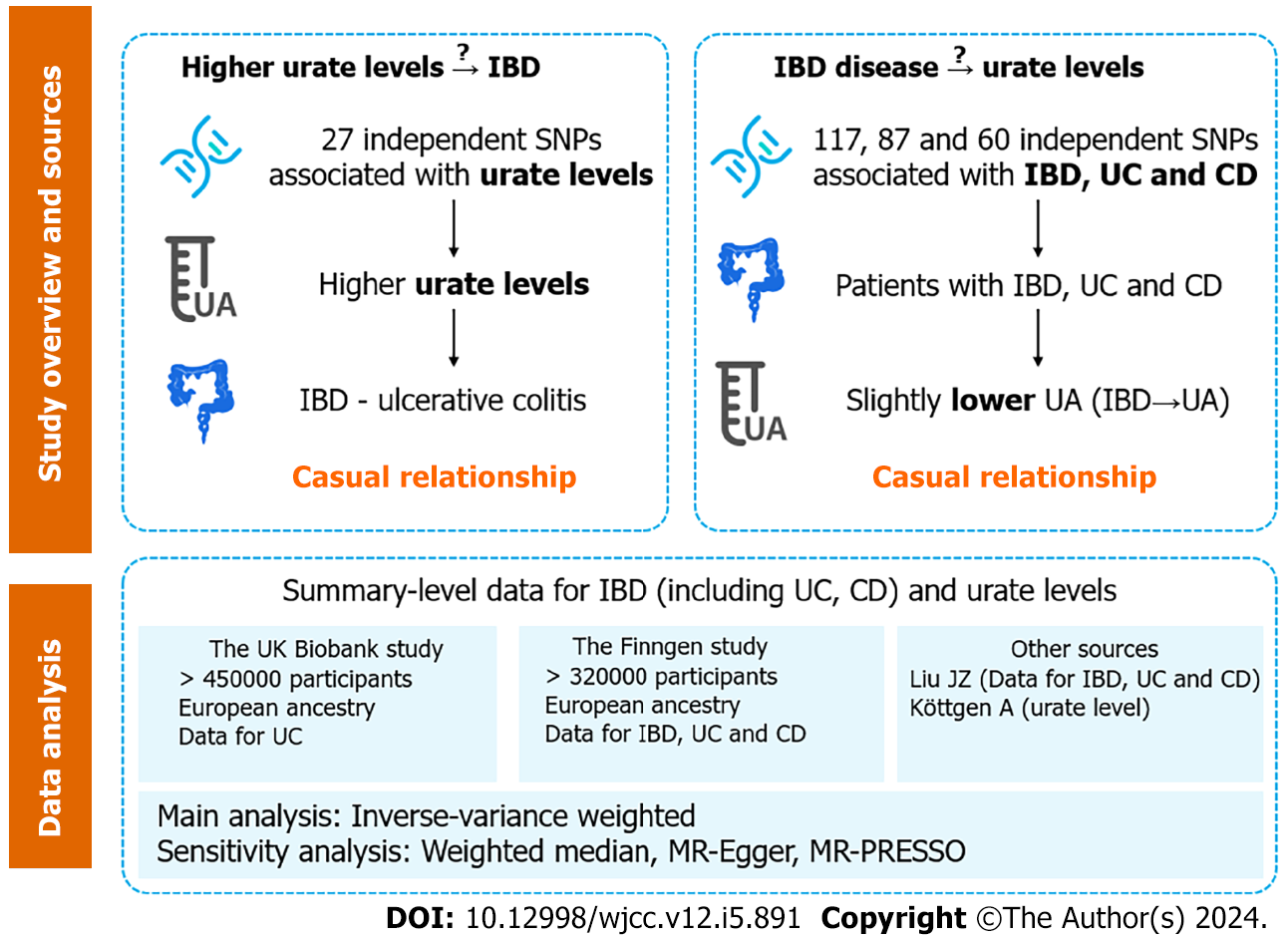
A bidirectional two-sample MR analysis was performed to assess the causal relationship between IBD and urate levels (Figure 1). SNPs associated with risk factors were selected as IVs. The MR study was based on three assumptions: (1) The SNPs used as IVs are strongly associated with exposure (urate level or IBD); (2) The SNPs are not associated with any confounder of exposure–outcome associations; and (3) The SNPs exert effects through exposure only. In combination with the three principles mentioned above, palindromic SNPs were identified and excluded in IV selection. All the data used in the current study were publicly available GWAS summary statistics; therefore, no additional ethical approval or informed consent was needed. GWAS summary statistics were searched to extract leading SNPs related to urate levels and IBD (including UC and CD) as IVs. Gene-outcome associations were retrieved from three databases: (1) A large-scale GWAS meta-analysis; (2) The Finngen database (version 7, https://r7.finngen.fi/); and (3) The UK Biobank (UKB).
SNPs related to urate levels were selected as IVs from a GWAS (Köttgen et al[18]), which included a total sample size of 110347 European individuals with various serum urate levels[18]. SNPs that were significantly associated with urate levels (P < 5 × 10-8) were extracted. A linkage disequilibrium (LD)-based clumping procedure was performed using the 1000 Genomes EUR reference panel (r2 < 0.01 and clump distance > 10000 kb) to ensure that each IV was independent. When SNPs related to exposure were absent in the outcome GWAS statistics, the proxy SNPs significantly associated with the variants of interest were selected (r2 > 0.8).
Summary statistics for IBD were obtained from the GWAS meta-analysis (Liu et al[19]), which included a total of 34652 participants of European ancestry (cases/controls for IBD: 12882/21770; UCs: 6968/20464; CDs: 5956/14927). Nearly 12 million SNPs were included in all three GWAS summary statistics. SNPs (P < 5 × 10-8) were selected and used for LD-based clumping. The proxy SNPs were extracted when SNPs related to exposure were absent. The IV selection procedure for IBD was the same as that for urate levels (described in the previous paragraph).
F-statistics, calculated as (beta/SE)2, were used to quantify the strength of each IV, and a value > 10 was considered sufficient[20]. In the present study, all F-statistics were greater than 10, indicating that there is little possibility of weak instrument bias based on summary statistics.
Summary-level data for urate levels were obtained from GWAS statistics (Köttgen et al[18]), as described in section 2.2. Gene-environment associations for IBD were obtained from three separate databases: (1) The GWAS meta-analysis from Liu et al[19]; (2) The Finngen database; and (3) The UKB (for UC data only). The Liu et al’s study has been described previously[19]. In the Finngen study, CD and UC were defined by their ICD codes, while IBD was a term consisting of CD, UC and indeterminate colitis. Among the patients and controls, 8966/312336 had IBD, 3243/318059 had CD, and 6803/314499 had UC. The UKB data for UC were extracted from a GWAS meta-analysis by Jiang et al[21], which included 2569 patients and 453779 controls. GWASs on IBD and CD were not available in the UKB.
The primary analysis method employed was the inverse-variance weighted (IVW) method, which assumes that all SNPs are valid and yields the most precise estimates[22]. In the presence of a sufficient sample size and absence of the pleiotropic effect of IVs, the IVW estimate is robust to confounding factors and approximates the true value[23]. A multiplicative random effect IVW model was applied when the heterogeneity significantly differed (P < 0.05).
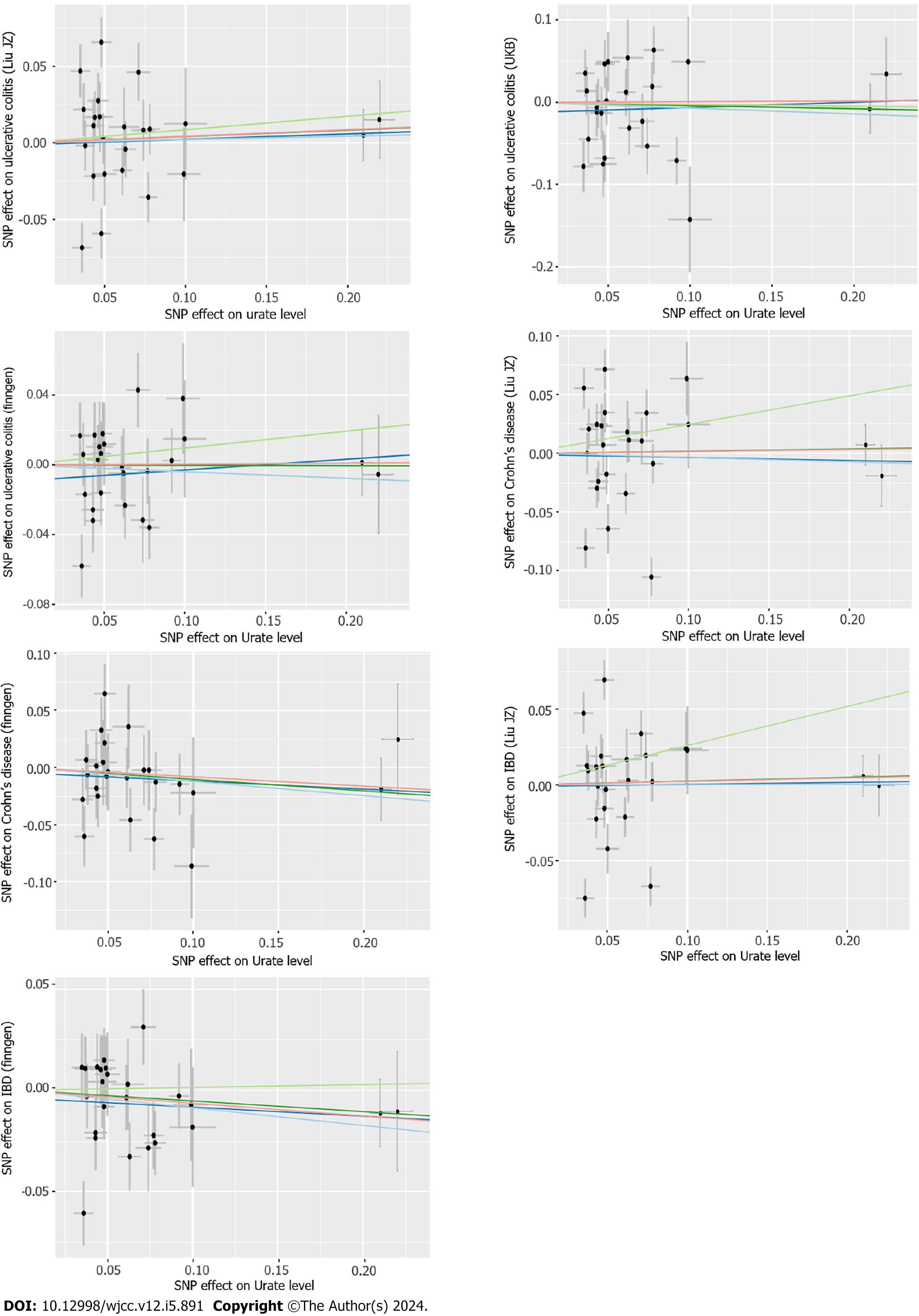
In addition to the IVW method, other robust methods (weighted median, MR-Egger and MR-PRESSO) were used to ensure the consistency and efficiency of the MR results. The weighted-median method could provide consistent causal estimates even when more than half of the IVs were invalid[23]. The MR-Egger estimates allowed the included IVs to demonstrate unbalanced pleiotropy[24]. The MR-PRESSO approach was used to detect horizontal pleiotropic outliers[25], and IVW estimates were performed to further investigate the causal relationship between exposure and outcome through outlier removal. Cochran’s Q test was applied to further examine the heterogeneity among all SNPs within each database. Leave-one-out analyses and scatter plots describing the causal relationship between serum urate levels and IBD were also generated.
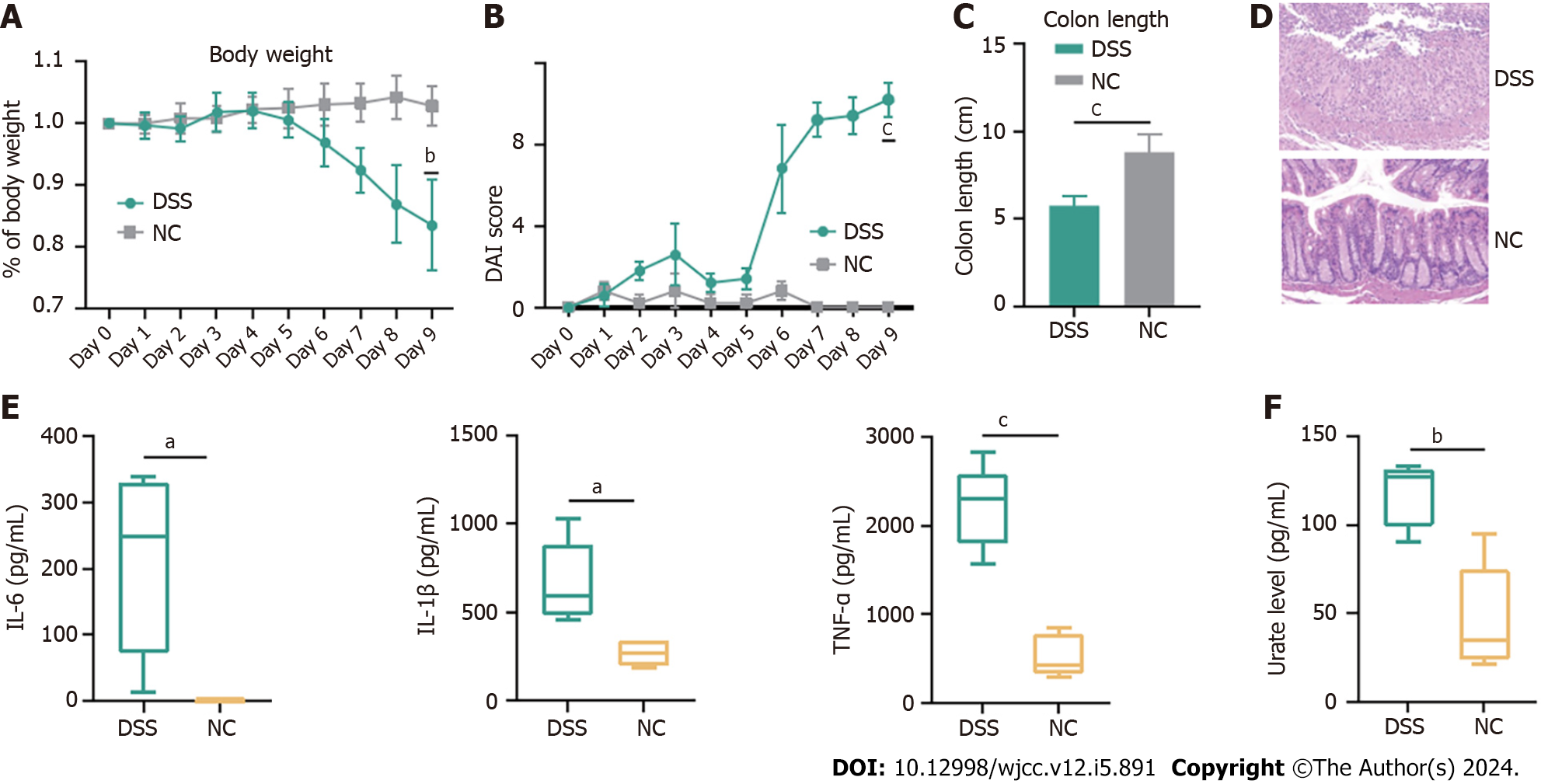
All animal experimental procedures were approved and conducted in accordance with the guidelines of the Animal Care Committee of Navy Medical University. C57BL/6 mice were kept under a 12-h light/dark cycle with free access to water and a standard rodent diet. Cohoused, seven-week-old male C57BL/6 mice (n = 5) were administered 2% dextran sulfate sodium (DSS) (36–50 kDa; MP Biomedicals) in their drinking water ad libitum for 7 consecutive days, followed by 2 d of normal water.
Body weight, the presence of occult bacteria per rectum, stool consistency, and colon length were documented. A scoring system was used to assess diarrhea and the presence of occult or overt blood in the stool. Changes in body weight are reported as the percentage loss of baseline body weight[26]. The ring of the rectum was harvested postmortem, fixed in 4% buffered formalin, and embedded in paraffin for subsequent HE staining.
Interleukin (IL)-6, IL-1β, tumor necrosis factor (TNF)-α and urate levels in the serum were quantified using commercial Enzyme-linked immunosorbent assay (ELISA) kits in accordance with the manufacturer’s instructions (Multi Sciences Ltd., Hangzhou, China).
MR results are presented as odds ratios (ORs) with 95% confidence intervals (CIs) of the outcome risk of a unit change in exposure. A two-sided P value < 0.05 was considered to indicate statistical significance. All the statistical analyses were performed mainly with R software (version 4.2.0, The R Foundation for Statistical Computing; TwoSampleMR and MR-PRESSO package) and SPSS 26.0.
Twenty-seven independent SNPs were identified as genetic IVs for urate levels, and the median (minimum, maximum) F statistic was 63.4 (35.4-1406.3) (Supplementary Table 1). Detailed information for urate-related SNPs is listed in Supplementary Table 2.
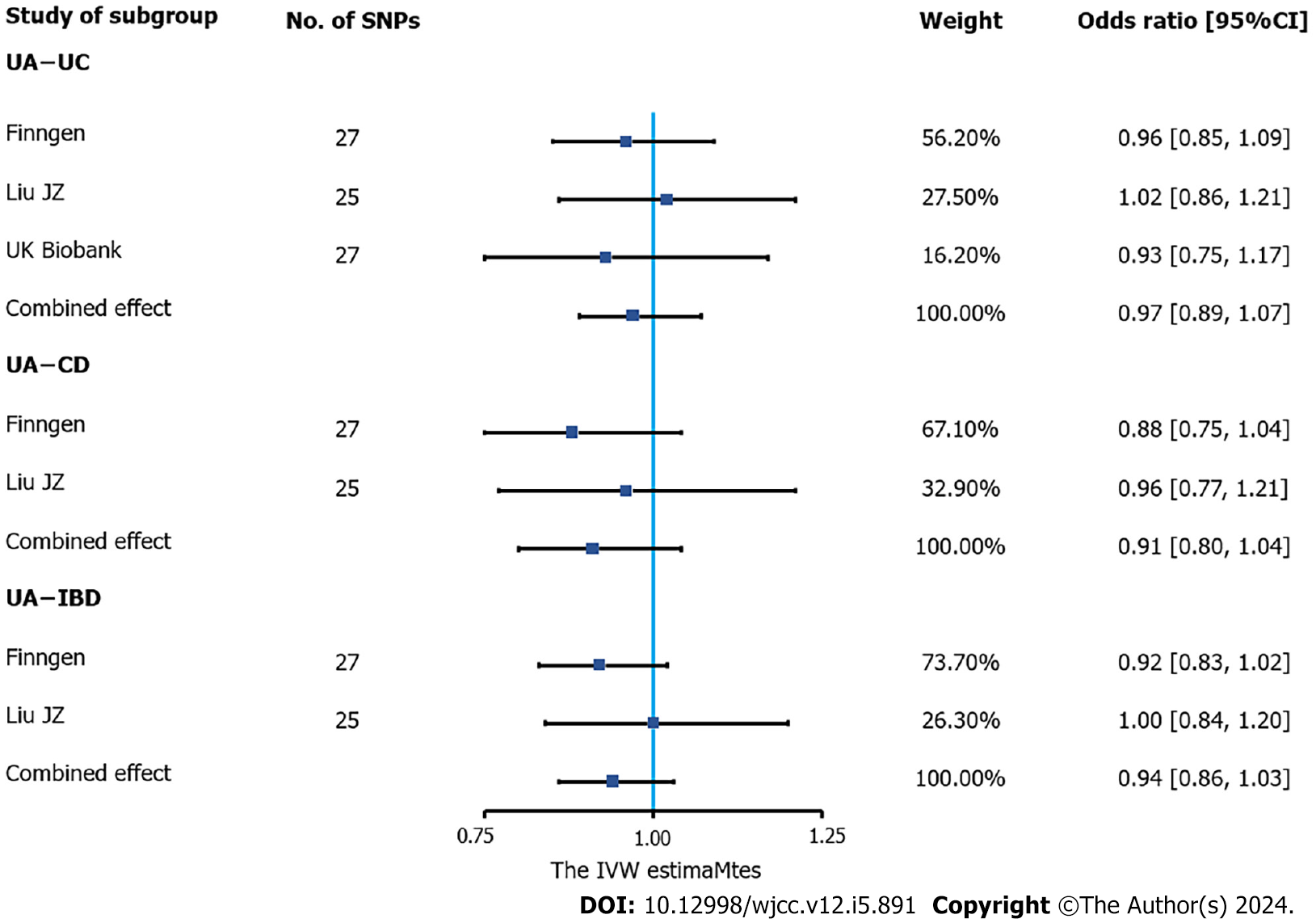
According to the meta-analysis of IVW estimates, the pooled ORs for IBD, UC and CD that were genetically predicted per log-OR increase in urate levels were 0.94 (95%CI: 0.86-1.03), 0.97 (95%CI: 0.89-1.07) and 0.91 (95%CI: 0.80-1.04), respectively (Figure 2).
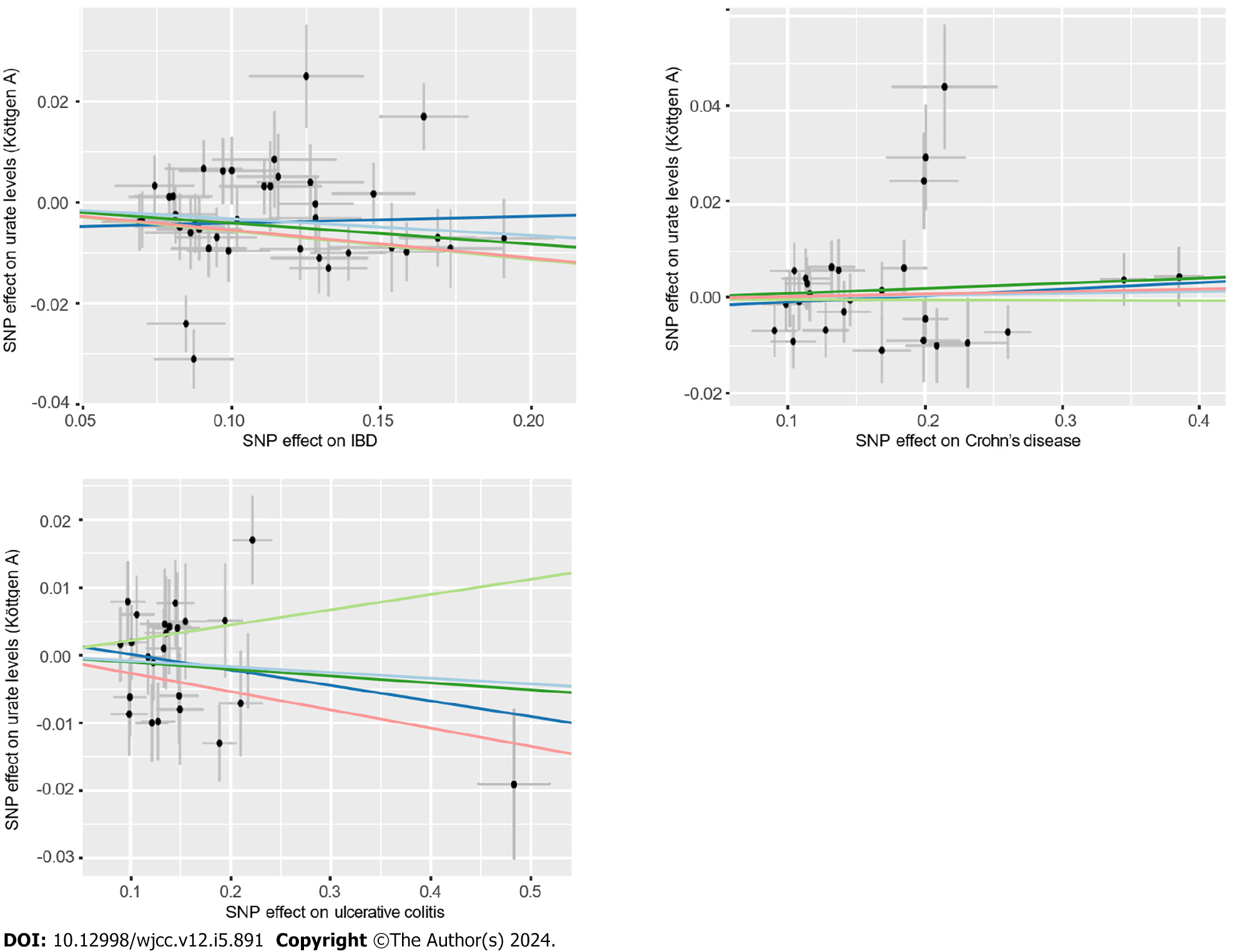
According to the sensitivity analysis (Supplementary Table 3), the three results were similar for the weighted-median estimator (Supplementary Figure 1). No pleiotropic effects were detected in any of the databases by MR-Egger estimation. Different outliers were identified by MR-PRESSO for IBD (n = 4), UC (n = 5) and CD (n = 5) in the GWAS meta-analysis by Liu et al[19] and UC (n = 3) in the UKB database, which resulted in potential pleiotropy assessed by global testing. Most of the results remained similar after outlier exclusion correction, except for IVs of urate levels on UC (UKB database) (before correction: OR = 0.93, 95%CI: 0.75-1.17; after correction: OR = 2.70, 95%CI: 2.54-2.87). Cochran’s Q test was performed after outlier exclusion to test heterogeneity. Among the urate level-related genetic IVs affecting IBD and CD identified by Liu et al[19] and UC (from the UKB database), a multiplicative random effect IVW model was used to evaluate the genetic estimate after heterogeneity was detected. A strongly positive causal relationship was detected between urate levels and UC after outlier exclusion and between urate levels and UC incidence according to a multiplicative random effects IVW estimate (OR = 1.95, 95%CI: 1.86-2.05). A scatter plot was generated to visualize the effect size of each MR method (Figure 3). Leave-one-out analysis indicated that the associations between urate levels and IBD incidence were unlikely to be driven by certain specific SNPs (Supplementary Figure 2).
A total of 117, 87, and 60 SNPs reached a genome-wide level of significance with IBD, UC and CD, respectively. A summary and detailed description of the variants are presented in Supplementary Tables 1 and 4.
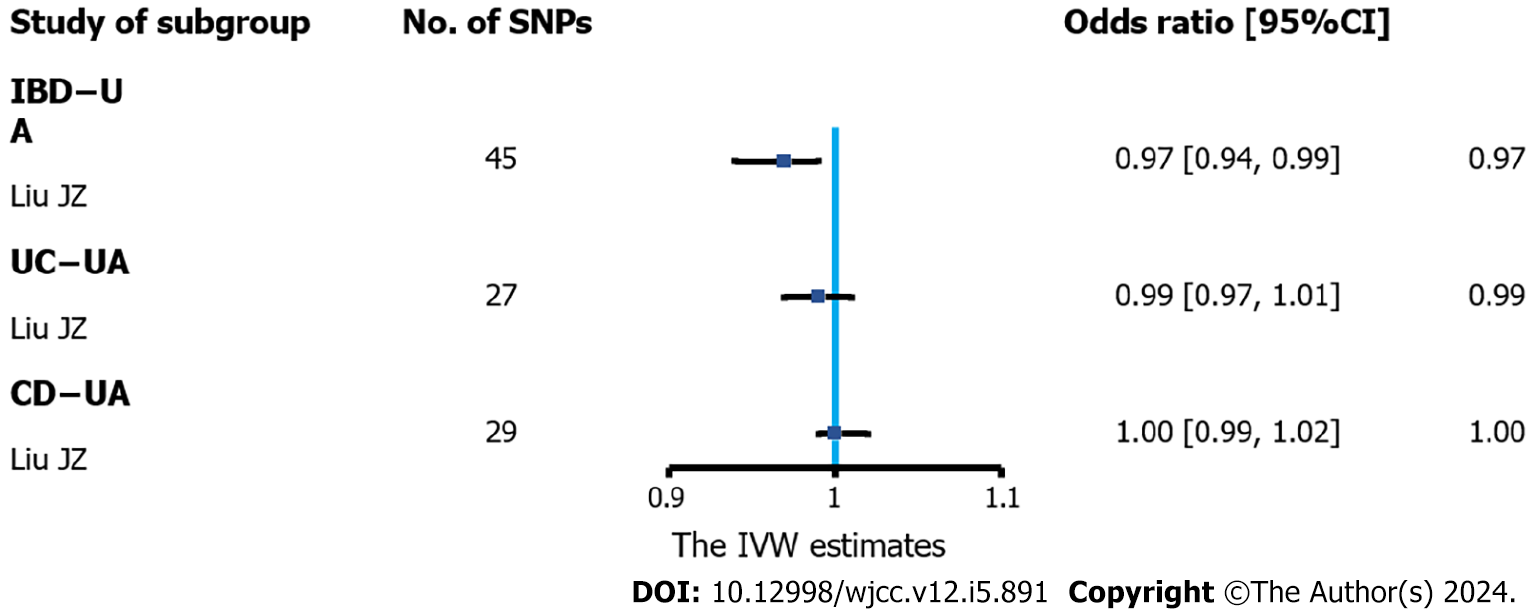
The results of IVW analysis demonstrated that IBD was negatively correlated with urate levels (OR = 0.97, 95%CI: 0.94-0.99) (Figure 4). However, no association between UC (or CD) and urate levels was observed. The combined ORs of UC and CD on urate levels were 0.99 (95%CI: 0.97-1.01) and 1.00 (95%CI: 0.99-1.02), respectively.
According to the sensitivity analysis (Supplementary Table 5), the weighted-median estimator showed comparable results to the estimates from the IVW analysis (Supplementary Figure 3). MR-Egger analysis demonstrated no evidence of pleiotropy, while the MR-PRESSO global test indicated that there were 7 outliers from the association between IBD and urate levels (P = 0.02) and 2 statistically nonsignificant outliers from the association between CD and urate levels (P = 0.006). Heterogeneity was detected from the association between IBD and urate levels after outlier correction by Cochran’s Q statistics. However, the results remained similar after correction for outliers and after the application of the multiplicative random effects IVW estimate (OR = 0.97, 95%CI: 0.94-0.99). A scatter plot was generated to visualize the effect size of each MR method (Figure 5). The results remained consistent in the leave-one-out analysis (Supple
To validate the positive association between serum urate levels and UC, 2% DSS was used to induce experimental colitis
In the current study, we evaluated the causal relationship between IBD and urate levels. We found evidence that genetic liability to urate levels was strongly associated with a higher risk of UC after outlier correction, and genetic liability to IBD was slightly anticorrelated to urate levels. Animal studies have confirmed the association between high urate levels and IBD. However, our study did not observe a causal relationship between CD and urate levels.
Previous observational studies have suggested that urate levels might be a risk factor for IBD. Zhu et al[13] included more than four hundred IBD patients and 51 non-IBD controls and reported that urate levels were significantly greater in IBD patients. Similarly, Tian et al[14] reported that increased urate levels were associated with UC in a retrospective case-control study. Moreover, IBD patients have an increased incidence of nephrolithiasis as well as urolithiasis[27]. To date, the only MR analysis conducted to investigate the causal relationship of urate levels with IBD has demonstrated that genetically predicted urate levels are not associated with the risk of CD or UC. In part, our study was consistent with previous reports in that we found a strong positive association between urate levels and UC but not with CD or IBD. Animal studies further demonstrated a positive association between urate levels and colitis incidence. In addition, IBD but not UC or CD was inversely correlated with urate levels.
The biological connection between IBD and urate levels has not been fully elucidated. Current studies suggest that intestinal inflammation (including oxidative stress) and dysbiosis of the gut microbiota are the main etiologies of IBD[6]. Increased urate levels mediate the exacerbation of mucosal colitis induced by DSS by enhancing intestinal permeability[15]. Treatment with allopurinol via gavage alleviated the pathogenic increase in proinflammatory cytokines and reduced oxidative stress biomarkers in patients with colitis[15,28]. A recent study reported that rhein significantly alleviated DSS-induced colitis and led to decreased urate levels, while the probiotic Lactobacillus was involved in regulating host metabolism[29]. These results support the idea of a relationship between serum urate levels and intestinal inflammation, suggesting that urate levels might be a therapeutic target for IBD. Our results supported previous results that urate levels were positively associated with an increased risk of UC but not with IBD or CD. One of the reasons could be the lack of association between urate levels and CD (a major subtype of IBD). Our results also confirmed that there was no bidirectional causal relationship between urate levels and CD incidence. Furthermore, we considered only the dichotomous IBD diagnosis rather than the IBD course or severity, which greatly influenced patients’ clinical manifestations. Further MR analysis should be conducted to investigate the causal relationship between urate levels and disease activity and course of IBD, as relevant GWAS data are available. Moreover, we found a slight inverse association between IBD incidence and urate levels. One possible explanation could be that including summary statistics from only one GWAS increased the heterogeneity and reduced the credibility of our results. A meta-analysis should be conducted once multiple data sources for urate levels are available.
There are three major strengths in the current study. First, the MR design is suitable for causal inference. As an alternative to randomized controlled trials, the MR method can partly avoid bias from confounding factors and reverse causation, which might increase the reliability of the results compared with those of observational studies. To our knowledge, this is the first bidirectional MR analysis investigating the causal relationship between IBD (and its subtypes) and urate levels. Second, we obtained summary-level data from large genetic consortia and GWASs, which included large sample sizes, with 110347 participants for urate levels, 355952 (21846 patients) for IBD, 805082 (16340 patients) for UC and 342185 (9199 patients) for CD. Third, population stratification bias was minimized because all GWAS summary statistics data in the current study were generated from the European population.
Nevertheless, potential limitations in our MR study should be considered. First, MR design can be biased by pleiotropic effects. The current study involved the implementation of various sensitivity analyses, which were performed based on distinct assumptions regarding the fundamental characteristics of pleiotropy, and most of the analyses showed stable results. Moreover, MR-Egger tests and MR-PRESSO analyses were conducted to explore horizontal pleiotropy[24,25]. After removing potential outlier SNPs, we observed a strong positive causal relationship between urate levels and UC, and most of the results were robust. Second, all participants included in the current study were European, which may limit the generalizability of our findings to other populations. Further MR analyses should be conducted to verify our findings in individuals of non-European descent. Third, in our present research, summary statistics for IBD were obtained from three databases, while data on urate levels were sourced solely from one large GWAS meta-analysis (Köttgen et al[18]). The utilization of data from a single source may compromise the reliability of the results. Therefore, once GWAS summary statistics from diverse sources become available, meta-analyses should be conducted to further verify our findings on the inverse association between IBD and urate levels.
The findings that serum urate levels increase the risk of UC add to the evidence from another MR analysis demonstrating a new risk factor for IBD. Recently, a meta-analysis based on large-scale cohorts demonstrated that the consumption of several types of food and drinks, for example, beer, wine, and beef, was associated with increased serum urate levels[30]; however, we are unaware of the risk related to the foods mentioned above. Moreover, many dietary approaches have been developed to reduce inflammation, prevent relapse, and manage the disease severity of IBD[31]. Our current study indicated that monitoring and managing urate levels in patients with IBD and accounting for diets that are associated with elevated urate levels in dietary therapy may provide additional benefits.
In summary, we systemically evaluated the potential causal relationship between IBD and urate levels. Our current MR analysis demonstrated that genetically predicted urate levels are causally associated with an elevated risk of UC, while IBD was inversely correlated with urate levels. Considering the close relationship between diet and urate levels, our study provides crucial new insight into treating and preventing IBD. These findings indicate that IBD patients may benefit from monitoring and reducing their serum urate levels.
Inflammatory bowel disease (IBD), mainly consisted of Crohn's disease (CD) and ulcerative colitis (UC), is a chronic inflammatory disease. As a vital antioxidant, urate can decrease oxidative stress in vivo, which may be associated with IBD state. However, the causality between IBD and urate levels has not been investigated.
Previous studies indicated uric acid-to-creatinine ratio and urate were positively correlated with the disease activity of CD and UC. Despite the existing findings demonstrated the bidirectional associations between urate levels and IBD, including UC and CD, the causality association between them remains unclear. This study seeks to investigate the causal association between IBD and urate through Mendelian randomization (MR) study, which may shed crucial new insight into treating and preventing IBD. In specific, IBD patients may benefit from monitoring and reducing serum urate levels.
The study aims to investigate the bidirectional causal relationship between urate levels and IBD by performing MR analysis, to better understand the gene susceptibility of urate levels and IBD.
Single nucleotide polymorphisms retrieved from genome-wide association studies (GWASs) was selected as instrument variants. Summary GWAS statistics for instrument-outcome associations were retrieved from three separate databases for IBD (UK Biobank, FinnGen database and a large GWAS meta-analysis) and one for urate levels (a large GWAS meta-analysis). Inverse-variance-weighted was performed to investigate the bidirectional causal relationship, and other sensitivity analysis were conducted to strengthen the results. Meta-analysis was conducted to merge the data from separate outcome databases using a fixed-effects model.
The current study found that the genetic susceptibility to urate levels was associated with increased UC risk [odds ratio (OR): 1.95, 95% confidence interval (CI): 1.86-2.05], and animal studies confirmed the positive association between urate levels and UC. Additionally, genetically predicted IBD was inversely related to urate levels (OR: 0.97, 95%CI: 0.94-0.99). However, no association was observed between genetically influenced UC or CD and urate levels.
This study identified urate levels might be risk factors for UC, whereas genetically predicted IBD was inversely associated with urate levels. The current results shed new insight into prevention and treatment of IBD.
Although the current study investigated the causal relationship between urate levels and UC, which was further verified by animal studies, the precise mechanism by which high urate levels affects the development of UC remains unknown. More basic and clinical studies should be conducted for identification of key regulators and molecules during the process.
We thank the UKB, the Finngen database and the IEU OpenGWAS project for sharing the summary-level data and all efforts from the researchers.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Vaishalli PM, India S-Editor: Fan JR L-Editor: A P-Editor: Yu HG
| 1. | Khalili H, Chan SSM, Lochhead P, Ananthakrishnan AN, Hart AR, Chan AT. The role of diet in the aetiopathogenesis of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2018;15:525-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 212] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 2. | Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769-2778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2677] [Cited by in RCA: 4060] [Article Influence: 507.5] [Reference Citation Analysis (110)] |
| 3. | Benchimol EI, Guttmann A, Griffiths AM, Rabeneck L, Mack DR, Brill H, Howard J, Guan J, To T. Increasing incidence of paediatric inflammatory bowel disease in Ontario, Canada: evidence from health administrative data. Gut. 2009;58:1490-1497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 297] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 4. | Frolkis AD, Dykeman J, Negrón ME, Debruyn J, Jette N, Fiest KM, Frolkis T, Barkema HW, Rioux KP, Panaccione R, Ghosh S, Wiebe S, Kaplan GG. Risk of surgery for inflammatory bowel diseases has decreased over time: a systematic review and meta-analysis of population-based studies. Gastroenterology. 2013;145:996-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 661] [Article Influence: 55.1] [Reference Citation Analysis (0)] |
| 5. | Huang H, Fang M, Jostins L, Umićević Mirkov M, Boucher G, Anderson CA, Andersen V, Cleynen I, Cortes A, Crins F, D'Amato M, Deffontaine V, Dmitrieva J, Docampo E, Elansary M, Farh KK, Franke A, Gori AS, Goyette P, Halfvarson J, Haritunians T, Knight J, Lawrance IC, Lees CW, Louis E, Mariman R, Meuwissen T, Mni M, Momozawa Y, Parkes M, Spain SL, Théâtre E, Trynka G, Satsangi J, van Sommeren S, Vermeire S, Xavier RJ; International Inflammatory Bowel Disease Genetics Consortium, Weersma RK, Duerr RH, Mathew CG, Rioux JD, McGovern DPB, Cho JH, Georges M, Daly MJ, Barrett JC. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 424] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 6. | Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1021] [Cited by in RCA: 1171] [Article Influence: 146.4] [Reference Citation Analysis (0)] |
| 7. | Buffinton GD, Doe WF. Depleted mucosal antioxidant defences in inflammatory bowel disease. Free Radic Biol Med. 1995;19:911-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 178] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 8. | Koutroubakis IE, Malliaraki N, Dimoulios PD, Karmiris K, Castanas E, Kouroumalis EA. Decreased total and corrected antioxidant capacity in patients with inflammatory bowel disease. Dig Dis Sci. 2004;49:1433-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 97] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 9. | Rahimian R, Fakhfouri G, Daneshmand A, Mohammadi H, Bahremand A, Rasouli MR, Mousavizadeh K, Dehpour AR. Adenosine A2A receptors and uric acid mediate protective effects of inosine against TNBS-induced colitis in rats. Eur J Pharmacol. 2010;649:376-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Bourgonje AR, Feelisch M, Faber KN, Pasch A, Dijkstra G, van Goor H. Oxidative Stress and Redox-Modulating Therapeutics in Inflammatory Bowel Disease. Trends Mol Med. 2020;26:1034-1046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 229] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 11. | Krishnan E. Inflammation, oxidative stress and lipids: the risk triad for atherosclerosis in gout. Rheumatology (Oxford). 2010;49:1229-1238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 12. | Waring WS. Uric acid: an important antioxidant in acute ischaemic stroke. QJM. 2002;95:691-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 86] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Zhu F, Feng D, Zhang T, Gu L, Zhu W, Guo Z, Li Y, Lu N, Gong J, Li N. Altered uric acid metabolism in isolated colonic Crohn's disease but not ulcerative colitis. J Gastroenterol Hepatol. 2019;34:154-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 14. | Tian S, Li J, Li R, Liu Z, Dong W. Decreased Serum Bilirubin Levels and Increased Uric Acid Levels are Associated with Ulcerative Colitis. Med Sci Monit. 2018;24:6298-6304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Chiaro TR, Soto R, Zac Stephens W, Kubinak JL, Petersen C, Gogokhia L, Bell R, Delgado JC, Cox J, Voth W, Brown J, Stillman DJ, O'Connell RM, Tebo AE, Round JL. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 173] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 16. | Chen J, Ruan X, Yuan S, Deng M, Zhang H, Sun J, Yu L, Satsangi J, Larsson SC, Therdoratou E, Wang X, Li X. Antioxidants, minerals and vitamins in relation to Crohn's disease and ulcerative colitis: A Mendelian randomization study. Aliment Pharmacol Ther. 2023;57:399-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 42] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 17. | Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3525] [Cited by in RCA: 2981] [Article Influence: 175.4] [Reference Citation Analysis (0)] |
| 18. | Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O'Seaghdha CM, Haller T, Yang Q, Tanaka T, Johnson AD, Kutalik Z, Smith AV, Shi J, Struchalin M, Middelberg RP, Brown MJ, Gaffo AL, Pirastu N, Li G, Hayward C, Zemunik T, Huffman J, Yengo L, Zhao JH, Demirkan A, Feitosa MF, Liu X, Malerba G, Lopez LM, van der Harst P, Li X, Kleber ME, Hicks AA, Nolte IM, Johansson A, Murgia F, Wild SH, Bakker SJ, Peden JF, Dehghan A, Steri M, Tenesa A, Lagou V, Salo P, Mangino M, Rose LM, Lehtimäki T, Woodward OM, Okada Y, Tin A, Müller C, Oldmeadow C, Putku M, Czamara D, Kraft P, Frogheri L, Thun GA, Grotevendt A, Gislason GK, Harris TB, Launer LJ, McArdle P, Shuldiner AR, Boerwinkle E, Coresh J, Schmidt H, Schallert M, Martin NG, Montgomery GW, Kubo M, Nakamura Y, Munroe PB, Samani NJ, Jacobs DR Jr, Liu K, D'Adamo P, Ulivi S, Rotter JI, Psaty BM, Vollenweider P, Waeber G, Campbell S, Devuyst O, Navarro P, Kolcic I, Hastie N, Balkau B, Froguel P, Esko T, Salumets A, Khaw KT, Langenberg C, Wareham NJ, Isaacs A, Kraja A, Zhang Q, Wild PS, Scott RJ, Holliday EG, Org E, Viigimaa M, Bandinelli S, Metter JE, Lupo A, Trabetti E, Sorice R, Döring A, Lattka E, Strauch K, Theis F, Waldenberger M, Wichmann HE, Davies G, Gow AJ, Bruinenberg M; LifeLines Cohort Study, Stolk RP, Kooner JS, Zhang W, Winkelmann BR, Boehm BO, Lucae S, Penninx BW, Smit JH, Curhan G, Mudgal P, Plenge RM, Portas L, Persico I, Kirin M, Wilson JF, Mateo Leach I, van Gilst WH, Goel A, Ongen H, Hofman A, Rivadeneira F, Uitterlinden AG, Imboden M, von Eckardstein A, Cucca F, Nagaraja R, Piras MG, Nauck M, Schurmann C, Budde K, Ernst F, Farrington SM, Theodoratou E, Prokopenko I, Stumvoll M, Jula A, Perola M, Salomaa V, Shin SY, Spector TD, Sala C, Ridker PM, Kähönen M, Viikari J, Hengstenberg C, Nelson CP; CARDIoGRAM Consortium; DIAGRAM Consortium; ICBP Consortium; MAGIC Consortium, Meschia JF, Nalls MA, Sharma P, Singleton AB, Kamatani N, Zeller T, Burnier M, Attia J, Laan M, Klopp N, Hillege HL, Kloiber S, Choi H, Pirastu M, Tore S, Probst-Hensch NM, Völzke H, Gudnason V, Parsa A, Schmidt R, Whitfield JB, Fornage M, Gasparini P, Siscovick DS, Polašek O, Campbell H, Rudan I, Bouatia-Naji N, Metspalu A, Loos RJ, van Duijn CM, Borecki IB, Ferrucci L, Gambaro G, Deary IJ, Wolffenbuttel BH, Chambers JC, März W, Pramstaller PP, Snieder H, Gyllensten U, Wright AF, Navis G, Watkins H, Witteman JC, Sanna S, Schipf S, Dunlop MG, Tönjes A, Ripatti S, Soranzo N, Toniolo D, Chasman DI, Raitakari O, Kao WH, Ciullo M, Fox CS, Caulfield M, Bochud M, Gieger C. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 627] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 19. | Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, Abedian S, Cheon JH, Cho J, Dayani NE, Franke L, Fuyuno Y, Hart A, Juyal RC, Juyal G, Kim WH, Morris AP, Poustchi H, Newman WG, Midha V, Orchard TR, Vahedi H, Sood A, Sung JY, Malekzadeh R, Westra HJ, Yamazaki K, Yang SK; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium, Barrett JC, Alizadeh BZ, Parkes M, Bk T, Daly MJ, Kubo M, Anderson CA, Weersma RK. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1898] [Cited by in RCA: 1842] [Article Influence: 184.2] [Reference Citation Analysis (0)] |
| 20. | Luo J, Xu Z, Noordam R, van Heemst D, Li-Gao R. Depression and Inflammatory Bowel Disease: A Bidirectional Two-sample Mendelian Randomization Study. J Crohns Colitis. 2022;16:633-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 92] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 21. | Jiang L, Zheng Z, Fang H, Yang J. A generalized linear mixed model association tool for biobank-scale data. Nat Genet. 2021;53:1616-1621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 341] [Article Influence: 85.3] [Reference Citation Analysis (0)] |
| 22. | Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1330] [Cited by in RCA: 3756] [Article Influence: 313.0] [Reference Citation Analysis (1)] |
| 23. | Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4015] [Cited by in RCA: 5667] [Article Influence: 629.7] [Reference Citation Analysis (0)] |
| 24. | Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2275] [Cited by in RCA: 6185] [Article Influence: 618.5] [Reference Citation Analysis (0)] |
| 25. | Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693-698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2495] [Cited by in RCA: 5313] [Article Influence: 759.0] [Reference Citation Analysis (0)] |
| 26. | Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2:541-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1063] [Cited by in RCA: 1253] [Article Influence: 69.6] [Reference Citation Analysis (0)] |
| 27. | van Sommeren S, Janse M, Karjalainen J, Fehrmann R, Franke L, Fu J, Weersma RK. Extraintestinal manifestations and complications in inflammatory bowel disease: from shared genetics to shared biological pathways. Inflamm Bowel Dis. 2014;20:987-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | El-Mahdy NA, Saleh DA, Amer MS, Abu-Risha SE. Role of allopurinol and febuxostat in the amelioration of dextran-induced colitis in rats. Eur J Pharm Sci. 2020;141:105116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 29. | Wu J, Wei Z, Cheng P, Qian C, Xu F, Yang Y, Wang A, Chen W, Sun Z, Lu Y. Rhein modulates host purine metabolism in intestine through gut microbiota and ameliorates experimental colitis. Theranostics. 2020;10:10665-10679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 259] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 30. | Major TJ, Topless RK, Dalbeth N, Merriman TR. Evaluation of the diet wide contribution to serum urate levels: meta-analysis of population based cohorts. BMJ. 2018;363:k3951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 143] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 31. | Halmos EP, Gibson PR. Dietary management of IBD--insights and advice. Nat Rev Gastroenterol Hepatol. 2015;12:133-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |