Published online Jun 6, 2024. doi: 10.12998/wjcc.v12.i16.2887
Revised: March 13, 2024
Accepted: April 11, 2024
Published online: June 6, 2024
Processing time: 115 Days and 9.9 Hours
We present a case of an EWSR1/FUS::NFATC2 rearranged sarcoma in the left fore
The patient is a 23-year-old woman. Microscopically, the tumor cells were me
Clinical imaging, immunohistochemistry, and molecular pathology should be considered to confirm the diagnosis.
Core Tip:EWSR1/FUS::NFATC2 rearranged sarcoma is a rare and aggressive malignancy of bone or soft tissue. We report a case of EWSR1/FUS::NFATC2 rearranged sarcoma of the left forearm. Immunohistochemistry and molecular detection play an important role in distinguishing this tumor from other tumors. It is clinically insensitive to classical Ewing's sarcoma chemotherapy, therefore it is very important to recognize its presence to avoid misdiagnosis. And more cases of EWSR1/ FUS::NFATC2 rearranged sarcomas need to be collected for clinicopathological evaluation and follow-up to explore its development, prognosis, and treatment options.
- Citation: Hu QL, Zeng C. Clinicopathological analysis of EWSR1/FUS::NFATC2 rearranged sarcoma in the left forearm: A case report. World J Clin Cases 2024; 12(16): 2887-2893
- URL: https://www.wjgnet.com/2307-8960/full/v12/i16/2887.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i16.2887
EWSR1-non-ETS fusion sarcomas involve gene partners unrelated to the ETS family. The reported incidence of EWSR1-non-ETS fusion sarcomas is 0.212 cases per million population[1]. EWSR1/FUS::NFATC2 rearranged sarcoma is the most common subtype of EWSR1-non-ETS fusion sarcoma. It is also a newly described class of solid tumors originating from bone or soft tissue with unique clinicopathological features[2-4]. EWSR1/FUS::NFATC2 rearranged sarcoma mainly occurs in the metaphysis or diaphysis of long bones and less often in soft tissues. In the long bones, the affected sites were femur, humerus, radius and tibia in descending order of frequency[5]. In soft tissues, it occurs mainly in limbs, head, neck and chest[6]. The median age of onset is approximately 32 years, and male-to-female ratio is 5:1[7-9]. Clinically, it usually presents with pain and locally destructive bone lesions that may invade the surrounding soft tissues. To date, a small number of EWSR1/FUS::NFATC2 rearranged bone tumors have been reported, many of which do not have detailed clinicopathological data[5-9]. Here, we report a case of EWSR1/FUS::NFATC2 rearranged sarcoma of the left forearm.
A 23-year-old female patient was admitted to the hospital 5 months after the recurrence of a tumor in her left forearm.
The patient had left forearm pain for 5 months, and magnetic resonance examination suggested the possibility of recurrence of malignant tumor in the left forearm.
The patient's left forearm tumor was diagnosed as a sclerosed epithelioid fibrosarcoma 10 years ago at a foreign hospital. Subsequently, the patient underwent tumor resection and bone grafting of the left distal radius. Chemotherapy was not administered, and no recurrence was observed in the regular review. Five months prior, pain in the dorsal left forearm recurred without an obvious trigger.
No personal or family history was available.
A surgical scar was seen on the left forearm, which was slightly swollen. There was dorsal tenderness in the distal left forearm.
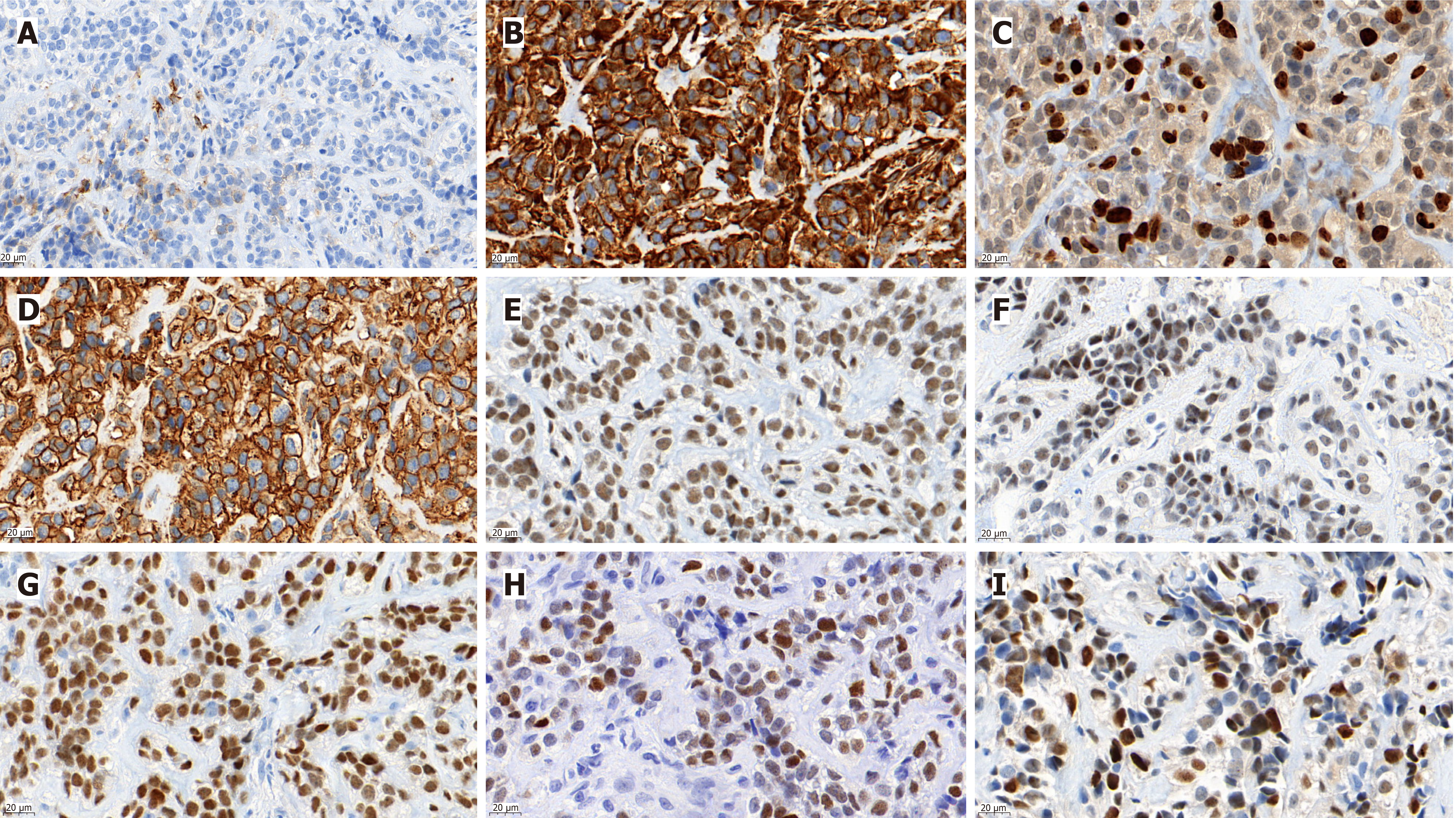
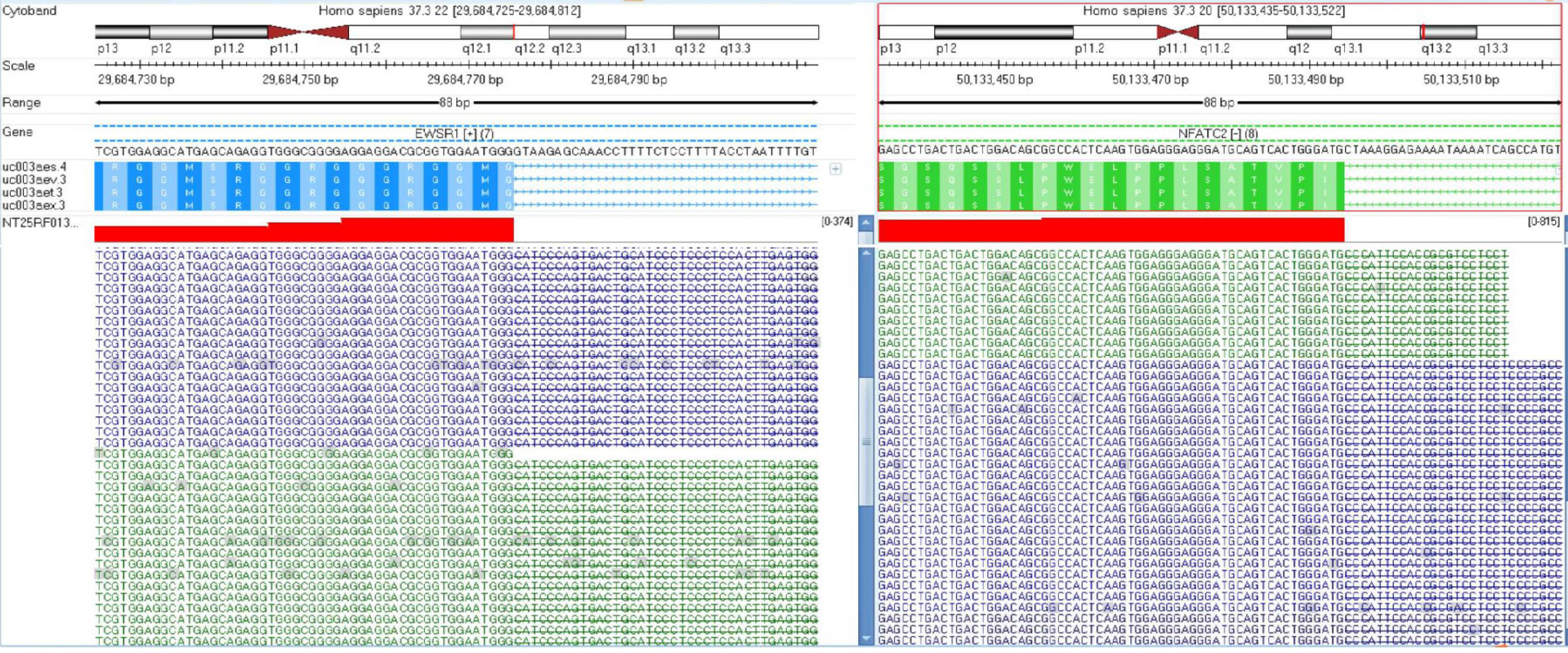
The tumor cells were positive for Vimentin, CD99, NKX2.2, NKX3.1, INI-1, CyclinD1, and FLI1, and focally positive for epithelial membrane antigen (EMA) (Figure 1). Approximately 25% of Ki67 hotspots were positive (Figure 1C). However, cytokeratin (CK)-pan, desmin, CD45-leukocyte common antigen, S-100, mouse double minute 2, myogenic differentiation 1, myogenin, special AT-rich2, mucoprotein-4 (MUC-4), CD34, signal transducer and activator of transcription 6, MelanomA, human melanoma black 45, CD56 (neural cell adhesion molecules), neuron specific enolase, synaptophysin, ETS-related gene, transducin-like enhancer of split 1, wilms tumor protein (WT1), calretinin, Pax-8, and CD68 were negative in this case. Next-generation sequencing revealed t(20;22)(q13.2;q12.2) EWSR1-NFATC2 gene fusion (Figure 2).
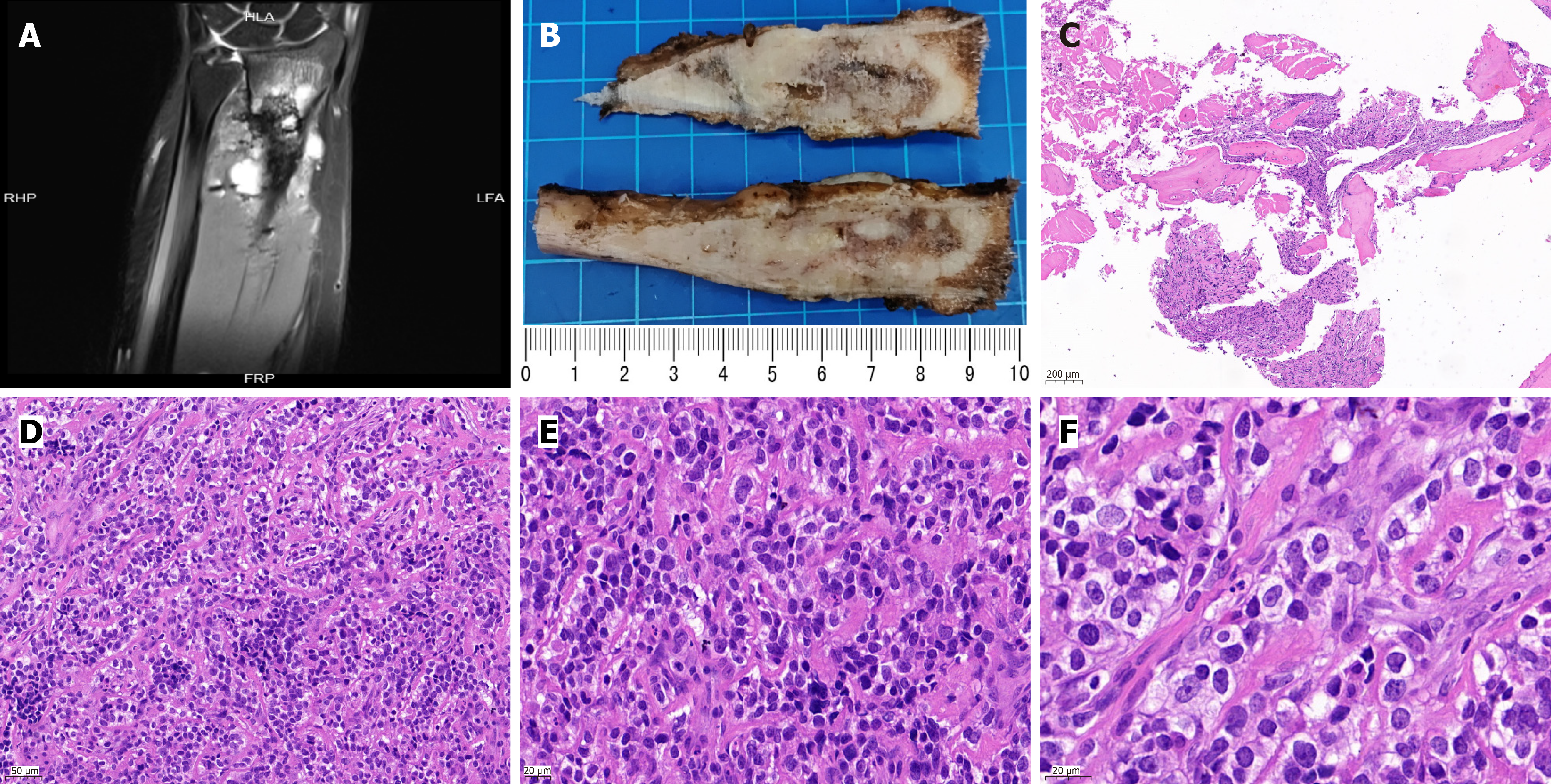
Plain magnetic resonance imaging (MRI) and enhanced (3.0T) examination of the unilateral upper limb showed multiple oval and patchy abnormal signal shadows in the original surgical area and multiple abnormal signal shadows in the distal and surrounding bones, with clear boundaries; the size of the largest one was approximately 19 mm × 14 mm (Figure 3A).
The surgical specimen consisted of a piece of left radial tissue, approximately 9.5 cm long and 1-2.5 cm in diameter. The local surface was raised, and the cut surface was grey-white and grey-red with an uneven texture (Figure 3B). Microscopically, medium-sized round cells were arranged in small nests between mature bone tissues (Figure 3C and D). The tumor cells had a clear cytoplasm, uniform nuclear morphology, dense chromatin, visible nucleoli, and rare mitotic figures (Figure 3E and F).
Finally, EWSR1/FUS::NFATC2 rearranged sarcoma was diagnosed.
The malignant tumor was treated with chemotherapy before surgery, and the left distal radius malignant tumor was resected and allogeneic radius was implanted under general anesthesia in September 2023.
Regular follow-up after surgery showed no tumor recurrence or metastasis.
This patient was a 23-year-old female who was found to have a tumor in the left forearm 10 years ago, which was diagnosed as sclerosing epithelioid fibrosarcoma in another hospital. The tumor was resected without chemotherapy, and no recurrence was found during regular review. Five months ago, back pain recurred in the left forearm, and MR examination suggested the possibility of recurrence of malignant tumor in the left forearm. Microscopically, there were medium-sized round cells arranged in small nests between the mature bone tissue. The cytoplasm of the tumor cells was clear, the nuclei were relatively uniform, the chromatin was dense, nucleoli were visible, and mitotic figures were rare. Immunohistochemical staining showed that the tumor cells were positive for Vimentin, INI-1, CD99, NKX2.2, CyclinD1, FLI1 and NKX3.1, and a few tumor cells were positive for EMA. Next generation sequencing found EWSR1 and NFATC2 gene fusion.
EWSR1/FUS::NFATC2 rearranged sarcomas are generally solid masses ranging from 4 to 18 cm with a yellowish-brown surface and hard or fleshy section[10]. Metastasis occurs mainly in the lungs and soft tissues. They mainly consist of relatively homogeneous round cells arranged in a diffuse, pseudoglandular, nested, or cord-like pattern against a fibrous or clear mucus background. The tumor nuclei are consistent in size and can have obvious pleomorphism, dense or vesicular chromatin, small or prominent nucleoli, tumor necrosis, or mitotic figures[9]. Immunohistochemically, the tumor cells were diffusely positive for CD99, PAX-7, and NKX2.2 in approximately half of the cases, and focally positive staining for cytokeratin AE1/AE3 was observed[9-12]. In addition, Aggercan and NKX3.1 are highly expressed in EWSR1/FUS::NFATC2 rearranged sarcomas, which can be used to distinguish EWSR1/FUS::NFATC2 rearranged sarcoma from Ewing's sarcoma[10,13,14]. In addition, EWSR1::NFATC2 rearranged sarcoma has a unique signal signature in FISH with the EWSR1 split probe, with frequent amplification of a single-probe (5' end) signal that is markedly different from other EWSR1-rearranged tumors. Molecular pathology showed that EWSR1-NFATC2 gene fusion is an important diagnostic marker for EWSR1::NFATC2 rearranged sarcomas. EWSR1/FUS::NFATC2 rearranged sarcoma has a potential risk of local recurrence, distant metastasis, and histological progression and is not sensitive to classical Ewing's sarcoma-specific chemotherapy[9,10,13]. Previously, EWSR1/FUS::NFATC2 rearranged sarcoma was likely to be diagnosed as Ewing sarcoma, myoepithelial tumor, ossifying fibromyxoid tumor, sclerosing epithelioid fibrosarcoma, extraskeletal myxoid chondrosarcoma, small cell osteosarcoma, etc.[3,6,15-18].
EWSR1/FUS::NFATC2 rearranged sarcoma is rare and should be distinguished from the following tumors: (1) Ewing’s sarcoma of the bone: Immunohistochemically, both sarcomas express CD99, NKX2.2, and PAX-7, which can lead to misdiagnosis, but NKX3.1, lymphoid, rhabdomyoblastic, neuroendocrine, and other markers are negative in Ewing’s sarcoma. In addition, all Ewing’s sarcomas harbor FET-ETS gene fusions[7,13,19]; (2) Myoepithelial tumors: Myoepithelial tumors of the bone are rare[20]. Histologically, the tumors were spindle-shaped with epithelioid, plasmacytoid, or clear cells. The stroma was myxochondroid, clear, or without obvious stroma. Immunohistochemically, the tumor cells were positive for myoepithelial markers such as cytokeratin, EMA, P63, alpha-smooth muscle actin (SMA), S-100, glial fibrillary acidic protein, and WT1. Some cases were positive for CD99 but negative or focally positive for NKX2.2. Approximately 50% of myoepithelial tumors harbor EWSR1-POU5F1 or PBX1 gene translocations[21,22]; (3) Ossifying fibromyxoid tumors are often well-circumscribed, lobulated, or multinodular. The tumor cells in the fibromyxoid stroma were uniform in size and bland in morphology, arranged in cords or nests, and accompanied by an intact or incomplete bone shell at the periphery. Immunohistochemically, the tumor cells were positive for S-100, Desmin, and CD99. The tumor cells were focally positive for MUC4, EMA, CK, and SMA but negative for NKX3.1 and NKX2.2. Molecular detection of the common PHF1 gene fusion, in addition to CORL1, CREBBP, and KDM2A gene fusion, has also been reported[23]; (4) Sclerosing epithelioid fibrosarcoma: Microscopically, epithelioid fibroblasts arranged in cords or nests in the sclerotic collagenous stroma, with pseudoacinar structure. The cells were uniform in size and had relatively mild morphology. Immunohistochemically, MUC4 was diffusely positive, EMA and SMA were expressed in some cases, and CK was negative. Molecular EWSR1-CREB3L1 rearrangement helps confirm the diagnosis, but additional molecular diagnostic testing is not required in cases with characteristic morphology and MUC4 expression[24,25]; (5) Extraskeletal myxoid chondrosarcoma: Primary bone lesions are rare. Histologically, the tumors were composed of ovoid, short-spindled, or rhabdoid cells arranged in cord-like or nest-like structures with myxochondroid stroma. CD99, NKX2.2 and NKX3.1 were not detected by immunohistochemistry[26]. Additionally, insulinoma-associated protein 1 is overexpressed in nearly 90% of extraskeletal myxoid chondrosarcomas. Molecular detection confirmed the presence of NR4A3 gene rearrangement[18]; and (6) Sarcomas with BCOR alterations or CIC-rearranged sarcomas: The category of undifferentiated small round cell malignant tumors is difficult to distinguish histologically, but both BCOR and CIC-rearranged sarcomas have typical molecular genetic features and lack the EWSR1 gene break and amplification signal.
EWSR1/FUS::NFATC2 rearranged sarcoma is a rare and aggressive malignancy of the bone or soft tissues. It predominantly occurs in the long bones of adult males and is clinically insensitive to chemotherapy for classical Ewing’s. EWSR1/FUS::NFATC2 rearranged sarcoma is relatively rare, and recognizing its presence is important to avoid misdiagnosis. Immunohistochemistry and molecular detection are important in distinguishing this tumor from other tumors. More cases of EWSR1/FUS::NFATC2 rearranged sarcomas should be collected for clinicopathological evaluation and follow-up to explore their development, prognosis, and treatment options.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s classification
Scientific Quality: Grade C
Novelty: Grade B
Creativity or Innovation: Grade C
Scientific Significance: Grade A
P-Reviewer: Laskin WB, United States S-Editor: Zheng XM L-Editor: A P-Editor: Zhao S
| 1. | de Pinieux G, Karanian M, Le Loarer F, Le Guellec S, Chabaud S, Terrier P, Bouvier C, Batistella M, Neuville A, Robin YM, Emile JF, Moreau A, Larousserie F, Leroux A, Stock N, Lae M, Collin F, Weinbreck N, Aubert S, Mishellany F, Charon-Barra C, Croce S, Doucet L, Quintin-Rouet I, Chateau MC, Bazille C, Valo I, Chetaille B, Ortonne N, Brouchet A, Rochaix P, Demuret A, Ghnassia JP, Mescam L, Macagno N, Birtwisle-Peyrottes I, Delfour C, Angot E, Pommepuy I, Ranchere D, Chemin-Airiau C, Jean-Denis M, Fayet Y, Courrèges JB, Mesli N, Berchoud J, Toulmonde M, Italiano A, Le Cesne A, Penel N, Ducimetiere F, Gouin F, Coindre JM, Blay JY; NetSarc/RePPS/ResSos and French Sarcoma Group- Groupe d’Etude des Tumeurs Osseuses (GSF-GETO) networks. Nationwide incidence of sarcomas and connective tissue tumors of intermediate malignancy over four years using an expert pathology review network. PLoS One. 2021;16:e0246958. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 169] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 2. | Szuhai K, Ijszenga M, de Jong D, Karseladze A, Tanke HJ, Hogendoorn PC. The NFATc2 gene is involved in a novel cloned translocation in a Ewing sarcoma variant that couples its function in immunology to oncology. Clin Cancer Res. 2009;15:2259-2268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 149] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 3. | Antonescu C. Round cell sarcomas beyond Ewing: emerging entities. Histopathology. 2014;64:26-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 136] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 4. | Baldauf MC, Orth MF, Dallmayer M, Marchetto A, Gerke JS, Rubio RA, Kiran MM, Musa J, Knott MML, Ohmura S, Li J, Akpolat N, Akatli AN, Özen Ö, Dirksen U, Hartmann W, de Alava E, Baumhoer D, Sannino G, Kirchner T, Grünewald TGP. Robust diagnosis of Ewing sarcoma by immunohistochemical detection of super-enhancer-driven EWSR1-ETS targets. Oncotarget. 2018;9:1587-1601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 5. | Diaz-Perez JA, Nielsen GP, Antonescu C, Taylor MS, Lozano-Calderon SA, Rosenberg AE. EWSR1/FUS-NFATc2 rearranged round cell sarcoma: clinicopathological series of 4 cases and literature review. Hum Pathol. 2019;90:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 6. | Cohen JN, Sabnis AJ, Krings G, Cho SJ, Horvai AE, Davis JL. EWSR1-NFATC2 gene fusion in a soft tissue tumor with epithelioid round cell morphology and abundant stroma: a case report and review of the literature. Hum Pathol. 2018;81:281-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 7. | Machado I, Yoshida A, Morales MGN, Abrahão-Machado LF, Navarro S, Cruz J, Lavernia J, Parafioriti A, Picci P, Llombart-Bosch A. Review with novel markers facilitates precise categorization of 41 cases of diagnostically challenging, "undifferentiated small round cell tumors". A clinicopathologic, immunophenotypic and molecular analysis. Ann Diagn Pathol. 2018;34:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 8. | Bode-Lesniewska B, Fritz C, Exner GU, Wagner U, Fuchs B. EWSR1-NFATC2 and FUS-NFATC2 Gene Fusion-Associated Mesenchymal Tumors: Clinicopathologic Correlation and Literature Review. Sarcoma. 2019;2019:9386390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 9. | Wang GY, Thomas DG, Davis JL, Ng T, Patel RM, Harms PW, Betz BL, Schuetze SM, McHugh JB, Horvai AE, Cho SJ, Lucas DR. EWSR1-NFATC2 Translocation-associated Sarcoma Clinicopathologic Findings in a Rare Aggressive Primary Bone or Soft Tissue Tumor. Am J Surg Pathol. 2019;43:1112-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 10. | Perret R, Escuriol J, Velasco V, Mayeur L, Soubeyran I, Delfour C, Aubert S, Polivka M, Karanian M, Meurgey A, Le Guellec S, Weingertner N, Hoeller S, Coindre JM, Larousserie F, Pierron G, Tirode F, Le Loarer F. NFATc2-rearranged sarcomas: clinicopathologic, molecular, and cytogenetic study of 7 cases with evidence of AGGRECAN as a novel diagnostic marker. Mod Pathol. 2020;33:1930-1944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 11. | Yau DTW, Chan JKC, Bao S, Zheng Z, Lau GTC, Chan ACL. Bone Sarcoma With EWSR1-NFATC2 Fusion: Sarcoma With Varied Morphology and Amplification of Fusion Gene Distinct From Ewing Sarcoma. Int J Surg Pathol. 2019;27:561-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Seligson ND, Maradiaga RD, Stets CM, Katzenstein HM, Millis SZ, Rogers A, Hays JL, Chen JL. Multiscale-omic assessment of EWSR1-NFATc2 fusion positive sarcomas identifies the mTOR pathway as a potential therapeutic target. NPJ Precis Oncol. 2021;5:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Yoshida KI, Machado I, Motoi T, Parafioriti A, Lacambra M, Ichikawa H, Kawai A, Antonescu CR, Yoshida A. NKX3-1 Is a Useful Immunohistochemical Marker of EWSR1-NFATC2 Sarcoma and Mesenchymal Chondrosarcoma. Am J Surg Pathol. 2020;44:719-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 14. | Syed M, Mushtaq S, Loya A, Hassan U. NKX3.1 a useful marker for mesenchymal chondrosarcoma: An immunohistochemical study. Ann Diagn Pathol. 2021;50:151660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Zhang M, Yu Y, Guan X, Yao X, Jia C, Hong E, Guo Y, He L. A group of sclerosing epithelioid fibrosarcomas with low-level amplified EWSR1-CREB3L1 fusion gene in children. Pathol Res Pract. 2022;230:153754. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 16. | Sadri N, Barroeta J, Pack SD, Abdullaev Z, Chatterjee B, Puthiyaveettil R, Brooks JS, Barr FG, Zhang PJ. Malignant round cell tumor of bone with EWSR1-NFATC2 gene fusion. Virchows Arch. 2014;465:233-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Makise N, Yoshida KI, Iijima T, Yoshida A, Ushiku T, Ishida T. Skeletal EWSR1-NFATC2 sarcoma previously diagnosed as Ewing-like adamantinoma: A case report and literature review emphasizing its unique radiological features. Pathol Int. 2021;71:614-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Stacchiotti S, Baldi GG, Morosi C, Gronchi A, Maestro R. Extraskeletal Myxoid Chondrosarcoma: State of the Art and Current Research on Biology and Clinical Management. Cancers (Basel). 2020;12:2703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 19. | Wang WL, Patel NR, Caragea M, Hogendoorn PC, López-Terrada D, Hornick JL, Lazar AJ. Expression of ERG, an Ets family transcription factor, identifies ERG-rearranged Ewing sarcoma. Mod Pathol. 2012;25:1378-1383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 20. | Song W, Flucke U, Suurmeijer AJH. Myoepithelial Tumors of Bone. Surg Pathol Clin. 2017;10:657-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Jo VY, Fletcher CD. Myoepithelial neoplasms of soft tissue: an updated review of the clinicopathologic, immunophenotypic, and genetic features. Head Neck Pathol. 2015;9:32-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 123] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 22. | Bishop JA, Alaggio R, Zhang L, Seethala RR, Antonescu CR. Adamantinoma-like Ewing family tumors of the head and neck: a pitfall in the differential diagnosis of basaloid and myoepithelial carcinomas. Am J Surg Pathol. 2015;39:1267-1274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 120] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 23. | Schneider N, Fisher C, Thway K. Ossifying fibromyxoid tumor: morphology, genetics, and differential diagnosis. Ann Diagn Pathol. 2016;20:52-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Tsuda Y, Dickson BC, Dry SM, Federman N, Suurmeijer AJH, Swanson D, Sung YS, Zhang L, Healey JH, Antonescu CR. Clinical and molecular characterization of primary sclerosing epithelioid fibrosarcoma of bone and review of the literature. Genes Chromosomes Cancer. 2020;59:217-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | Warmke LM, Meis JM. Sclerosing Epithelioid Fibrosarcoma: A Distinct Sarcoma With Aggressive Features. Am J Surg Pathol. 2021;45:317-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 26. | Giner F, López-Guerrero JA, Machado I, Rubio-Martínez LA, Espino M, Navarro S, Agra-Pujol C, Ferrández A, Llombart-Bosch A. Extraskeletal myxoid chondrosarcoma: p53 and Ki-67 offer prognostic value for clinical outcome - an immunohistochemical and molecular analysis of 31 cases. Virchows Arch. 2023;482:407-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |