Published online Jan 26, 2024. doi: 10.12998/wjcc.v12.i3.503
Peer-review started: October 31, 2023
First decision: November 28, 2023
Revised: December 12, 2023
Accepted: January 4, 2024
Article in press: January 4, 2024
Published online: January 26, 2024
Processing time: 78 Days and 17.6 Hours
Angelman syndrome (AS) is caused by maternal chromosomal deletions, imprinting defects, paternal uniparental disomy involving chromosome 15 and the ubiquitin-protein ligase UBE3A gene mutations. However the genetic basis remains unclear for several patients.
To investigate the involvement of UBE3A gene in AS and identifying new potential genes using exome sequencing.
We established a cohort study in 50 patients referred to Farhat Hached University Hospital between 2006 and 2021, with a strong suspicion of AS and absence of chromosomal aberrations. The UBE3A gene was screened for mutation detection. Two unrelated patients issued from consanguineous families were subjected to exome analysis.
We describe seven UBE3A variants among them 3 none previously described including intronic variants c.2220+14T>C (intron14), c.2507+43T>A (Exon15) and insertion in Exon7: c.30-47_30-46. The exome sequencing revealed 22 potential genes that could be involved in AS-like syndromes that should be investigated further.
Screening for UBE3A mutations in AS patients has been proven to be useful to confirm the diagnosis. Our exome findings could rise to new potential alternative target genes for genetic counseling.
Core Tip: Angelman syndrome (AS) is caused by maternal chromosome 15q11q13 deletions, imprinting defects, paternal uniparental disomy 15, and ubiquitin-protein ligase E3A (UBE3A) gene mutations. UBE3A is a brain-specific imprinting gene that encodes a ubiquitin-protein ligase. Here, we describe the variants in the UBE3A coding region detected by sequencing analysis in 50 AS Tunisian individuals with a normal bi-parental inheritance and methylation pattern of 15q11q13. Seven polymorphisms were found in our patients, including three novel variants. To identify bi-allelic recessive mutations that give rise to AS-like phenotypes, we considered consanguineous families, as they are more likely to develop such a recessive disease.
- Citation: Manoubi W, Mahdouani M, Hmida D, Kdissa A, Rouissi A, Turki I, Gueddiche N, Soyah N, Saad A, Bouwkamp C, Elgersma Y, Mougou-Zerelli S, Gribaa M. Genetic investigation of the ubiquitin-protein ligase E3A gene as putative target in Angelman syndrome. World J Clin Cases 2024; 12(3): 503-516
- URL: https://www.wjgnet.com/2307-8960/full/v12/i3/503.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i3.503
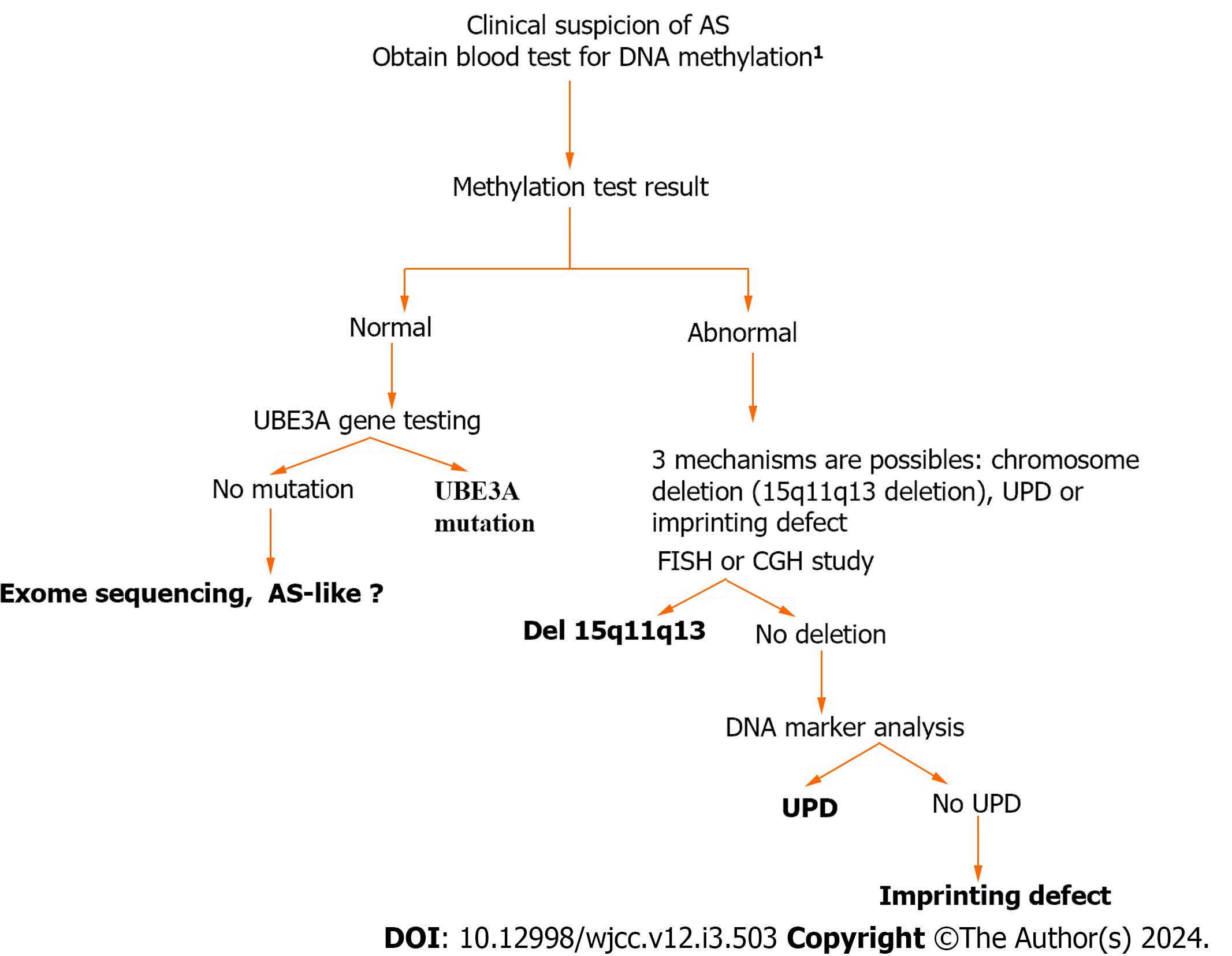
Angelman syndrome (AS) (OMIM-105830) is a neurodevelopmental disorder with a frequency of approximately 1 in 15000 births[1]. It is characterized by severe intellectual disability, lack of speech, easy-provoked smiling and laughter, a happy disposition, ataxia, sleep disorder, an electroencephalographic background, epilepsy, and a distinct behavioral profile. Patients with AS love water and have a fascination for reflective surfaces, plastic, and balloons[1]. This pathology is related to the genomic imprinting of chromosomal region 15q11–q13, which contains several genes, including ubiquitin-protein ligase E3A (UBE3A). A loss of function of the maternally expressed UBE3A protein causes the AS phenotype[2-4]. During gametogenesis in the parents, the 15q11_q13 region is subject to differential gene silencing via methylation, which is controlled by an adjacent imprinting center. Four different types of genetic defects have been identified in individuals with AS: Maternal deletions involving the chromosome 15q11_q13 region (accounting for approximately 70% of cases), paternal uniparental disomy (UPD) of chromosome 15 (occurring in 3% to 5% of cases), imprinting defects (occurring in 2% to 6% of cases), and mutations in the UBE3A gene (found in 4% to 23% of cases)[5-7]. An algorithm for genetic testing is shown in Figure 1. UBE3A mutations can be identified in around 75% of familial patients based on Sanger sequencing screening (Figure 1)[1,5,8,9].
Numerous mutations have been documented[2,3], and while some of them have been identified in multiple patients[2,8-11], most mutations are unique to individual cases. Additionally, various studies have reported different polymo
In the present study, screening for the UBE3A gene was performed in 50 patients referred with a strong suspicion of AS and for whom classical molecular diagnostic tests had failed to provide the diagnosis. We described the difficulties encountered in determining the genetic etiologies of AS and suggested some solutions.
Fifty patients (33 males and 17 females, aged 1-7 years) with a probable diagnosis of AS were referred to the Laboratory of Human Cytogenetics, Molecular Genetics, and Reproductive Biology of Farhat Hached University Hospital between 2006 and 2021. These patients showed severe mental retardation, severe speech impairment, epileptic seizures, abnormal electroencephalogram (EEG) findings, and dysmorphic facial features. The clinical characteristics of these patients are presented in Table 1.
| Number of patients | Percentage (%) | |
| Gender | 33 male; 17 female | |
| Hypotonia | 100 | |
| Neck support | (32/50) | 64 |
| Walk without support | (21/50) | 42 |
| Sitting without support | (38/50) | 76 |
| Absent speech | (40/50) | 80 |
| Developmental delay | 50/50) | 100 |
| Severe mental retardation | (50/50) | 100 |
| Microcephaly | (44/50) | 88 |
| Macrostomia | (40/50) | 80 |
| Clinical seizures | (44/50) | 88 |
| Occipital groove | (45/50) | 90 |
| Protruding tongue | (43/50) | 86 |
| Wide-spaced teeth | (35/50) | 70 |
| Prognathism | (40/50) | 80 |
| Unusually light hair or skin color | (13/50) | 26 |
| Easily provoked laughter | (50/50) | 100 |
| Hyperactivity | (48/50) | 96 |
| Gastro-esophageal reflux | (40/50) | 80 |
| Ataxic movements | (48/50) | 96 |
| Frequent drooling | (47/50) | 94 |
| History of sleep difficulties | (45/50) | 90 |
| Fascination with water | (40/50) | 80 |
| Autistic behavior | (14/50) | 28 |
Clinical geneticists evaluated all patients. The blood samples were collected after obtaining written consent from the parents and after the approval of the ethics committee of Farhat Hached University Hospital.
Combined cytogenetic, fluorescence in situ hybridization, Methylation-Specific Multiplex Ligation-Dependent Probe Amplification (MS-MLPA), and microsatellite analyses excluded deletions, UPDs, and imprintin 222 g defects (IDs) in all patients (data not shown).
Genomic DNA was extracted and purified from peripheral blood leukocytes using the Qia-amp DNA blood mini kit (Qiagen, Valencia, CA, and United States). Protocol was performed according to the manufacturer’s instructions. DNA concentration and purity were determined using a NanoDrop 1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States).
Exon 7 to exon 16 of the UBE3A gene (Table 2) were amplified by polymerase chain reaction (PCR) of a 25 µL reaction mixture containing: 1 µL of genomic DNA (150 ng/µL), 1 µL of forward primer (20 pg/µL), 1 µL of reverse primer (20 pg/µL), 0.2 µL of Taq DNA polymerase recombinant (Invitrogen), 5 µL MgCl2 in the Taq buffer (MgCl2 = 7.5 mmol/L), and 16.8 µL of deionized water. PCR was performed in thermocycler Gene Amp PCR System 9700 from Applied Biosystems, Foster City, CA.
| Exon and primers | Forward and reverse primers (5’->3’) | Region | Annealing temperature (℃) |
| 7 | |||
| Ex7F | GCC ACC TGA TCT GAC CAC T | Intron | 52 |
| Ex7R | GCA GTC TAG GGC AAC TCA AA | Intron | |
| 8 | |||
| Ex8AF | GCC TTG ATG ATA TGT TGA GC | Intron | 55 |
| Ex8AR | AAT TCT AGC GCC TTT CTT GT | Exon | |
| Ex8BF | GCC TGC ACG AAT GAG TTT TGT | Exon | 55 |
| Ex8BR | AGT TAT TAT TCC TGT CCG TTA CC | Intron | |
| 9 | |||
| Ex9AF | TGT TTG GCT GTT TTA CTT TTA GA | Intron | 55 |
| Ex9AR | GGC ATC AAT ATC CAC AGA CAC A | Exon | |
| Ex9BF | AGA AGC ATC TTC CTC AAG G | Exon | 55 |
| Ex9BR | CAC TTC CCC TCC CAC TAC | Exon | |
| Ex9CF | CAA TGA ATT TAA CAG TCG A | Exon | 55 |
| Ex9CR | CAT CAT CTA TGA TAT GGT CAC G | Exon | |
| Ex9DF | CGC ATG TAC AGT GAA CGA AGA A | Exon | 55 |
| Ex9DR | TGC ACA GGA ACA ACA AAA GTA T | Intron | |
| 10 | |||
| Ex10F | GTT TGC TTT CTG TTT CCA TTT AC | Intron | 52 |
| Ex10R | ATC CTT CTT TTG CTG CTC TTC | Intron | |
| 11 | |||
| Ex11F | CAA TGT TGC ATG CCT AAT TAC A | Intron | |
| GGT ACT TCG GTC AGA TTA AAA C | Intron | ||
| 12 | |||
| Ex12F | GGG GAC TGG AGG GAT ACT GT | Intron | 55 |
| ACA TGC TTT GAA AGT GTT AAT G | Intron | ||
| 13 | |||
| Ex13F | GAA ATT GTT AAG AAG TAG GTG | Intron | 52 |
| Ex13R | ATA TGT CTT AGT TAT CTG CTA | Intron | |
| 14 | |||
| Ex14F | AGG TGT CTG CAA AAA GTC | Intron | 55 |
| Ex14R | TTA GCT CTG AAA AAT GGT G | Intron | |
| 15 | |||
| Ex15F | ATA ATG AAT GCC AAA CTG AA | Intron | 55 |
| Ex15R | ATA TGT ATG TGA CGA GGA ATG | Intron | |
| 16 | |||
| Ex16F | CCC ATG ACT TAC AGT TTT CCT G | Intron | 55 |
| Ex16R | AAG AAG GGA GGC ACA GAC AT | Intron |
The UBE3A gene was analyzed by direct sequencing of exons 7 to 16 and flanking exon/intron boundaries in patients and their parents (Transcript: ENST00000232165). The primers and conditions are summarized in Table 2.
The amplicons were purified and directly sequenced. Sequencing reactions were prepared using an ABI Big Dye Terminator v3.1 cycle sequencing kit and separated by a 3500 Genetic Analyzer 16-Capillary Array. Sequencing data were analyzed with the Seqscape V2.0 software (Applied Biosystems) and compared to the reference sequence of the exon UBE3A gene downloaded from the Genome Browser Gateway (http://www.genome.ucsc.edu/). In-silico softwares such as PolyPhen-2 and Mutation Taster were used to calculate variable effects.
We used Illumina Human OmniExpress 700 K single nucleotide polymorphism (SNP) arrays for both linkage and copy number analysis with DNA isolated from venous blood, assuming an autosomal recessive model. Linkage analysis was aimed to identify chromosomal regions shared by all affected family members. Allegro embedded in EASY Linkage was used to perform linkage analysis to identify chromosomal regions shared by all affected family members.
Copy number variations analysis was performed using NEXUS Discovery Edition, version 7 (Biodiscovery, El Segundo, CA). Whole-exome sequencing was performed on affected patients and their unaffected parents (Figure 2).
The exome sequencing was performed using the in-use capture (Agilent SureSelect V4 Human 50 MB kit, Agilent Technologies) and paired-end sequencing on an Illumina Hi-Seq 2000 sequencer. Reads were aligned to the human reference genome version 19 using the Burrows-Wheels Aligner. Genome Analysis Toolkit was used for SNPs and indels. We used Cartagenia software (Cartagenia Bench Lab, Agilent Technologies) to filter the variants.
We filtered the heterozygous variants based on the following criteria: (1) Present within the shared genomic regions; (2) predicted to affect protein-coding (nonsense, missense, splice site, frameshift); and (3) have a minor allele frequency of < 0.1% in the more recent databases (10000G, ExAC).
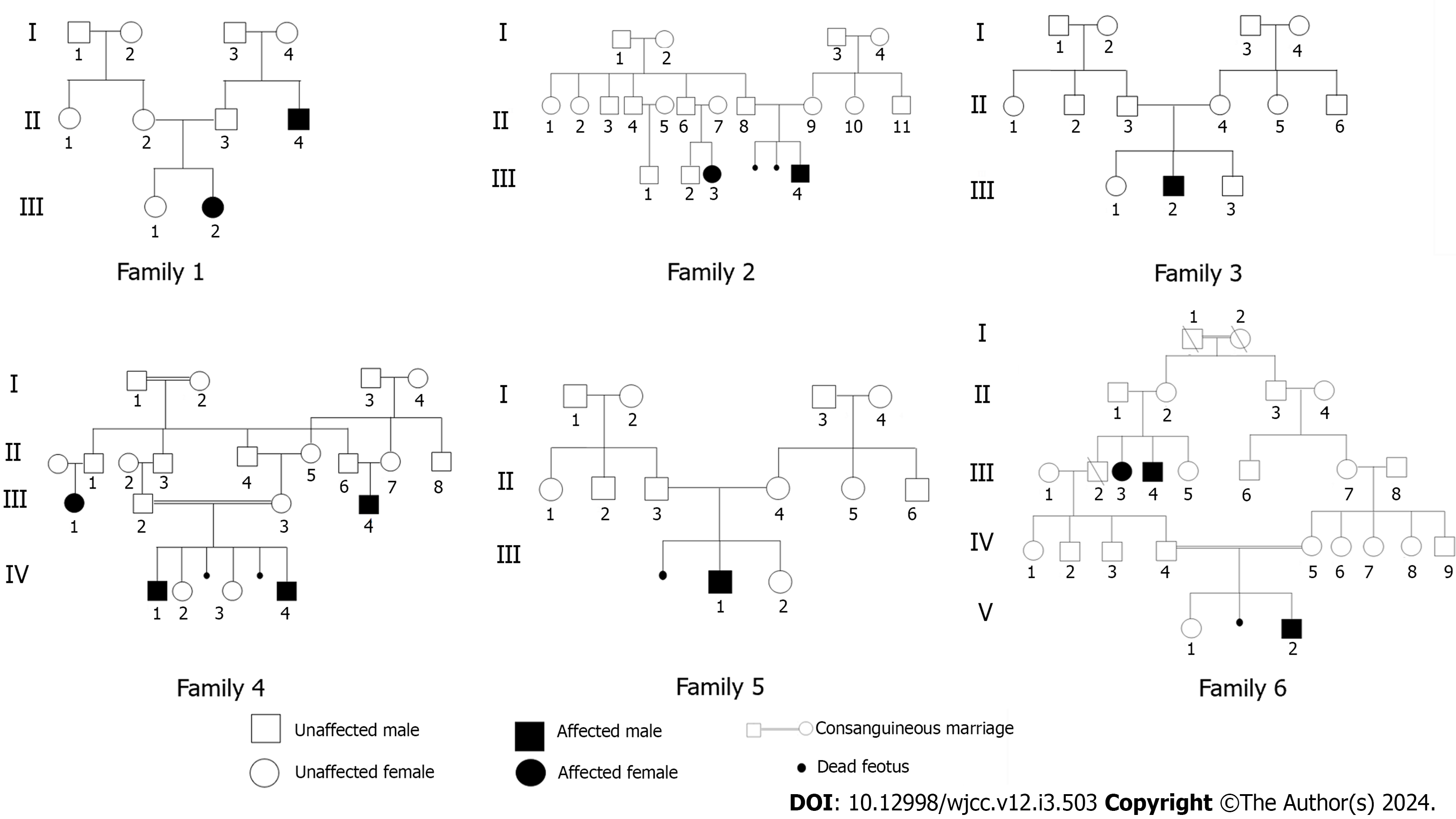
All our patients exhibited significant traits, including severe mental retardation, inability to speak, abnormal EEG findings, epileptic seizures and dysmorphic facial features (Table 1). The consanguinity was observed in four families. Additionally, we examined patients from six unrelated families, originating from different regions in Tunisia. To identify rare coding variants associated with the disease within these families, we performed linkage analysis in conjunction with exome sequencing in carefully selected consanguineous patients.
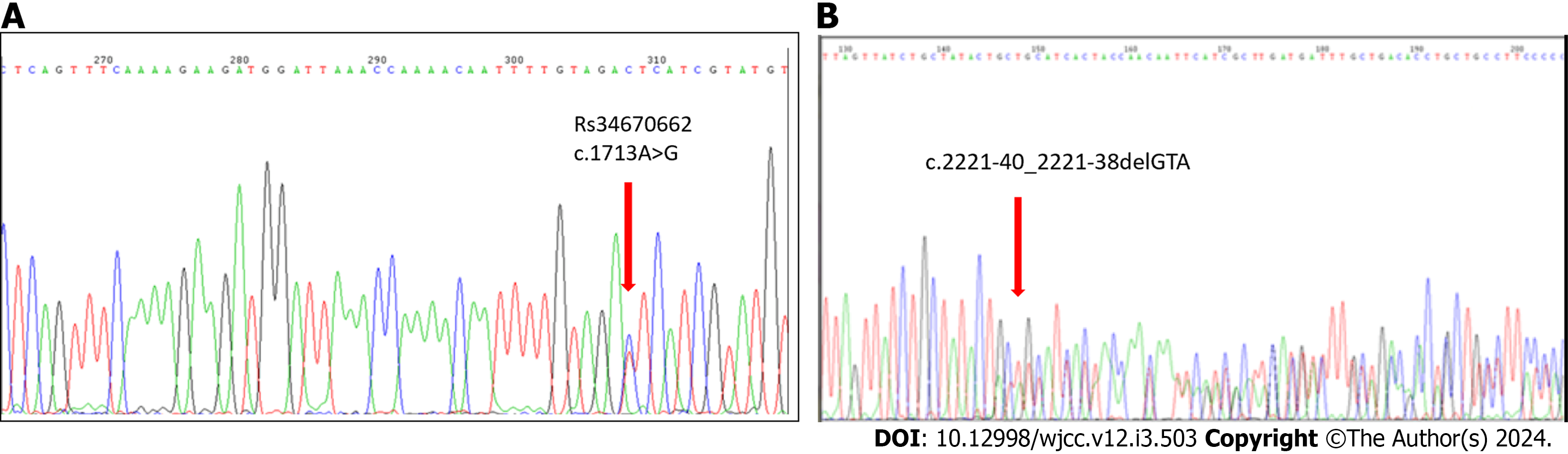
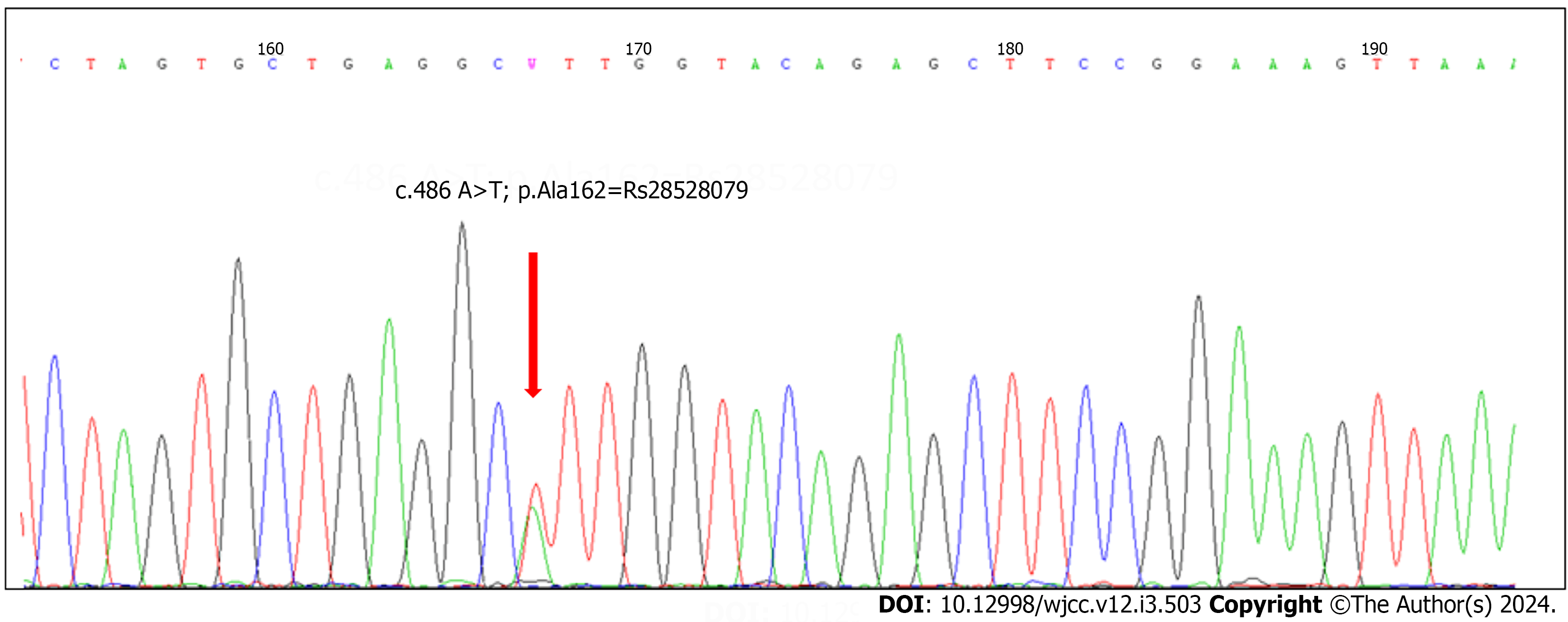
To investigate mutations in the coding regions of UBE3A, we examined 50 cases diagnosed with probable AS. Based on clinical observations, the patients displayed characteristics consistent with the AS phenotype. We ruled out deletion, UPD, and imprinting defects in these individuals (data not provided). After sequencing all ten coding exons of UBE3A, we identified seven polymorphisms. There were four previously known mutations and three novel mutations, but none of them was identified as causal (Table 3).
| Four common polymorphisms | New polymorphisms |
| c.2064+9T>C → rs79328837 (intro13) | c.2220+14T>C (intron14) |
| c.1713A>G → RS34670662 (exon 11) | c.30-47_30-46 insT (Exon7) |
| c.2221-40_2221-38delGTA, RS149854051 (intron 14) | c.2507+43T>A (Exon15) |
| c.486A>T; p.Ala162 =, RS28528079 (exon9A) |
The six suspected AS patients were from sporadic families, with sometimes one affected member, such as an uncle or cousin, suffering from a learning disability with an unknown syndrome (Figure 3).
The analysis conducted using NEXUS software did not identify any copy number variants associated with the disease. Shared genomic regions on chromosomes 4, 6, 10, and 11 (around 50 Mb) were detected using the parametric linkage analysis under an autosomal recessive model. Exome sequencing was employed to investigate variant discovery in all shared regions.
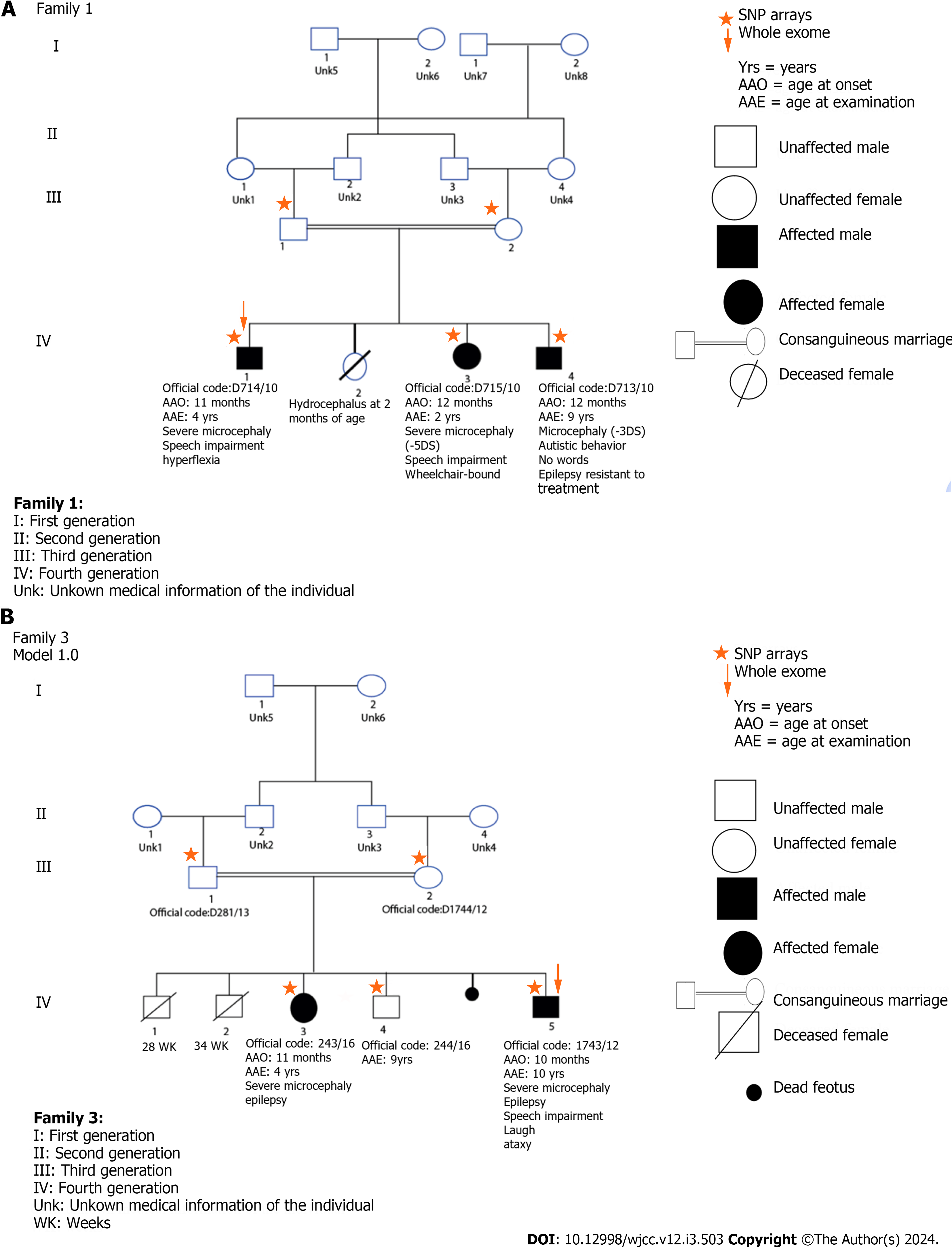
Exome sequencing: Exome sequencing was done for two separate families; every family had a consanguineous relationship with their relatives. The two families had more than one affected member (Figure 3).
The analysis of the exome sequencing data did not reveal any homozygous variants, but it did reveal the presence of compound heterozygous variants in six genes for the patient within the selected family (Figure 3, Family 1) (Table 4): SHPRH (c.4331C>T p.A1444V), SLC30A9 (c.528-7T>C), HBS1L (c.2043+5T>G), TAAR6 (c.865C>Tp.P289S), TAAR2 (c.467C>T p.T156I), SASH1 (c.1126C>T p.P376S), LOC100287896 (c.4C>T p.R2C), PCF11 (c.3355C>T p.H1119Y), ANKRD42 (c.676A>G p.N226D), PDGFD (c.7C>G p.R3G), and DIXDC1 (c.226G>A p.G76S). Due to the high polymorphism observed in the SLC30A9 and HBS1L genes, we did not consider these variants as plausible candidates in our analysis.
| Gene (gene) | SHPRH | SLC30A9 | HBS1L | TAAR6 | TAAR2 | SASH1 | LOC100287896 | PCF11 | ANKRD42 | PDGFD | DIXDC1 |
| Chromosome | 6 | 4 | 6 | 6 | 6 | 6 | 11 | 11 | 11 | 11 | 11 |
| Location (varLocation) | Exonic | Intronic | intronic | Exonic | Exonic | Exonic | Exonic | Exotic | Exonic | Exonic | Exonic |
| Effect (codingEffect) | Nonsynonymous | Nonsynonymous | Nonsynonymous | Nonsynonymous | Nonsynonymous | Nonsynonymous | Nonsynonymous | Nonsynonymous | Nonsynonymous | ||
| cDNA (cNomen) | c.4331C>T | c.528-7T>C | c.2043+5T>G | c.865C>T | c.467C>T | c.1126C>T | c.4C>T | c.3355C>T | c.676A>G | c.7C>G | c.226G>A |
| Protein (pNomen) | p.A1444V | p.P289S | p.T156I | p.P376S | p.R2C | p.H1119Y [Histidine (His)- Tyrosine (Tyr)] | p.N226D [Asparagine (Asn)- Aspartic Acid (Asp)] | p.R3G [Arginine (Arg)- Glycine (Gly)] | p.G76S [Glycine (Gly)- Serine (Ser)] | ||
| Pathogenicity | 09/11 | This mutation probably has no impact on splicing | Activation of an intronic cryptic donor site. | 07/11 | 10/11 | 09/11 | 08/11 | 09/11 | 03/Nov | 0/11 | |
| Potential alteration of splicing | |||||||||||
| GME Variome (%) | Not available | Not available | Not available | 0.1 | Not available | Not available | Not available | 0.1 | 0.1 | 0.1 | Not available |
| Gnomad browser (%) | 0.0008 | 0.04 | 0.2 | 0.006 | 0.001 | 0.001 | 0.4 | 0.2 | 0.3 | 0.01 | 0.002 |
The analysis of the exome sequencing for the patient in the second consanguineous family (Figure 3, Family 2) revealed the presence of the following variants: KMT5A [c.904T>Cp. (C302R)], KMT5A[c.995T>Cp. (L332)], PSTK36[c.2516G>Ap. (R839Q)], PIK3CB[c.2150A>Gp. (N717S)], GPR149[c.1404A>Cp. (R468S)], RARRES1[c.230C>Tp. (P77L)], KPNA4[c.1103A>Gp.(N368S)], NOS1[c.1922C>Tp. (A641V)], CAMKK2[c.1612_1614dupAAA p. (K538dup)], WDR66[c.196_197insAGAAAGAGGAGGAGG p. (E65G66insEKEEE)], and SBNO1(c.3220+5C>G).
AS is a severe neurodevelopmental disorder that affects 1 in 20000 children. There is no effective treatment for cognitive defects, and the treatment of seizures is often ineffective. Lack of insight into the fundamental mechanisms underlying AS hinders drug development for the condition. This syndrome is caused by the lack of a functional maternal UBE3A gene. This gene encodes a protein known as UBE3A or E6AP (E6-associated protein), which can modify other proteins through a process called ubiquitination. However, it is unclear which proteins are modified by UBE3A. We sought to identify these proteins, referred to as ‘targets’, which is a crucial stage in the development of treatments. In this study, we examined 50 patients from unrelated Tunisian families who presented with a profile consistent with AS phenotype.
The patients were referred to different pediatric departments, due to unidentified etiology of severe mental retardation, abnormal EEG findings or epileptic seizures, severe speech impairment and dysmorphic facial characteristics, and the genetic anomaly was confirmed for all of them.
Cytogenetic analysis demonstrated a 46, XY and 46, XX karyotype in all analyzed cells from the patients. The parent’s karyotypes were found to be normal as well. After that, microsatellite marker PCR analysis was conducted using the conventional methodology, employing polymorphic markers situated within the 15q11q13 region. The purpose was to authenticate the duplication and ascertain the parental source of the duplicated chromosome 15. All our patients were normal. We performed MS-MLPA for diagnosis of AS associated with deletions, UPD15, or rare duplications. After all these tests, the deletion, UPD, and imprinted defects were excluded in all 50 patients.
Molecular analysis by direct sequencing (exons 7 to 16 and flanking exon/intron boundaries) of the UBE3A gene performed on all patients revealed negative results.
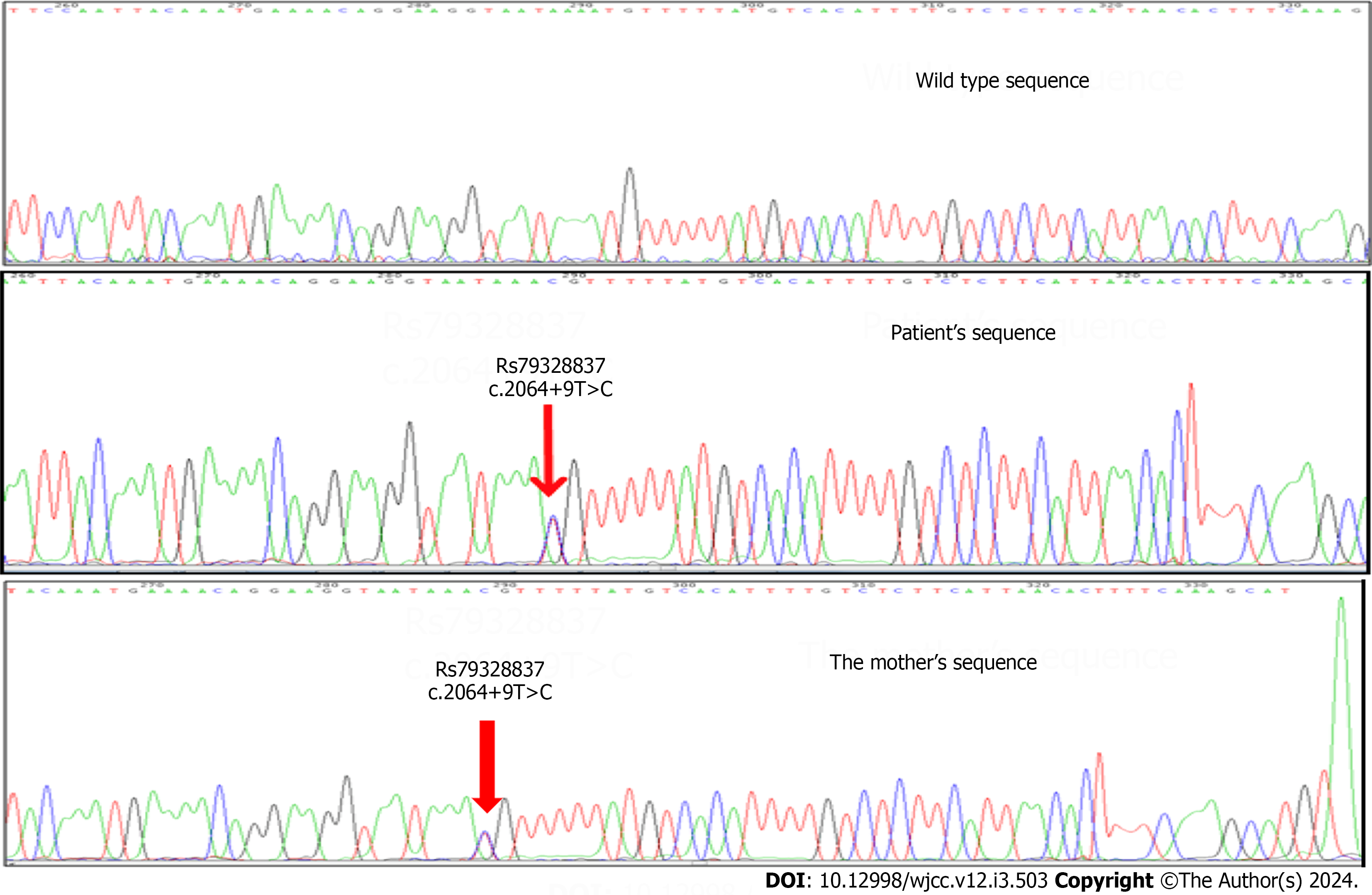
We found seven polymorphisms, three were novel, and four had been described in the literature. In Family 1 (Figure 2), we found a variation in intron 13 (c.2064+9T>C), which is known in the NCBI database as rs79328837 (Figure 4). This variant was described by Sadikovic et al[12] 2014. It manifests at a poorly conserved position in the protein. The variant is anticipated to be benign according to multiple in silico algorithms, and its population frequency is incongruent with the disease.
The second patient (Figure 2, Family 2) showed two known polymorphisms (Figure 5). The third patient (Figure 2, Family 3) showed a single nucleotide variation in exon 9 with an uncertain significant allele (Figure 6). The patient presented with severe microcephaly (-5 SD), aggressive behavior, and an abnormal EEG.
To our knowledge, there is limited information in the literature about the c.2220+14 T>C (intron 14), c.30-47_30-46 insT (Exon 7), and c.2507+43 T>A (Exon 15) variants. It is unclear whether these mutations directly caused AS or they were just non-synonymous polymorphisms with low effect.
The allele frequency of the c.2064+9T >C variant in UBE3A is 0.4% in gnomAD, a level deemed high enough to classify it as benign, according to thresholds set by the ClinGen Rett/Angelman-like Expert Panel for Rett/AS-like conditions. Splice prediction analysis, employing multiple computational biology tools does not indicate an impact on splicing. In summary, the c.2064+9T>C variant in UBE3A is classified as benign based on the American College of Medical Genetics and Genomics /Association for Molecular Pathology criteria.
The alteration c.1713A>G (RS34670662) is deemed benign through a comprehensive assessment, considering several factors. These include population frequency, absence of segregation with the disease, intact protein function, co-occurrence, analysis of RNA, in silico models, amino acid conservation, absence of disease association in case-control studies, incongruence with a known cause of pathogenicity in terms of the mechanism of disease or impacted region
According to the Global Variome shared Leiden Open Variation Database for UBE3A, the variant c.2221-40_2221-38delGTA (RS149854051) in intron 14 marks the initiation of clinical classification. For the polymorphism, c.486A > T; p.Ala162 [RS28528079 (exon9A)], the clinical significance was considered benign, as described by Nykamp et al[13].
Approximately 10% of patients clinically diagnosed with AS or AS-like conditions do not exhibit an identifiable molecular defect[14]. Some of these patients have genetic variations that exhibit overlapping features with AS. Hence, we need next-generation sequencing (NGS) as one of the fastest techniques to screen other genes responsible for the development of these disorders[15]. The integration of whole-exome and whole-genome sequencing, along with high-throughput genotyping and linkage analysis, may contribute to the identification of new genes linked to AS-like syndromes. Here, we aimed to use a genetic approach to identify putative UBE3A targets. We hypothesize that a mutation in a UBE3A-target protein may result in a syndrome resembling AS. However, it is likely that such a syndrome only arises when a mutated gene is inherited from both the father and mother.
The results of exome sequencing (Tables 4 and 5), revealed that patients in the first family (Figure 3, Family 1) present several gene mutations, such as TAAR6 (TRAR4), that are expressed in low abundance in various human brain tissues, especially frontal cortex, substantia nigra, amygdala, and hippocampus. The gene expression for the first family was detailed in Table 6. The exome sequencing of the second patient in family 2 (Figure 2) gave different genes. The gene expression for the second family was detailed in Table 6.
| Gene (gene) | KMT5A | KMT5A | STK36 | PIK3CB | GPR149 | RARRES1 | KPNA4 | NOS1 | CAMKK2 | WDR66 | SBNO1 |
| Chromosome | 12 | 12 | 2 | 3 | 3 | 3 | 3 | 12 | 12 | 12 | 12 |
| Location (varLocation) | Exonic | Exonic | Exonic | Exonic | Exonic | Exonic | Exonic | Exonic | Exonic | Exonic | Intronic |
| Effect (codingEffect) | Non-synonymous | Non-synonymous | Non-synonymous | Non-synonymous | Non-synonymous | Non-synonymous | Non-synonymous | Non-synonymous | Non-synonymous | Non-synonymous | Non-synonymous |
| cDNA (cNomen) | c.904>C | c.995>C | c.2516 A | c.2150>G | c.1404A>C | c.230C>T | c.1103>G | c.1922>T | c.1612_1614dupAAA | c.196_197insAGAAAGAGGAGGAGG | c.3220+5C>G |
| Protein (pNomen) | p.C302R | p.L332P | p.R839Q | p.N717S | p.R468S | p.P77L | p.N368S | p.A641V | p.K538dup | p.E65_G66insEKEEE | No significant splicing motif alteration detected. This mutation probably has no impact on splicing |
| Pathogenicity | 03/11 | 10/11 | 06/11 | 01/11 | 01/11 | 10/11 | 03/11 | 09/11 | 01/11 | 01/11 | Not available |
| GME Variome (%) | Not available | Not available | 0.05 | 0.9 | 0.1 | Not available | Not available | Not available | Not available | Not available | 0.01 |
| Gnomad browser (%) | Not available | Not available | 0.7 | 0.1 | 0.04 | 0.004 | 0.003 | Not available | 12 | Not available | 0.01 |
| Gene | Function |
| Gene expression in Family 1 | |
| TAAR2 | Relevance to brain function and behavior, including schizophrenia, depression, anxiety, and drug addiction |
| CITED2 | Gene is identified in human endothelial cells and neonatal brain. It is implicated in the development of the heart and neural tube |
| HPRH | Ubiquitously expressed protein that contains motifs characteristics of several DNA repair proteins, transcription factors, and helicases. A possible candidate for the tumor suppressor gene |
| SASH1 | Highly expressed in brain, heart, lung, ovary, and kidney. It is also identified as a candidate tumor suppressor gene in breast cancer |
| DIXDC1 | Highly expressed in the brain and in specific brain regions; important in embryonic cortical development |
| SLC30A9 | Shows ubiquitous expression in various human tissues, with high expression in the fetal brain, cerebellum, skeletal muscle, and kidney. Sub-cellular localization studies suggested that it is expressed in the vesicular cytosolic compartment, possibly in the endoplasmic reticulum |
| Gene expression in Family 2 | |
| KMT5A | Is a lysine methyl-transferase that predominantly mono-methylates lysine-20 (K20) of histone H4 |
| SETD8 | Influences transcriptional regulation, heterochromatin formation, genomic stability, cell cycle progression, and development |
| STK36 | Important for brain development |
| PIK3CB | Plays a role in systemic insulin-like growth factor (IGF1) regulation and human longevity |
| RARRES1 | Implicated in hyperproliferation, inflammatory skin diseases and, prostate cancer |
| GPR149 | Increases fertility in mice, and causes prostatic cancer |
| NOS1 | Important for the brain and peripheral nervous system |
| SBNO1 | Implicated in cognition and psychoses, essential roles in vertebrate brain development |
We identified several consanguineous families where multiple children present symptoms of AS, but no mutation was identified. In this project, we propose to use state-of-the-art genetic technologies to identify mutated genes. The identified genes in these families will be studied for their pathological effect on neural development in laboratory cell-culture experiments and in mouse models. Moreover, we will test if these genes are targets of UBE3A. By using this approach, we hope to generate fundamental insight into the neurobiology underlying AS and AS-like syndromes, which may lead to the development of therapeutic interventions. The 50 patients studied had been previously found negative for 15q11q13 deletions, paternal UPD, and in imprinting defects.
Their clinical characteristics with suspected AS are presented in Table 1. All patients demonstrated severe speech impairments or a total absence of speech, behavioral abnormalities, movement difficulties and severe developmental delays.
Thirty-eight of them (76%) had the capacity to sit unsupported, 21 (42%) could walk with support, and 29 were unable to walk. Forty of them (80%) couldn’t speak, while 10 (20%) could speak a few meaningful words. Dysmorphic facial characteristics like protruding tongue, occipital groove, prognathism and wide-spaced teeth were most common in our patients (Table 1). All patients had an ataxic gait. Moreover, all patients were on antiepileptic therapy because of the presence of seizures. Epileptic attacks were completely controlled in five patients, and partially in one.
The clinicians performed to all patients the cranial magnetic resonance imaging. Only two patients showed minimal cerebral atrophy, and eight were normal. Metabolic screening tests showed normal results for every patient.
The genetic etiologies of AS are unknown in many populations[16]. However, implementing a NGS based approach will reduce cost reduction, increased efficiency, save time, and facilitate the exploration of novel pathways that contribute to the pathophysiology of AS.
The introduction of NGS technology, which enables the comprehensive reading of all protein-coding nucleotides in the genome (the exome), or the entire genome, has brought about a revolution in the field of human molecular genetics. When combined with family-based linkage analysis, this technological advancement has played a pivotal role in identifying numerous novel variants responsible for rare Mendelian forms of human diseases. He present study describes genetic analyses in three families suspected to have AS with a high incidence of intellectual disability and genetic disorder. In our study, we conducted linkage analysis alongside exome sequencing in specific patients to pinpoint rare coding variants that segregate with the disease within the family.
AS still lacks a cure. It is possible to generate pluripotent stem cells (iPSCs) line derived from skin fibroblasts of AS patients. These iPSC models for genomic imprinting disorders will enable the exploration of AS processes. Moreover, they provide an opportunity to explore the developmental mechanism and timing of UBE3A repression in human neurons[17,18].
The rapid advancement of iPSC technology has turned these cells into versatile tools for both basic and clinical research. Several studies have already developed this method, which may be very interesting to investigate for our patients.
Subsequent research on iPSC holds the promise of advancing drug discovery, enhancing cell therapy, and introducing novel diagnostic approaches for neurogenetic disorders.
The most important goal in our study is to investigate all the genes that may be responsible for the Angelman-like syndrome, since all the samples present the clinical features of Angelman syndrome (AS) without any genetic ab
Exome sequencing of patients suspect to have AS, showed the presence of different genes that may be responsible for the disease clinicians that may have patients suspect to have AS, cannot look into ubiquitin-protein ligase E3A (UBE3A) gene but also to other genes already described and that may be responsible for AS.
The student of AS cohort patients is to our knowledge the first research study in the Tunisian patients, this study may help physicians to know how to diagnose the patients in case of the absence of all genetic alterations for AS and to look further for the Angelman-like syndromes. It help also to do functional studies that may be interesting for further treatment in the future.
Patient with a strong suspicion of AS were assigned from pediatric departments and referred to Farhat Hached University Hospital. The chromosomal aberrations were assessed using constitutional investigations (karyotype, fluorescence in situ hybridization). The UBE3A gene was screened for mutation detection using Sanger method sequencing. The exome investigation was established using Illumina Hi-Seq 2000 sequencer. The exome data were analyzed using Genome Analysis Toolkit and Cartagenia software.
Sanger sequencing revealed several variants from which 3 novel ones not previously described. An interesting insertion involving exon 7 c.30-47_30-46 could be pathogenic and should be investigated further trough functional studies. The 22 potential genes displayed trough the exome sequencing brought to light new candidate genes to be investigated further for both AS and AS-Like syndromes.
The physicians, geneticists and researchers have to investigate very carefully the suspected AS patients. in case of all the molecular and cytogenetics tests were negative for AS, they must go further with exome sequencing and think more about AS-like syndromes that may be responsible for the disease in the patients.
However, additional evidence is required to clarify the developmental mechanism and timing of UBE3A repression in human neurons using cellular modeling by generating pluripotent stem cells (iPSCs) line derived from skin fibroblasts of AS patients. Subsequent research on iPSC holds the promise of advancing drug discovery, enhancing cell therapy, and introducing novel diagnostic approaches for neurogenetic disorders.
We thank the members of the genetics laboratory at Farhat Hached Hospital in Sousse, Tunisia, as well as the patients and their families.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: Tunisia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Upadhya D S-Editor: Qu XL L-Editor: A P-Editor: Cai YX
| 1. | Clayton-Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet. 2003;40:87-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 377] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 2. | Kishino T, Wagstaff J. Genomic organization of the UBE3A/E6-AP gene and related pseudogenes. Genomics. 1998;47:101-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 3. | Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 616] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 4. | Nicholls RD, Saitoh S, Horsthemke B. Imprinting in Prader–Willis and Angelman syndromes. Trends Genet. 1998;14:194-200. [RCA] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 269] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 5. | Williams CA. Neurological aspects of the Angelman syndrome. Brain Dev. 2005;27:88-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 109] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 6. | Russo S, Cogliati F, Viri M, Cavalleri F, Selicorni A, Turolla L, Belli S, Romeo A, Larizza L. Novel mutations of ubiquitin protein ligase 3A gene in Italian patients with Angelman syndrome. Hum Mutat. 2000;15:387. [PubMed] [DOI] [Full Text] |
| 7. | Liljelund P, Handforth A, Homanics GE, Olsen RW. GABAA receptor beta3 subunit gene-deficient heterozygous mice show parent-of-origin and gender-related differences in beta3 subunit levels, EEG, and behavior. Brain Res Dev Brain Res. 2005;157:150-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Malzac P, Webber H, Moncla A, Graham JM, Kukolich M, Williams C, Pagon RA, Ramsdell LA, Kishino T, Wagstaff J. Mutation analysis of UBE3A in Angelman syndrome patients. Am J Hum Genet. 1998;62:1353-1360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 95] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Fang P, Lev-Lehman E, Tsai TF, Matsuura T, Benton CS, Sutcliffe JS, Christian SL, Kubota T, Halley DJ, Meijers-Heijboer H, Langlois S, Graham JM Jr, Beuten J, Willems PJ, Ledbetter DH, Beaudet AL. The spectrum of mutations in UBE3A causing Angelman syndrome. Hum Mol Genet. 1999;8:129-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 102] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 10. | Tsai TF, Raas-Rothschild A, Ben-Neriah Z, Beaudet AL. Prenatal diagnosis and carrier detection for a point mutation in UBE3A causing Angelman syndrome. Am J Hum Genet. 1998;63:1561-1563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 11. | Moncla A, Malzac P, Livet MO, Voelckel MA, Mancini J, Delaroziere JC, Philip N, Mattei JF. Angelman syndrome resulting from UBE3A mutations in 14 patients from eight families: clinical manifestations and genetic counselling. J Med Genet. 1999;36:554-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 12. | Sadikovic B, Fernandes P, Zhang VW, Ward PA, Miloslavskaya I, Rhead W, Rosenbaum R, Gin R, Roa B, Fang P. Mutation Update for UBE3A variants in Angelman syndrome. Hum Mutat. 2014;35:1407-1417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 13. | Nykamp K, Anderson M, Powers M, Garcia J, Herrera B, Ho YY, Kobayashi Y, Patil N, Thusberg J, Westbrook M; Invitae Clinical Genomics Group, Topper S. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19:1105-1117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 542] [Article Influence: 67.8] [Reference Citation Analysis (0)] |
| 14. | Tan WH, Bird LM, Thibert RL, Williams CA. If not Angelman, what is it? A review of Angelman-like syndromes. Am J Med Genet A. 2014;164A:975-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 15. | Aguilera C, Gabau E, Laurie S, Baena N, Derdak S, Capdevila N, Ramirez A, Delgadillo V, García-Catalan MJ, Brun C, Guitart M, Ruiz A. Identification of a de novo splicing variant in the Coffin-Siris gene, SMARCE1, in a patient with Angelman-like syndrome. Mol Genet Genomic Med. 2019;7:e00511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Williams CA, Beaudet AL, Clayton-Smith J, Knoll JH, Kyllerman M, Laan LA, Magenis RE, Moncla A, Schinzel AA, Summers JA, Wagstaff J. Angelman syndrome 2005: updated consensus for diagnostic criteria. Am J Med Genet A. 2006;140:413-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 399] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 17. | Wang H, Doering LC. Induced pluripotent stem cells to model and treat neurogenetic disorders. Neural Plast. 2012;2012:346053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Sabitha KR, Shetty AK, Upadhya D. Patient-derived iPSC modeling of rare neurodevelopmental disorders: Molecular pathophysiology and prospective therapies. Neurosci Biobehav Rev. 2021;121:201-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |