Published online Jun 26, 2024. doi: 10.12998/wjcc.v12.i18.3385
Revised: April 23, 2024
Accepted: May 10, 2024
Published online: June 26, 2024
Processing time: 103 Days and 0.8 Hours
Endometrial cancer (EC) is a common gynecological malignancy that typically requires prompt surgical intervention; however, the advantage of surgical management is limited by the high postoperative recurrence rates and adverse outcomes. Previous studies have highlighted the prognostic potential of circu
To develop and validate an optimized ctDNA-based model for predicting short-term postoperative EC recurrence.
We retrospectively analyzed 294 EC patients treated surgically from 2015-2019 to devise a short-term recurrence prediction model, which was validated on 143 EC patients operated between 2020 and 2021. Prognostic factors were identified using univariate Cox, Lasso, and multivariate Cox regressions. A nomogram was crea
Based on the regression analysis and the nomogram created, patients with post
The nomogram accurately predicted RFS after EC surgery at 1, 1.5, and 2 years. This model will help clinicians personalize treatments, stratify risks, and enhance clinical outcomes for patients with EC.
Core Tip: This study introduces a predictive nomogram for endometrial cancer recurrence postsurgery, incorporating circulating tumor DNA, carcinoembryonic antigen 125, and tumor grade. The model, validated through receiver operating characteristic and decision curve analysis, accurately forecasts short-term recurrence-free survival and aids in risk stratification.
- Citation: Liu Y, Lu XN, Guo HM, Bao C, Zhang J, Jin YN. Development and validation of a circulating tumor DNA-based optimization-prediction model for short-term postoperative recurrence of endometrial cancer. World J Clin Cases 2024; 12(18): 3385-3394
- URL: https://www.wjgnet.com/2307-8960/full/v12/i18/3385.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i18.3385
Endometrial cancer (EC) is one of the most prevalent gynecological malignancies across the world, with notably rising incidence and mortality rates. According to the Global Cancer Statistics report of 2020, EC ranks sixth among female malignancies worldwide[1]. Currently, the standard surgical protocol for early-stage EC is complete hysterectomy and bilateral salpingo-oophorectomy coupled with lymph node dissection; however, about 10%-15% of patients with early-stage disease and 40% of those with late-stage disease experience recurrence or metastasis[2,3]. Therefore, accurately identifying populations that are at a high risk of postoperative recurrence is essential to customize postoperative treatment regimens for recurrence prevention.
Currently, patients at a high risk of EC recurrence and those with suspected recurrence are primarily evaluated through imaging examinations, including transvaginal ultrasound, magnetic resonance imaging, computed tomography (CT), or positron emission tomography-CT[4-6]. However, residual tumor lesions or cells often escape detection via conventional clinical and imaging approaches. Molecular residual disease (MRD), also known as minimal residual disease, refers to the presence of a few cancer cells persisting within an organism following anticancer therapy[7]. MRD may be one of the most significant factors underlying recurrence and metastasis. Circulating tumor DNA (ctDNA), a frequently used biomarker for MRD detection, holds substantial value for assessing the efficacy and prognosis of tumor treatment. ctDNA comprises fragments of tumor cell DNA that circulate freely in the bloodstream and can reflect tumor burden[8]. Previous studies have demonstrated that ctDNA monitoring can help detect recurrence earlier than traditional imaging, thereby enabling timely therapeutic intervention and improving the prognosis[9].
Multiple studies have shown that postoperative ctDNA level serves as a robust prognostic indicator for disease-free survival (DFS) and recurrence-free survival (RFS) in nonsmall cell lung cancer[10-12]. However, evidence supporting the predictive value of ctDNA in RFS following EC resection remains scarce. In this study, we developed a novel nomogram that incorporated the presence of ctDNA to accurately predict the risk of short-term recurrence following EC surgery. We optimized variable selection by combining Cox and Lasso regression methods and identified the predominant factors that influence EC recurrence. Through this optimization process, we successfully reduced the number of variables required for the prediction model. Additionally, risk stratification was done based on the nomogram which helped us predict RFS in patients with EC belonging to different risk subgroups.
For creating a model training set, we retrospectively searched the hospital’s database to identify patients who underwent surgical resection for EC at the First Affiliated Hospital of Kunming Medical University from January 2015 to December 2019. The following inclusion criteria were used for recruiting patients: (1) Confirmed diagnosis of primary EC using histopathological examination; (2) patients undergoing hysterectomy along with bilateral adnexectomy and pelvic lymphadenectomy; (3) absence of distant metastasis based on preoperative imaging assessment; and (4) availability of complete follow-up data. The study was approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University; the committee waived the need for obtaining informed consent from the patients.
The validation set comprised data from patients with EC who underwent surgical resection between June 2020 and July 2021. The inclusion criteria were the same as above, with an additional prerequisite of signed informed consent. Follow-up evaluations were conducted quarterly for the first two years, and then semi-biannually until recurrence occurred. For follow-up patients without recurrence, data from the last follow-up until June 2023 were censored at the final follow-up time point.
The exclusion criteria were as follows: (1) Past medical history of malignant neoplasms; (2) severe dysfunction of the heart, liver, or kidney; (3) history of receiving systemic therapies, such as chemotherapy or radiotherapy, before the surgical procedure; (4) absence of imaging or pathology records during follow-up; and (5) patients who died or were lost to follow-up during the study period.
Ten milliliters of venous blood were collected from all patients within 4-10 wk post-surgery. The blood was immediately centrifuged (within 2 h of collection) at 3000 r/min for 10 min. The resultant upper plasma sample was transferred to a centrifuge tube and subjected to a second round of centrifugation at the same speed and duration; this step ensured the removal of the blood cell components. Finally, the purified upper plasma was stored in a frozen storage tube until further examination. The extraction and detection of ctDNA were performed using the QIAamp Circulating Nucleic Acid kit (QIAGEN, Germany). A NanoDrop 2000 spectrophotometer (Thermo Fisher, United States) was used to determine the concentration and purity of the isolated DNA samples. ctDNA detection and quantification were performed using the Droplet Digital polymerase chain reaction technique. For the bioinformatic analysis, the GATK software was used to compare sequencing read information with the mutant allele fraction (MAF) in the hg19 human reference genome. In cases where multiple gene mutations were identified, the mutation with the highest MAF was selected for ctDNA analysis. Differences in the MAF distribution were evaluated using a permutation test, and the results were categorized using a threshold of P = 0.1 as ctDNA-positive (P < 0.1) or ctDNA-negative[13].
The following clinical and demographic data were recorded for all patients - age, history of smoking, hypertension, diabetes, pathological features of EC including histological type, differentiation grade, International Federation of Gynecology and Obstetrics (FIGO) stage, myometrial invasion, lymph node metastasis, postoperative carcinoembryonic antigen 125 (CA125) and ctDNA level, and postoperative adjuvant therapy.
For registering recurrence, EC recurrence was defined as per the criteria set by FIGO: (1) Suspicious lesions on imaging examination with subsequent pathological confirmation of tumor recurrence; and (2) In the absence of pathological verification, two consecutive imaging examinations at minimum 4-wk intervals demonstrating enlarged lesions compared to previous scans. Typical imaging findings from one examination alone were also sufficient to define a re
RFS was defined as the time interval from the date of surgical intervention to the documentation of tumor recurrence or metastasis, or to the date of the patient’s latest follow-up examination if no recurrence was observed.
All statistical computations and data analyses were performed using R statistical software (version 4.0.1). First, each collected variable was assigned a value for subsequent statistical evaluation; these variables were then subjected to univariate Cox and Lasso regression analyses in the training set. The univariate Cox regression analysis was performed to evaluate the individual relationship of each factor with the outcome of recurrence and screen variables with significant associations. The chosen explanatory variables were subsequently input into a multivariate Cox regression model. Next, a predictive nomogram was formulated using the salient explanatory variables to calculate the probability of 1-, 1.5-, and 2-year RFS outcomes in the studied EC cohort. Statistical significance was defined using a P value of < 0.05.
Further, the prognostic accuracy of the constructed nomogram was evaluated using calibration analysis to compare predicted and observed outcomes - a receiver operating characteristic (ROC) curve analysis for quantifying discrimination ability and decision curve analysis (DCA) for determining the clinical utility of the prediction model. Risk probability values for each patient were generated using a predictive nomogram model. According to this model, patients were classified into low, moderate, and high recurrence risk strata through computational analysis; the X-tile software was used to determine optimal cutoff points based on nomogram-derived risk scores. Finally, survival differences and cumulative recurrence hazards were examined across the three risk subgroups.
A total of 437 patients were included in the study - 294 in the training set and 143 in the validation set. Table 1 sum
| Variables | Levels | Training set (n = 294) | Validation Set (n = 143) | χ2/Z | P value |
| Recurrence | No | 243 (82.7) | 123 (86.0) | 0.799 | 0.372 |
| Yes | 51 (17.3) | 20 (14.0) | |||
| Follow up (months) | Median (IQR) | 19.10 (10.40, 29.38) | 18.23 (9.97, 29.03) | -1.007 | 0.314 |
| Age (yr) | < 60 | 116 (39.5) | 55 (38.5) | 0.040 | 0.842 |
| ≥ 60 | 178 (60.5) | 88 (61.5) | |||
| Histological type | Ⅰ | 241 (82.0) | 120 (83.9) | 0.253 | 0.615 |
| Ⅱ | 53 (18.0) | 23 (16.1) | |||
| Smoking | No | 275 (93.5) | 135 (94.4) | 0.125 | 0.724 |
| Yes | 19 (6.5) | 8 (5.6) | |||
| Hypertension | No | 244 (83.0) | 121 (84.6) | 0.184 | 0.668 |
| Yes | 50 (17.0) | 22 (15.4) | |||
| ctDNA | Negative | 222 (75.5) | 112 (78.3) | 0.422 | 0.516 |
| Positive | 72 (24.5) | 31 (21.7) | |||
| Diabetes | No | 267 (90.8) | 127 (88.8) | 0.436 | 0.509 |
| Yes | 27 (9.2) | 16 (11.2) | |||
| Myometrial invasion | < 1/2 | 119 (40.5) | 63 (44.1) | 0.507 | 0.476 |
| ≥ 1/2 | 175 (59.5) | 80 (55.9) | |||
| Grade | G1 | 98 (33.3) | 54 (37.8) | 0.885 | 0.642 |
| G2 | 145 (49.3) | 67 (46.9) | |||
| G3 | 51 (17.4) | 22 (15.3) | |||
| FIGO | Ⅰ | 88 (29.9) | 44 (30.8) | 0.719 | 0.869 |
| Ⅱ | 71 (24.1) | 32 (22.4) | |||
| Ⅲ | 94 (32.1) | 50 (35.0) | |||
| Ⅳ | 41 (13.9) | 17 (11.8) | |||
| Postoperative adjuvant therapy | No | 90 (30.6) | 48 (33.6) | 0.389 | 0.533 |
| Yes | 204 (69.4) | 95 (66.4) | |||
| Lymph node metastasis | No | 173 (58.8) | 81 (56.6) | 0.191 | 0.662 |
| Yes | 121 (41.2) | 62 (43.4) | |||
| Postoperative CA125 (U/mL) | < 19 | 174 (59.2) | 84 (58.7) | 0.008 | 0.930 |
| ≥ 19 | 120 (40.8) | 59 (41.3) |
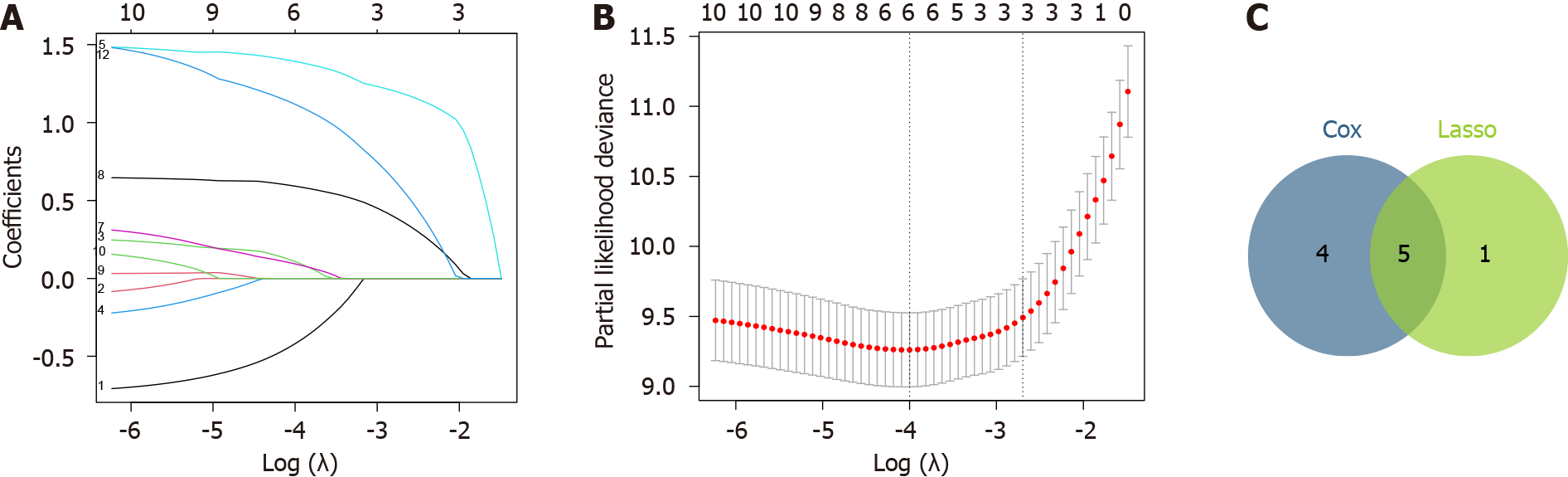
Results of the univariate Cox regression analysis revealed significant associations between the EC recurrence and the following factors: Histological type, smoking, ctDNA, myometrial invasion, tumor grade, FIGO stage, postoperative adjuvant therapy, lymph node metastasis, and postoperative CA125 levels (P < 0.05 each; Table 2). Next, Lasso regression modeling was done to refine the selection of prognostic factors associated with RFS and identify the optimal subset of predictive variables. Accordingly, the following six variables were identified, which had non-zero characteristic coefficients within a certain range of variance: Age, smoking, ctDNA, myometrial invasion, tumor grade, and postoperative CA125 (Figure 1A and B).
| Variables | HR | 95%CI | P value |
| Age | 0.803 | 0.457-1.413 | 0.447 |
| Histological type | 3.508 | 2.018-6.099 | 0.001 |
| Smoking | 3.409 | 1.593-7.298 | 0.002 |
| Hypertension | 1.227 | 0.597-2.524 | 0.578 |
| ctDNA | 13.009 | 6.655-25.431 | 0.001 |
| Diabetes | 0.524 | 0.163-1.684 | 0.278 |
| Myometrial invasion | 3.762 | 1.879-7.533 | 0.001 |
| Grade | 5.652 | 3.465-9.218 | 0.001 |
| FIGO | 2.015 | 1.507-2.694 | 0.001 |
| Postoperative adjuvant therapy | 0.487 | 0.280-0.848 | 0.011 |
| Lymph node metastasis | 5.612 | 2.874-10.959 | 0.001 |
| Postoperative CA125 | 8.024 | 3.896-16.529 | 0.001 |
We compared the significant factors identified by each technique and selected a final predictive model comprising the five most robust prognostic variables as depicted in the variable selection flow diagram (Figure 1C).
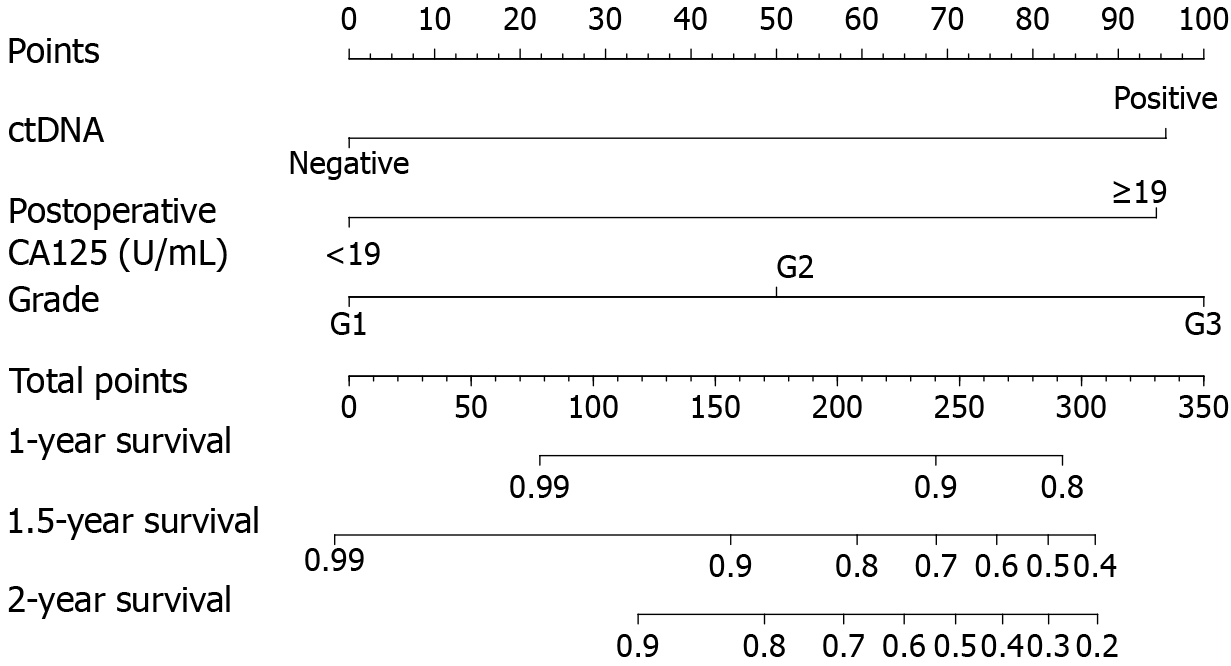
Finally, a multivariate Cox regression model was constructed using the following five variables: Smoking, ctDNA, myometrial invasion, tumor grade, and postoperative CA125 (Table 3). Three of these variables were identified as independent prognostic factors for RFS: ctDNA [hazard ratio (HR) = 3.864, 95%CI: 1.576-9.472], tumor grade (HR = 1.976, 95%CI: 1.120-3.485), and postoperative CA125 (HR = 3.740, 95%CI: 1.739-8.046). Using these variables, we constructed a prognostic nomogram capable of estimating the probability of 1-, 1.5-, and 2-year postoperative RFS in EC patients. The corresponding point value for ctDNA positivity was 96, while postoperative CA125 levels ≥ 19 U/mL corresponded to 94 points. In addition, the point values for G2 and G3 were 50 and 100, respectively (Figure 2).
| Variables | HR | 95%CI | P value |
| Smoking | 1.125 | 0.501-2.530 | 0.775 |
| ctDNA | 3.864 | 1.576-9.472 | 0.003 |
| Myometrial invasion | 1.477 | 0.713-3.062 | 0.294 |
| Grade | 1.976 | 1.120-3.485 | 0.019 |
| Postoperative CA125 | 3.740 | 1.739-8.046 | 0.001 |
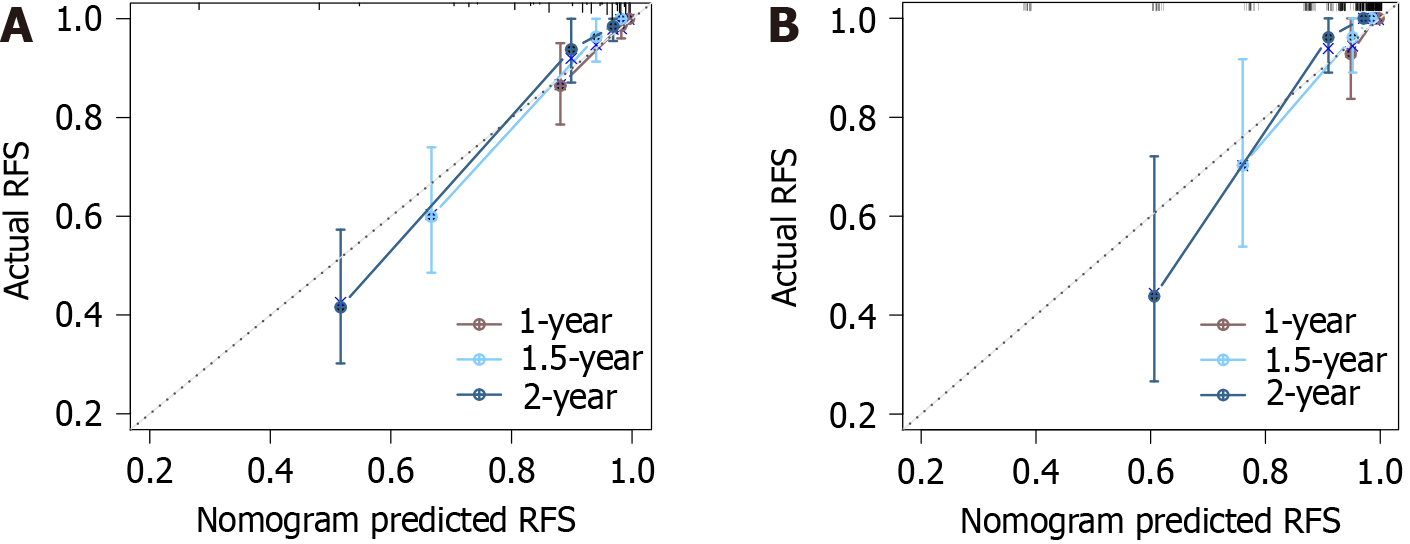
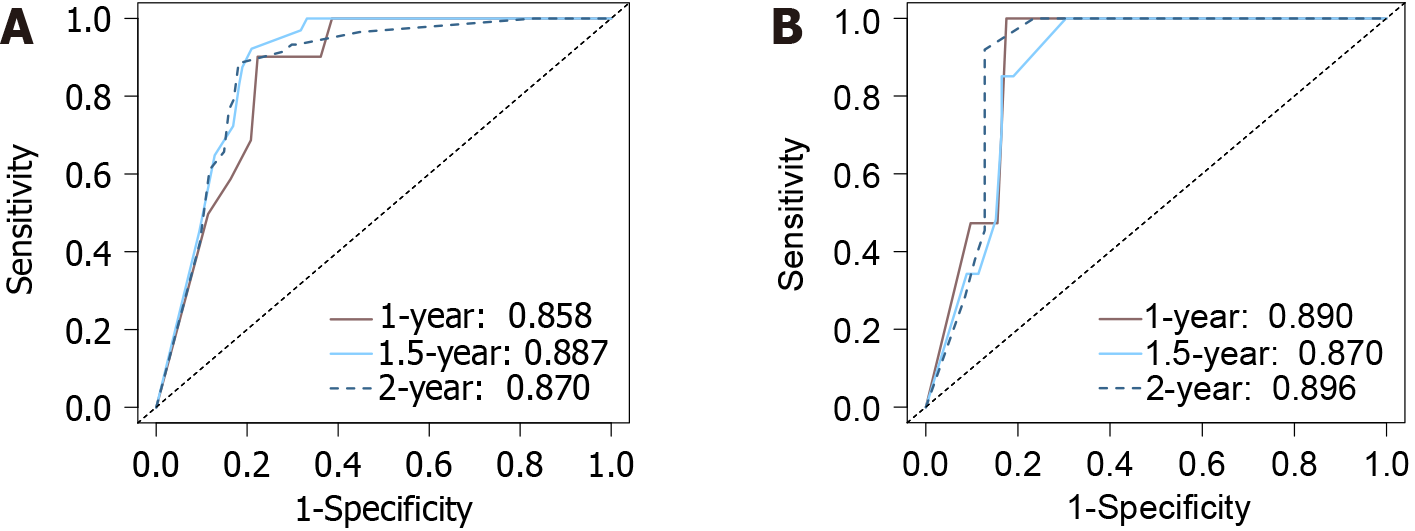
The calibration curves depicting the agreement between the predicted and observed RFS at 1, 1.5, and 2 years exhibited high concordance, illustrating the predictive accuracy of the nomogram model. This was further confirmed by a C-index of 0.870 (95%CI: 0.831-0.910) for the model, indicating an excellent predictive performance. Additionally, the ROC curves depicted the nomogram’s area under the curve (AUC) values as 0.858, 0.887, and 0.870 for the training set at 1 year, 1.5 years, and 2 years, respectively (Figure 3A). For the validation set, the AUCs were 0.890, 0.870, and 0.968, respectively, over the same period (Figure 3B).
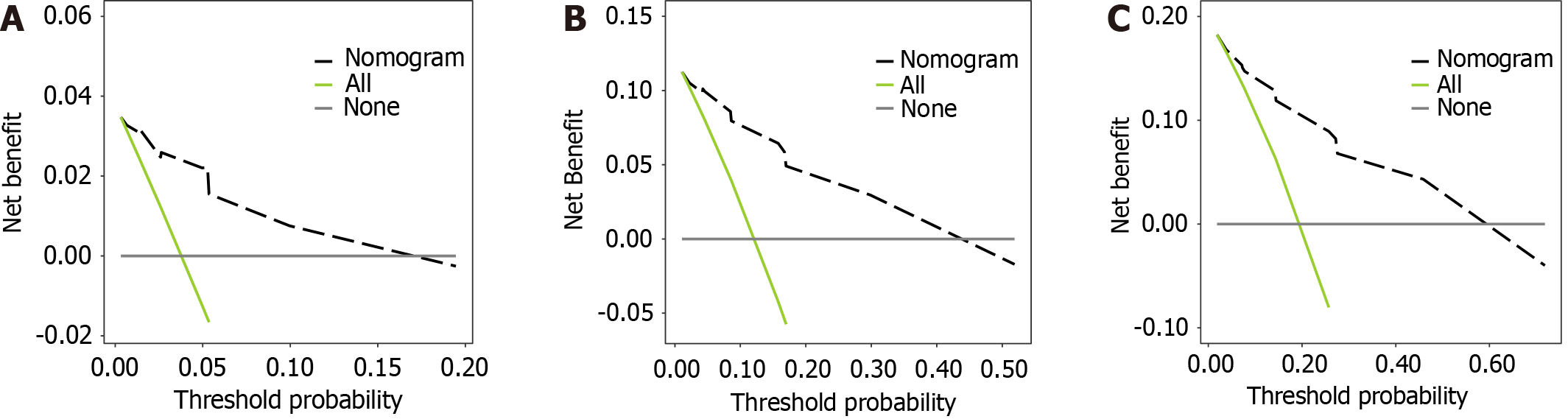
A DCA was conducted to quantify the net benefit under various threshold probabilities, determining the clinical utility of the model (Figure 4). The model provided a significant clinical net benefit compared to “none” (treat none) and “all” (treat all) when the 1-year recurrence risk threshold was 0.003-0.195. Also, the model demonstrated a good net benefit for a 1.5-year recurrence risk threshold of 0.011-0.519 and a significant net benefit relative to the comparators when the 2-year recurrence risk threshold was 0.019-0.718. As the time horizon increased, the applicability of the model improved, covering more patients with a higher recurrence risk (Figure 5).
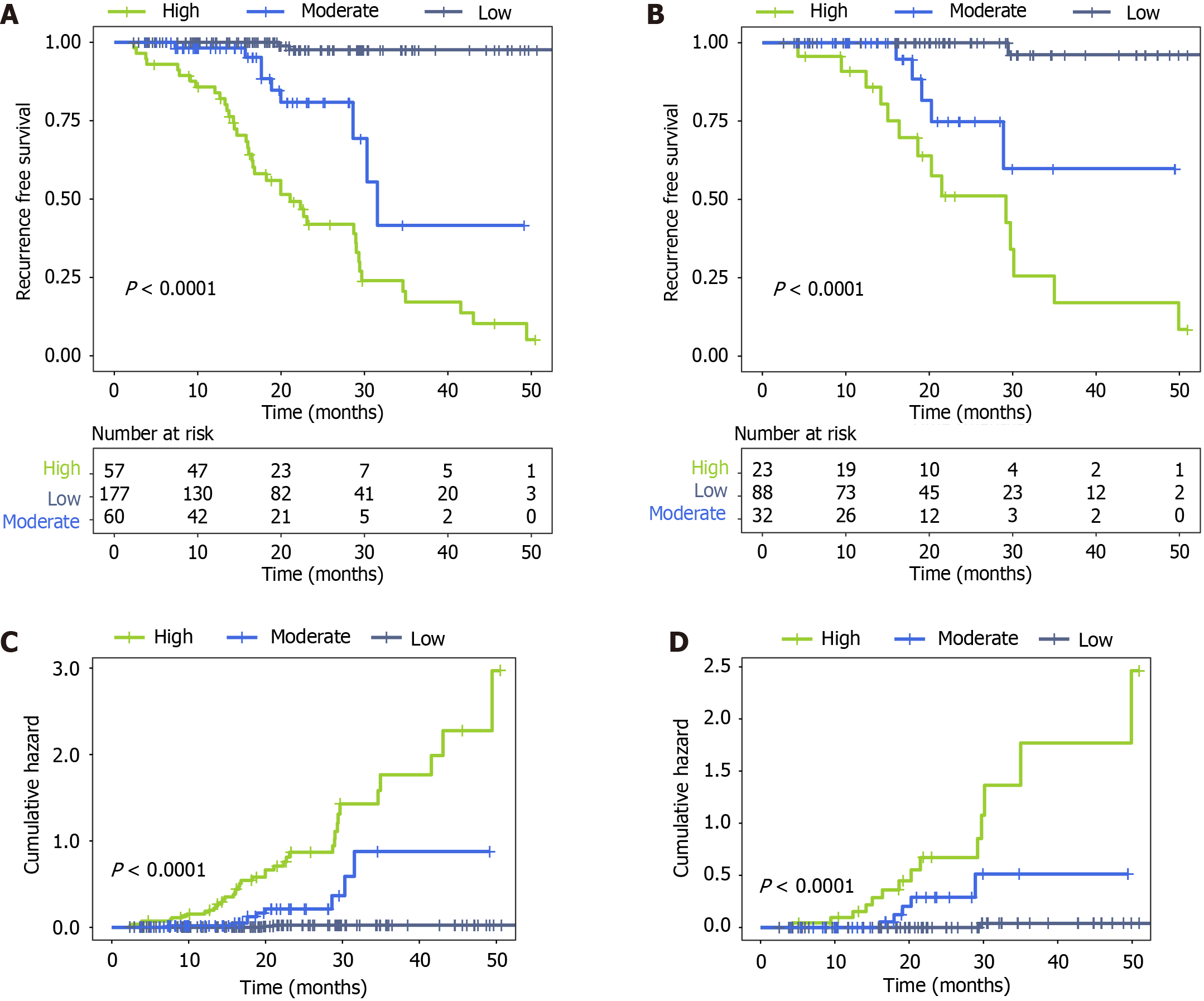
Based on the nomogram-predicted risk scores, patients were stratified into low-risk (< 96 points), moderaterisk (96-190 points), and high risk (> 190 points) subgroups. Kaplan-Meier estimates of RFS curves depicted a distinct prognostic separation among the three risk strata, with significantly improved survival rates in the low-risk cohort relative to moderate and high risk patients (P < 0.001; Figure 6A and B). Specifically, at 24 months postoperatively, patients categorized as moderate or high risk demonstrated substantially inferior RFS outcomes relative to those stratified as low-risk, evidenced by the considerable separation in Kaplan-Meier curves. This was further corroborated by the cumulative hazard curves (Figure 6C and D), which demonstrated considerably higher relapse risk for the medium- and high risk vs low-risk groups (P < 0.001).
EC is a common gynecological malignancy associated with considerable disease-specific mortality. Although early-stage diagnosis is possible and patients generally have a favorable prognosis, approximately 10%-40% of patients experience disease recurrence within two years of the initial treatment[2,3]. With the recent advent of liquid biopsy modalities, there have been remarkable advances in the molecular characterization and longitudinal monitoring of cancers. Of note, ctDNA detection is particularly relevant as a promising biomarker for the diagnosis, treatment, and prognostic evaluation of tumors. This study evaluated ctDNA as a prognostic indicator of early recurrence following surgery in patients with EC and developed an optimized prognostic model to predict postoperative recurrences in the short term. We found that ctDNA levels, postoperative CA125 Levels, and tumor grade are significant prognostic factors for short-term recurrence in patients with EC. Furthermore, the calibration curves and ROC analyses of the nomogram model showed that the model had high accuracy and discrimination. The DCA revealed that the nomogram provided a significant net clinical benefit, indicating that it could effectively guide clinical decision-making to achieve optimal outcomes compared to either treating no patients or all patients. After stratifying patients using the nomogram-predicted risk scores, we demonstrated its ability to discriminate recurrence outcomes, whereby patients classified as low-risk exhibited markedly improved RFS compared to the moderate and high risk groups.
Typically, a total or extensive/subextensive hysterectomy is the primary treatment for patients with EC. Several prognostic systems have been developed for this population, including FIGO staging, lymph node metastasis, preoperative CA125 Level, and imaging omics, which are considered among the most predictive factors. Tejerizo-García et al[14] conducted a retrospective study on 276 surgically treated EC patients and revealed that a higher FIGO stage and grade G2 histopathology served as independent predictors of inferior DFS and overall survival. However, cancer grading and staging in EC are performed after surgical resection, resulting in several limitations to prognostication, such as difficulties in sample preservation and poor reproducibility. Consequently, there is a need to identify readily accessible and highly reproducible biomarkers that accurately estimate the risk of recurrence. These biomarkers can be used as criteria in postoperative decisions to optimize individualized monitoring and treatment strategies for patients.
ctDNA originates primarily from primary tumors, metastases, and circulating tumor cells, and reflects the tumor burden and mutation status in real time. It is a sensitive and specific biomarker for cancer recurrence. Furthermore, ctDNA detection rates differ by cancer type. Previous studies have reported ctDNA positivity rates of 78.3% in metastatic pancreatic cancer patients[15], and 21% and 68% in stage I-II and stage III gastric cancer patients, respectively[16]. The current study found a 23.6% (n = 103/437) ctDNA positivity rate in EC patients. Tie et al[17] showed that postoperative ctDNA positivity was independently associated with worse RFS (HR = 3.8) in colon cancer, which is consistent with our findings (HR = 3.864). Chaudhuri et al[18] evaluated the prognosis of patients with lung cancer by detecting post
CA125 is a known predictor of tumor diagnosis, response to treatment, and even survival. A high preoperative level of CA125 indicates worse tumor differentiation and shorter overall survival, while postoperative CA125 Levels are suggestive of tumor recurrence and poor prognosis. Gong et al[19] reported that CA125 Levels decreased at initial treatment and increased again with EC recurrence, with a cutoff value of 26.4 U/mL as a significant predictor of EC recurrence. Brennan et al[20] selected 35 U/mL as the critical CA125 level, although this was not statistically significant for evaluating EC recurrence. In our study, a postoperative CA125 level of ≥ 19 U/mL was found to be a risk factor for short-term EC recurrence after surgery. There are considerable variations among the reported critical values for CA125, which may be due to differences in the sample size and study endpoints. Therefore, large-sample studies are warranted to attain consistency in the critical values of CA125 for early diagnosis.
The tumor differentiation grade reflects the similarity between tumor cells and normal tissue cells. Poor differentiation is associated with higher malignancy because poorly differentiated cancer cells have greater invasive and metastatic potential, thereby increasing the risk of tumor spread[21]. Our study found that EC patients with moderate and poor differentiation (G2 and G3) had an increased risk of short-term recurrence after surgery. The recurrence prediction model established using ctDNA levels, postoperative CA125 Levels, and tumor grade demonstrated a good predictive ability and can be used to prognosticate patient RFS and identify recurrence risk in time.
Our study had some limitations. First, the high cost and technical complexity associated with ctDNA detection resulted in a relatively small sample size. Future research should explore more cost-effective methods for ctDNA analysis and innovative study designs to increase sample size and enhance the statistical power of the study. Second, the administration of adjuvant therapy may have confounded ctDNA detection outcomes. Subsequent studies should consider ctDNA monitoring at different stages of treatment and develop novel biomarkers to distinguish between therapeutic effects and disease recurrence. Additionally, as our study data were derived from a single institution, there is a potential lack of generalizability. Therefore, future studies should include multicenter samples and involve prospective investigations to bolster the external validity of the prognostic model. Lastly, while we have highlighted the necessity for further ctDNA surveillance, there is no well-defined monitoring regimen for this. Subsequent efforts should focus on establishing the optimal frequency and timing for ctDNA monitoring and integrating these findings into clinical practice to refine treatment decisions.
We developed a novel prognostic nomogram capable of estimating the individualized risk of recurrence of EC in patients undergoing surgical resection. Patients with negative ctDNA, postoperative CA125 Levels of < 19 U/mL, and grade G1 tumors had improved RFS. The nomogram demonstrated an excellent ability to predict patient RFS; however, dynamic ctDNA monitoring is needed in this population to comprehensively analyze and elucidate the clinical utility of ctDNA for predicting recurrence and progression of EC.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64589] [Article Influence: 16147.3] [Reference Citation Analysis (176)] |
| 2. | Tran DN, Rozen V, Hunter MI, Kim TH, Jeong JW. ARG1 is a potential prognostic marker in metastatic and recurrent endometrial cancer. Res Sq. 2023;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Gallos ID, Yap J, Rajkhowa M, Luesley DM, Coomarasamy A, Gupta JK. Regression, relapse, and live birth rates with fertility-sparing therapy for endometrial cancer and atypical complex endometrial hyperplasia: a systematic review and metaanalysis. Am J Obstet Gynecol. 2012;207:266.e1-266.12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 232] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 4. | Maheshwari E, Nougaret S, Stein EB, Rauch GM, Hwang KP, Stafford RJ, Klopp AH, Soliman PT, Maturen KE, Rockall AG, Lee SI, Sadowski EA, Venkatesan AM. Update on MRI in Evaluation and Treatment of Endometrial Cancer. Radiographics. 2022;42:2112-2130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 52] [Reference Citation Analysis (0)] |
| 5. | Ryu SY, Kim K, Kim Y, Park SI, Kim BJ, Kim MH, Choi SC, Lee ED, Lee KH, Kim BI. Detection of recurrence by 18F-FDG PET in patients with endometrial cancer showing no evidence of disease. J Korean Med Sci. 2010;25:1029-1033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Silva C, Carneiro C, Cunha TM. Role of Imaging in the Management of High-Risk Endometrial Cancer. Cureus. 2021;13:e19286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 7. | Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16:409-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 722] [Article Influence: 120.3] [Reference Citation Analysis (0)] |
| 8. | Amoozgar Z, Jaymand M, Jahanban-Esfahlan R. Editorial: Circulating molecular biomarkers: next-generation tools for monitoring minimal residual disease in cancer patients. Front Oncol. 2023;13:1226974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 9. | Bellone S, McNamara B, Mutlu L, Demirkiran C, Hartwich TMP, Harold J, Yang-Hartwich Y, Siegel ER, Santin AD. Monitoring Treatment Response, Early Recurrence, and Survival in Uterine Serous Carcinoma and Carcinosarcoma Patients Using Personalized Circulating Tumor DNA Biomarkers. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 10. | Chidharla A, Rapoport E, Agarwal K, Madala S, Linares B, Sun W, Chakrabarti S, Kasi A. Circulating Tumor DNA as a Minimal Residual Disease Assessment and Recurrence Risk in Patients Undergoing Curative-Intent Resection with or without Adjuvant Chemotherapy in Colorectal Cancer: A Systematic Review and Meta-Analysis. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 23] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 11. | Tie J, Wang Y, Cohen J, Li L, Hong W, Christie M, Wong HL, Kosmider S, Wong R, Thomson B, Choi J, Fox A, Field K, Burge M, Shannon J, Kotasek D, Tebbutt NC, Karapetis C, Underhill C, Haydon A, Schaeffer J, Ptak J, Tomasetti C, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: A prospective cohort study. PLoS Med. 2021;18:e1003620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 12. | Jung HA, Ku BM, Kim YJ, Park S, Sun JM, Lee SH, Ahn JS, Cho JH, Kim HK, Choi YS, Choi YL, Shin SH, Jeong BH, Um SW, Kim H, Kim K, Ahn MJ, Kim J. Longitudinal Monitoring of Circulating Tumor DNA From Plasma in Patients With Curative Resected Stages I to IIIA EGFR-Mutant Non-Small Cell Lung Cancer. J Thorac Oncol. 2023;18:1199-1208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 38] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 13. | Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, Kosmider S, Skinner I, Wong R, Steel M, Tran B, Desai J, Jones I, Haydon A, Hayes T, Price TJ, Strausberg RL, Diaz LA Jr, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 1037] [Article Influence: 129.6] [Reference Citation Analysis (0)] |
| 14. | Tejerizo-García A, Jiménez-López JS, Muñoz-González JL, Bartolomé-Sotillos S, Marqueta-Marqués L, López-González G, Gómez JF. Overall survival and disease-free survival in endometrial cancer: prognostic factors in 276 patients. Onco Targets Ther. 2013;9:1305-1313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 15. | Hata T, Mizuma M, Iseki M, Takadate T, Ishida M, Nakagawa K, Hayashi H, Morikawa T, Motoi F, Unno M. Circulating tumor DNA as a predictive marker for occult metastases in pancreatic cancer patients with radiographically non-metastatic disease. J Hepatobiliary Pancreat Sci. 2021;28:648-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 16. | Yang J, Gong Y, Lam VK, Shi Y, Guan Y, Zhang Y, Ji L, Chen Y, Zhao Y, Qian F, Chen J, Li P, Zhang F, Wang J, Zhang X, Yang L, Kopetz S, Futreal PA, Zhang J, Yi X, Xia X, Yu P. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 2020;11:346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 118] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 17. | Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, Wong R, Kosmider S, Ananda S, McKendrick J, Lee B, Cho JH, Faragher I, Jones IT, Ptak J, Schaeffer MJ, Silliman N, Dobbyn L, Li L, Tomasetti C, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019;5:1710-1717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 451] [Article Influence: 112.8] [Reference Citation Analysis (0)] |
| 18. | Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, Khodadoust MS, Esfahani MS, Liu CL, Zhou L, Scherer F, Kurtz DM, Say C, Carter JN, Merriott DJ, Dudley JC, Binkley MS, Modlin L, Padda SK, Gensheimer MF, West RB, Shrager JB, Neal JW, Wakelee HA, Loo BW Jr, Alizadeh AA, Diehn M. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7:1394-1403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 742] [Article Influence: 92.8] [Reference Citation Analysis (0)] |
| 19. | Gong S, Quan Q, Meng Y, Wu J, Yang S, Hu J, Mu X. The value of serum HE4 and CA125 levels for monitoring the recurrence and risk stratification of endometrial endometrioid carcinoma. Heliyon. 2023;9:e18016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 20. | Brennan DJ, Hackethal A, Mann KP, Mutz-Dehbalaie I, Fiegl H, Marth C, Obermair A. Serum HE4 detects recurrent endometrial cancer in patients undergoing routine clinical surveillance. BMC Cancer. 2015;15:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Satta S, Dolciami M, Celli V, Di Stadio F, Perniola G, Palaia I, Pernazza A, Della Rocca C, Rizzo S, Catalano C, Capuani S, Manganaro L. Quantitative diffusion and perfusion MRI in the evaluation of endometrial cancer: validation with histopathological parameters. Br J Radiol. 2021;94:20210054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |