Published online Jan 16, 2024. doi: 10.12998/wjcc.v12.i2.383
Peer-review started: September 19, 2023
First decision: November 20, 2023
Revised: December 1, 2023
Accepted: December 22, 2023
Article in press: December 22, 2023
Published online: January 16, 2024
Processing time: 114 Days and 4.9 Hours
The SETD1B gene is instrumental in human intelligence and nerve development. Mutations in the SETD1B gene have been linked in recent studies to neurodevelopmental disorders, seizures, and language delay.
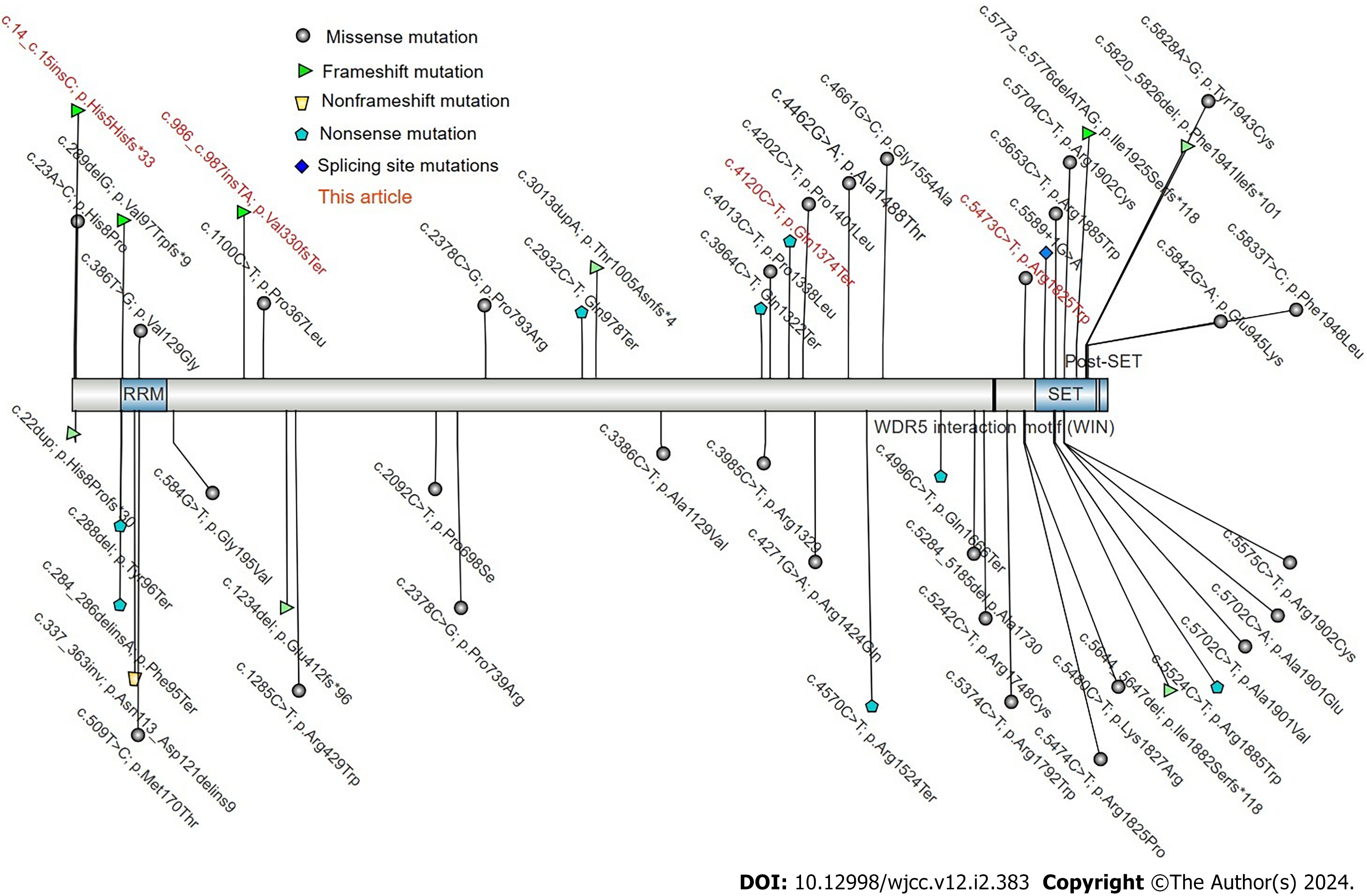
This study aimed to analyze the clinical manifestations and treatment of three patients suffering from mental retardation, epilepsy, and language delay resulting from a new mutation in the SETD1B gene. Three individuals with these symptoms were selected, and their clinical symptoms, gene test results, and treatment were analyzed. This article discusses the impact of the SETD1B gene mutation on patients and outlines the treatment approach. Among the three patients (two females and one male, aged 8, 4, and 1, respectively), all exhibited psychomotor retardation, attention deficit, and hyperactivity disorder, and two had epilepsy. Antiepileptic treatment with sodium tripolyvalproate halted the seizures in the affected child, although mental development remained somewhat delayed. Whole exome sequencing revealed new mutations in the SETD1B gene for all patients, specifically with c.5473C>T (p.Arg1825trp), c.4120C>T (p.Gln1374*, 593), c.14_15insC (p.His5Hisfs*33).
Possessing the SETD1B gene mutation may cause mental retardation accom
Core Tip: This study identified three novel mutations in the SETD1B gene: c.5473C>T (p.Arg1825trp), c.4120C>T (p.Gln1374*, 593), c.14_15insC (p.His5Hisfs*33). The findings suggest that these mutations may result in overall developmental delays and intellectual disabilities, thereby broadening the known phenotypic spectrum of the SETD1B gene. Additionally, the study highlighted the potential pathological connection between SETD1B abnormalities and neurodevelopmental retardation associated with epilepsy.
- Citation: Ding L, Wei LW, Li TS, Chen J. Mental retardation, seizures and language delay caused by new SETD1B mutations: Three case reports. World J Clin Cases 2024; 12(2): 383-391
- URL: https://www.wjgnet.com/2307-8960/full/v12/i2/383.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i2.383
Mental retardation with seizures and language delay (IDDSELD) (OMIM: 619000) typically manifests within the first few years of a child’s life. This genetic disease, inherited in an autosomal dominant (AD) manner, is marked by general developmental delay with accompanying speech and language impairments. Several affected individuals also display behavioral anomalies, including the most common ones, like autism spectrum disorder and anxiety. Other observable characteristics could include facial abnormalities, slender fingers, and changes in skin pigmentation[1]. Scientific studies have shown that the rare variations in the coding region of SETD1B, along with deletions of large SETD1B fragments caused by copy number variations, are connected to neurodevelopmental retardation. Specifically, IDDSELD results from a heterozygous mutation of the SETD1B gene[2,3]. Patients with a microdeletion in SETD1B exhibit similar phenotypes-related symptoms, such as seizures, mental retardation, language difficulties, and bradykinesia[2-5].
Case 1: An 8-year-old girl was brought to the Department of Neurology at the Children’s Hospital affiliated with Nanjing Medical University due to “repeated episodes of confusion” (Table 1).
| Child No. | Child 1 | Child 2 | Child 3 |
| Age | Eight years old | Four years and eight months | one year and one month |
| Gender | Female | Male | Female |
| Gene | SETD1B | SETD1B | SETD1B |
| Nationality, ethnic group | Chinese, Han | Chinese, Han | Chinese, Han |
| Exon | exon 14 | exon 11 | exon 1 |
| Parental consanguinity | No | No | No |
| Nucleotide change | c.5473C>T | c.4120C>T | c.14_15insC |
| Amino acid change | p.Arg1825Trp | p.Gln1374*,593 | p.His5Hisfs*33 |
| Parental consanguinity | No | No | No |
| Main clinical phenotypes | Seizure, EEG abnormality, attention deficit hyperactivity disorder, delayed ability to walk, epileptic encephalopathy | Impaired social interactions, delayed speech and language development, global developmental delay, motor delay, dyskinesia | Seizure, motor delay, myoclonus, fair hair, EEG abnormality, abnormal dermatoglyphics, delayed ability to walk |
Case 2: A boy aged four years and eight months presented to our department because of delays in their nervous system development (Table 1).
Case 3: A girl aged one year and one month demonstrated motor retardation, seizure, but appeared to have normal intelligence (Table 1).
Case 1: Currently, the mental development of child 1 is slightly lagging, her progress is slow, and she is willing to engage with others, but her concentration is inconsistent, and she displays little emotion. Her motor development is normal. The DSM-IV hyperactivity diagnostic scale scored 3 points for hyperactivity and 9 points for attention deficit, with an IQ of 52 points (Table 1).
Case 2: Child 2 could only utter single words and occasionally sought help but were reluctant to engage with others (Table 1).
Case 3: Child 3 had a history of seizures with upward rolling of the eyes, occurring 0-10 times per day. At 10 mo old, the child was able to sit up, and at 1 year old, the child started to crawl. Currently, the child can walk with support (Table 1).
Case 1: Child 1 was diagnosed with epilepsy, characterized by typical absence seizures, and was started on sodium valproate (VPA) antiepileptic therapy on May 22, 2017. However, the seizures continued six months into the treatment. Upon the addition of lamotrigine (LTG), seizures persisted during the drug titration phase but ceased after three months.
Case 2: Child 2 started walking independently at the age of two. He was capable of expressing his own needs and recognizing basic facial features.
Case 3: Child 3 had a history of seizure.
Case 1: The personal history of child 1 includes being the first child born full-term through an elective cesarean section G1P1 without history of labor complications or hypoxia and no family history of genetic or similar disorders. Her last recorded visit was on May 12, 2022. The parents were not blood relatives.
Case 2: Child 2 had a two-year-old brother who only occasionally speaks in single words. The parents had no family history, and were not blood relatives.
Case 3: The parents of child 3 had no family history, and were not blood relatives.
Case 1: Physical examination of child 1 showed a weight of 32 kg, a height of 112 cm, and a blood pressure of 95/60 mmHg. She was conscious and mentally responsive, with no abnormalities detected in the heart, lungs, or abdomen. Limb muscle tension and strength were normal.
Case 2: Child 2 exhibited unusual hand movements and a tendency to talk to themselves. His eye contact was fair, but he was non-responsive to questions.
Case 3: Child 3 was conscious, with a stable breathing pattern, soft neck, normal muscle strength and muscle tone in her limbs. Her hair was fair, and her skin striations on both lower limbs are asymmetric.
Case 1: The nervous system examination of child 1 did not find any exceptions. Her auxiliary examinations, including tests for urine, blood, stool, liver and kidney function, and electrolytes, revealed no abnormalities.
Case 2: Child 2 slept well and had normal urinary and bowel functions.
Case 3: The laboratory test results of child 3 were all normal.
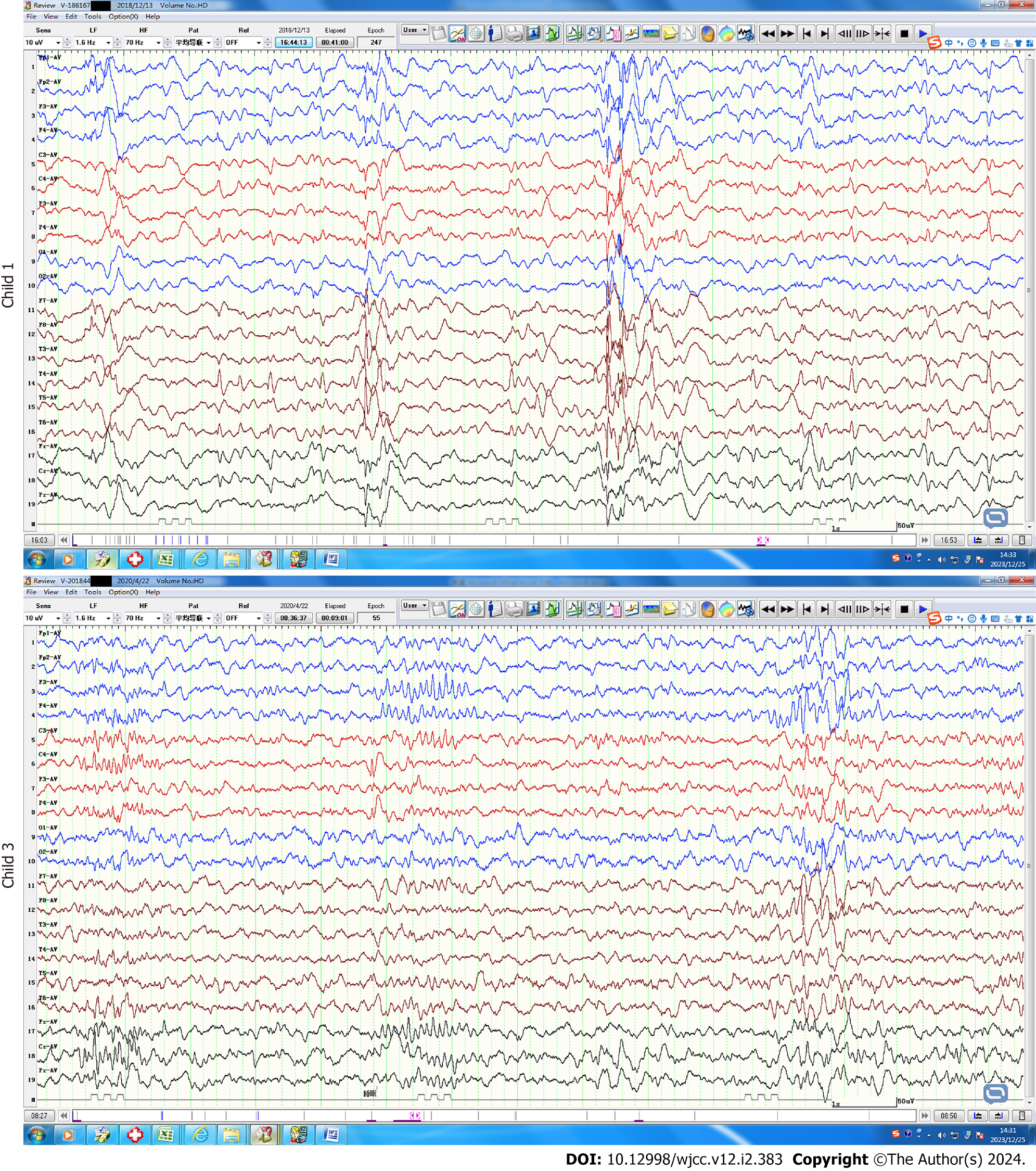
Case 1: The electroencephalogram examination of child 1 revealed abnormalities, including widespread 3 Hz spikes and slow waves, and a typical absence seizure was detected. Five video-electroencephalogram examinations during the course of her illness again showed widespread 3 Hz spike and slow waves, with a pointed slow wave distribution in the right Rolandic region (Figure 1).
Case 2: The brain magnetic resonance imaging results of child 2 showed no significant abnormalities.
Case 3: During video electroencephalogram examinations of child 3, epileptic waves were detected and myoclonic seizures were observed more than ten times (Figure 1). She was susceptible to convulsions, sometimes accompanied by a bilateral upward turn of the eyes.
DNA was extracted from the peripheral blood of the patients and their parents. DNA was submitted to Chigene (Beijing) Translational Medical Research Center Co. Ltd for trio (including the proband and both parents) whole-exome sequencing. Protein-coding exome enrichment was performed using xGen Exome Research Panel v2.0 (IDT, Iowa, United States), which consists of 429826 individually synthesized and quality-controlled probes targeting 39 Mb protein-coding region (19396 genes) of the human genome, covering 51 Mb end-to-end tiled probe space. High-throughput sequencing was performed by MGISEQ-T7 series sequencer, and not less than 99% of target sequence were sequenced. The sequencing process was performed by Chigene (Beijing) Translational Medical Research Center Co. Ltd.
The Fastp software was used to perform quality control on the raw data, removing adapter sequences and low-quality reads. The Burrows-Wheeler Aligner software was used to align the sequencing sequence with the Ensemble reference genome GRCh37/hg19. Then, the GATK software was used to analyze the single nucleotide polymorphisms (SNPs) and indels. Finally, the detected SNPs and indels were filtered and screened based on sequencing depth and mutation quality to obtain high-quality and reliable mutations. In this filtering strategy, a single whole exome sequencing (WES) sample contains approximately 13.5 G-14 G of raw data, which is then filtered to produce approximately 12 G-12.5 G of clean data. The self-developed variant annotation software of Chigene company was used to annotate high-quality variants detected with major databases (such as dbSNP, 1000 Genomes, gnomAD, OMIM, HGMD, ClinVar, etc). Provean, SIFT, Revel and other software were used for protein hazard prediction, while MaxEntScan, spliceAI and other software were used for splicing impact prediction. Variants that may have harmful effects on protein structure were ultimately screened out.
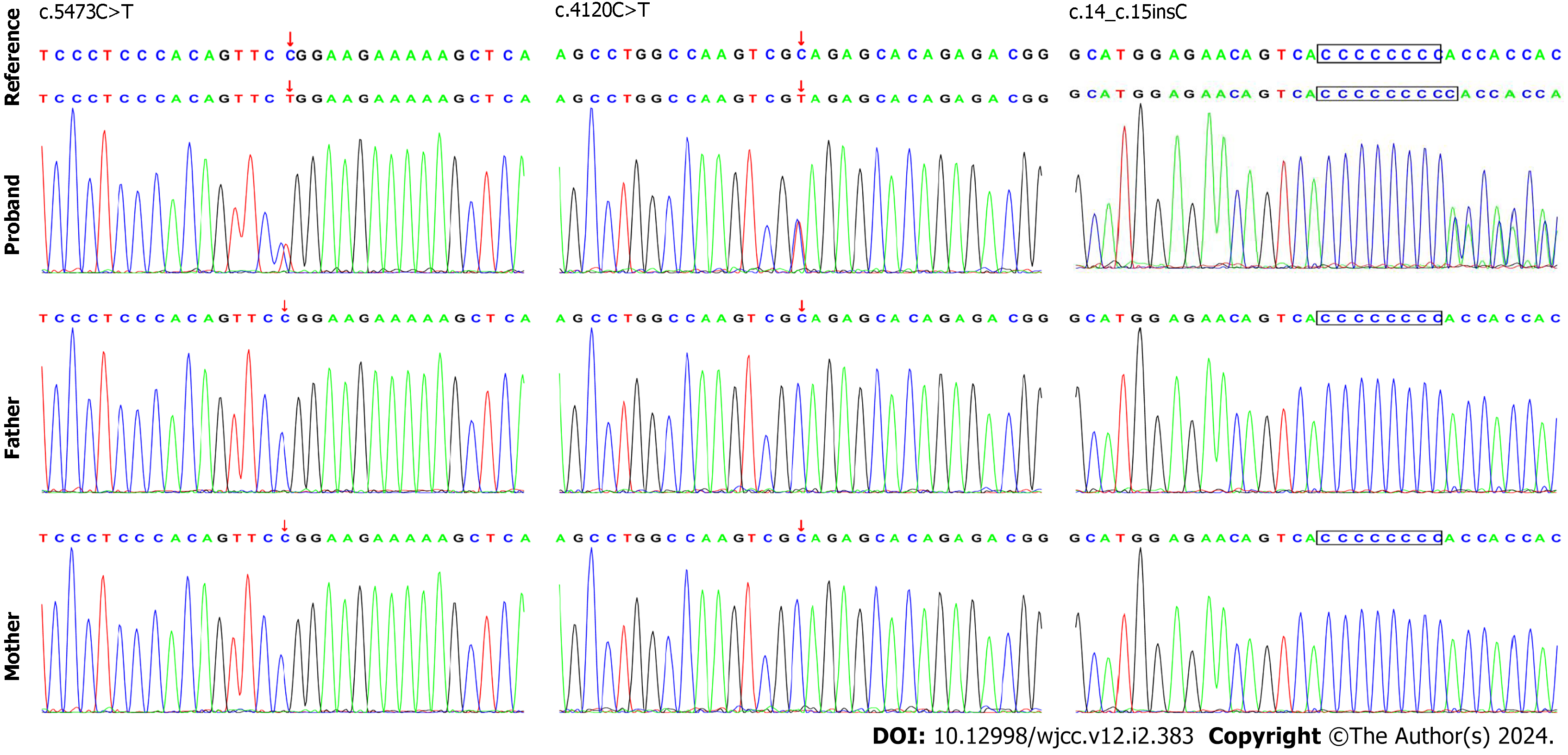
Following the signing of informed consent by the children’s family members, 3 mL of peripheral venous anticoagulant was extracted from each child and their parents and sent to the Beijing Full Spectrum Medical Laboratory for WES. The results revealed that the first child had a heterozygous variant c.5473C>T in the SETD1B gene, leading to the conversion of arginine to tryptophan at position 1825 (p.Arg1825Trp). The second child possessed the heterozygous variant c.4120C>T, resulting in early termination at the 1374th amino acid (p.Gln1374*,593). The third child carried a heterozygous variant c.14_15insC, leading to premature termination at the fifth amino acid position (p.His5Hisfs*33).
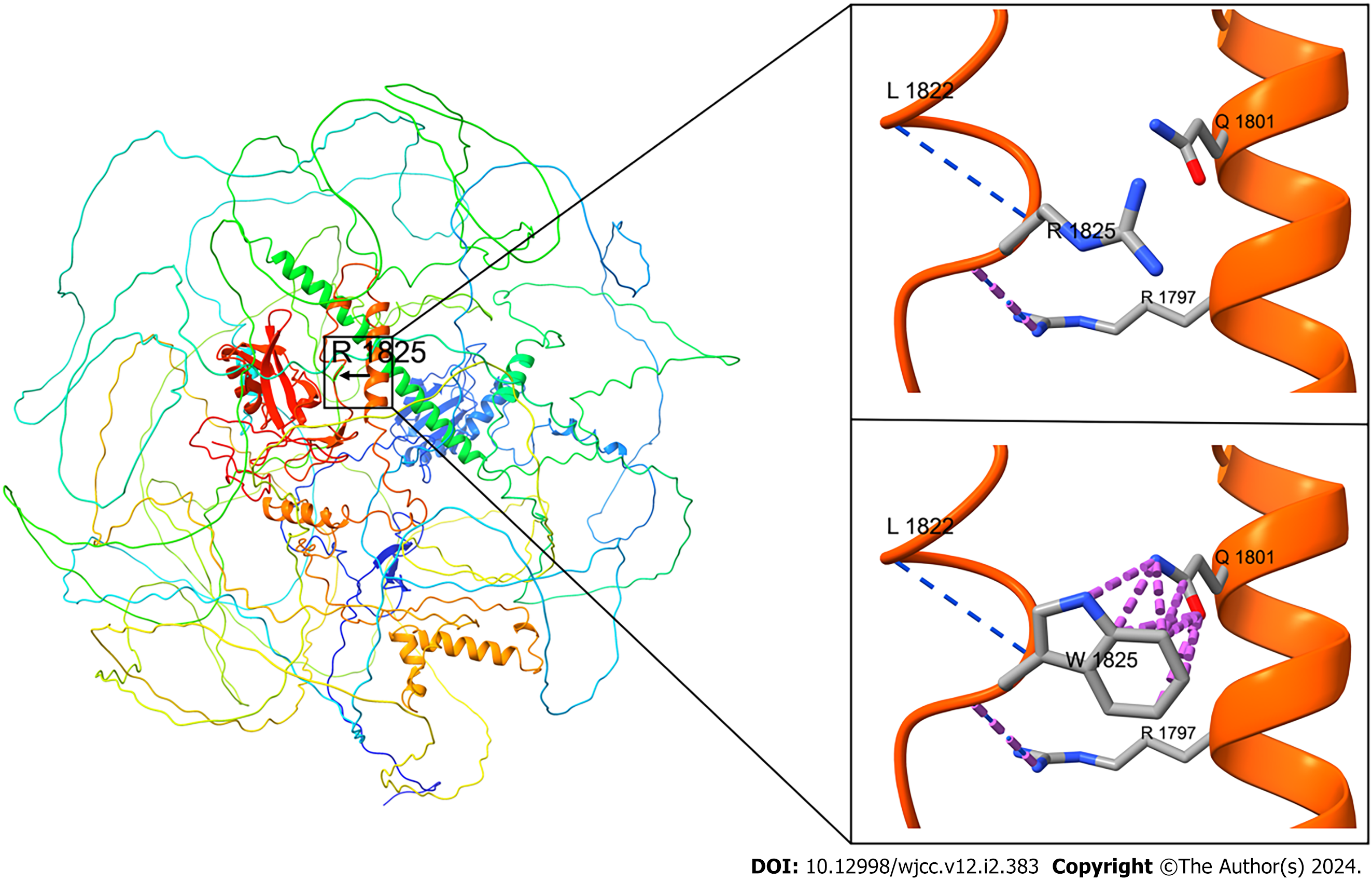
The WES findings were confirmed by Sanger sequencing (Figure 2) and simulated using protein structure software (Figure 3). These mutations were not found in the children’s parents, indicating that they were new heterozygous mutations, consistent with the pathogenesis of AD diseases. The phenotype and genotype of the proband and family members aligned with co-segregation.
None of the three mutation sites had been reported in the existing literature, nor were they included in the ClinVar and HGMD databases. The allele frequencies of the three variants detected were not recorded in dbSNP, 1000 Genomes and gnomAD databases. According to the clinical characteristics of the study subjects and the analysis of the WES test results, the disease-causing genes that can lead to similar clinical phenotypes have not mutated, thus excluding the possibility of other diseases. The parental consanguinity was not present in these cases.
The three sites were assessed according to the American Society for Medical Genetics and Genomics guidelines (201) (Figure 4). The ratings were: c.5473C>T as causative (PS2+PM1+PM2+PP2+PP3); c.4120C>T (PVS1+PM2+PM6); c.14_15insC (PVS1+PS2). All three locations were of pathogenicity grade, in line with the rules for AD inheritance with gene mutation. In conjunction with the clinical diagnosis, mental retardation with epilepsy and language delay were identified as a result of the SETD1B mutation.
The WES results showed that three patients had pathogenic mutations in the SETD1B gene and were diagnosed with intellectual developmental disorder with seizures and language delay (OMIM: 619000).
Child 1 is currently being treated with oral LTG 50 mg, bid + VPA 5 mL, bid.
Treatment information for child 2 is not available.
Child 3 is undergoing medication treatment. She takes oral sodium VPA oral solution (Depakine) twice a day, with a dosage of 6 mL each time; simultaneously, she takes oral topiramate tablets (Topamax) twice a day, with a dosage of 37.5 mg each time.
The symptoms of child 1 are well controlled and there have been no seizures.
The information on the outcome and follow-up of child 2 is not obtained.
Child 3 has good epilepsy control and has not had seizures for 2 years.
The SETD1B gene is found in the region of 12q24.31 and includes 18 exons. It encodes the SET1B protein, part of Histone methyltransferases, and features a C-terminal SET domain with an N-SET domain and a post-SET peptide on either side[2,6]. At the N-terminal, there is a tandem RNA recognition motif, which facilitates the trimethylation (H3K4me3) of the lysine site of the third histone subunit. H3K4me3 functions as a histone marker to identify gene promoters and is involved in the epigenetic regulation of gene transcription[6-8].
Methylation modification of H3K4me3 is generally linked to gene transcription activation[9]. During DNA damage, H3K4me3 at the damaged site becomes demethylated[10,11] to enhance DNA repair. Additionally, the mutation c.5473C>T (p.Arg1825Trp) of the SETD1B gene, in this case, is situated in the low complexity domain (LCD) of the protein, and LCD is a sequence enriched in particular amino acids, crucial for the function of transcription factors. Aberrant accumulation of low-complexity sequences is highly correlated with degenerative diseases[12].
Currently, the HGMD database contains only 20 SETD1B gene mutations, comprising 12 missense mutations, 1 splice site mutation, 3 frameshift mutations, 2 large fragment deletions, and 2 nonsense mutations (Figure 4).
Children 1 and 3 were diagnosed with typical absence seizures of epilepsy, displaying mental and motor development delays, slow progress, a lack of focus, and numerous minor movements, which aligns with descriptions found in the literature. Child 2 on the other hand, only exhibited generalized developmental delay, delayed motor development, and delayed speech and language development, with no seizures observed so far.
The pathogenic variation of SETD1B leads to a conservative phenotype encompassing intellectual disability and childhood-onset, treatment-resistant epileptic seizures[1], a finding that matches the phenotype of our patients. However, some individuals with this variation have also presented facial and ocular phenotypic abnormalities[13] in our group of patients. It is important to recognize that in certain cases documented in the literature, even without epilepsy, variations in SETD1B can have a severe impact on neurodevelopment, suggesting that SETD1B is associated with developmental encephalopathy[14,15].
In the case of an AD pathogenic abnormality in the SETD1B gene, Weerts MJA documents an instance of transmission from an afflicted mother to her affected child. Thus, upon identification of the causative variant of SETD1B in family members showing symptoms, prenatal genetic testing becomes essential. The phenotypic traits linked to SETD1B-related neurodevelopmental abnormalities are not distinct, so the differential diagnosis should consider all scenarios involving intellectual developmental disorders without any other apparent symptoms. Genetic testing stands as the most effective approach in these cases.
Currently, there is no specific treatment for epilepsy connected to SETD1B, and all available methods are symptomatic treatments aimed at controlling epileptic seizures. In the case of child 1, seizures were effectively managed using a combination of sodium VPA and LTG. Over time, some patients may have an increased risk of cancer and other complex diseases. As a result, it is necessary to establish efficient follow-up care, and more findings from longitudinal studies are anticipated.
In conclusion, this study identified three novel mutations in the SETD1B gene: c.5473C>T (p.Arg1825trp), c.4120C>T (p.Gln1374*, 593), c.14_15insC (p.His5Hisfs*33). The findings suggest that these mutations may result in overall developmental delays and intellectual disabilities, thereby broadening the known phenotypic spectrum of the SETD1B gene. Additionally, the study highlighted the potential pathological connection between SETD1B abnormalities and neurodevelopmental retardation associated with epilepsy.
We are grateful to the patients and their families for participation in the study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Malik S, Pakistan S-Editor: Wang JJ L-Editor: A P-Editor: Li X
| 1. | Roston A, Evans D, Gill H, McKinnon M, Isidor B, Cogné B, Mwenifumbo J, van Karnebeek C, An J, Jones SJM, Farrer M, Demos M, Connolly M, Gibson WT; CAUSES Study; EPGEN Study. SETD1B-associated neurodevelopmental disorder. J Med Genet. 2021;58:196-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 2. | Labonne JD, Lee KH, Iwase S, Kong IK, Diamond MP, Layman LC, Kim CH, Kim HG. An atypical 12q24.31 microdeletion implicates six genes including a histone demethylase KDM2B and a histone methyltransferase SETD1B in syndromic intellectual disability. Hum Genet. 2016;135:757-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 3. | Palumbo O, Palumbo P, Delvecchio M, Palladino T, Stallone R, Crisetti M, Zelante L, Carella M. Microdeletion of 12q24.31: report of a girl with intellectual disability, stereotypies, seizures and facial dysmorphisms. Am J Med Genet A. 2015;167A:438-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Chouery E, Choucair N, Abou Ghoch J, El Sabbagh S, Corbani S, Mégarbané A. Report on a patient with a 12q24.31 microdeletion inherited from an insulin-dependent diabetes mellitus father. Mol Syndromol. 2013;4:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | Krzyzewska IM, Maas SM, Henneman P, Lip KVD, Venema A, Baranano K, Chassevent A, Aref-Eshghi E, van Essen AJ, Fukuda T, Ikeda H, Jacquemont M, Kim HG, Labalme A, Lewis SME, Lesca G, Madrigal I, Mahida S, Matsumoto N, Rabionet R, Rajcan-Separovic E, Qiao Y, Sadikovic B, Saitsu H, Sweetser DA, Alders M, Mannens MMAM. A genome-wide DNA methylation signature for SETD1B-related syndrome. Clin Epigenetics. 2019;11:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 6. | Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282:13419-13428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 200] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 7. | Hiraide T, Hattori A, Ieda D, Hori I, Saitoh S, Nakashima M, Saitsu H. De novo variants in SETD1B cause intellectual disability, autism spectrum disorder, and epilepsy with myoclonic absences. Epilepsia Open. 2019;4:476-481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 8. | Shinsky SA, Monteith KE, Viggiano S, Cosgrove MS. Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J Biol Chem. 2015;290:6361-6375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 9. | Xu R, Li C, Liu X, Gao S. Insights into epigenetic patterns in mammalian early embryos. Protein Cell. 2021;12:7-28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 10. | Gong F, Clouaire T, Aguirrebengoa M, Legube G, Miller KM. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J Cell Biol. 2017;216:1959-1974. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 131] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 11. | Hendriks IA, Treffers LW, Verlaan-de Vries M, Olsen JV, Vertegaal ACO. SUMO-2 Orchestrates Chromatin Modifiers in Response to DNA Damage. Cell Rep. 2015;10:1778-1791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 112] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 12. | Zhou X, Lin Y, Kato M, Mori E, Liszczak G, Sutherland L, Sysoev VO, Murray DT, Tycko R, McKnight SL. Transiently structured head domains control intermediate filament assembly. Proc Natl Acad Sci U S A. 2021;118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 13. | Hiraide T, Nakashima M, Yamoto K, Fukuda T, Kato M, Ikeda H, Sugie Y, Aoto K, Kaname T, Nakabayashi K, Ogata T, Matsumoto N, Saitsu H. De novo variants in SETD1B are associated with intellectual disability, epilepsy and autism. Hum Genet. 2018;137:95-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 14. | Baple E, Palmer R, Hennekam RC. A microdeletion at 12q24.31 can mimic beckwith-wiedemann syndrome neonatally. Mol Syndromol. 2010;1:42-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Qiao Y, Tyson C, Hrynchak M, Lopez-Rangel E, Hildebrand J, Martell S, Fawcett C, Kasmara L, Calli K, Harvard C, Liu X, Holden JJ, Lewis SM, Rajcan-Separovic E. Clinical application of 2.7M Cytogenetics array for CNV detection in subjects with idiopathic autism and/or intellectual disability. Clin Genet. 2013;83:145-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |