Published online Jun 6, 2024. doi: 10.12998/wjcc.v12.i16.2780
Revised: March 19, 2024
Accepted: April 9, 2024
Published online: June 6, 2024
Processing time: 143 Days and 3.7 Hours
Existing evidence suggests that gut microbiota represent a significant environmental risk factor for various forms of dementia, including Alzheimer's dementia, vascular dementia, and dementia in other diseases classified elsewhere. However, the exact causal relationships between gut microbiota and the different forms of dementia or their subtypes remain unclear.
To investigate putative causal relationships between gut microbiota and dementia or its subtypes using Mendelian randomization (MR) analysis.
A bidirectional, two-sample, MR analysis was conducted utilizing publicly available gut microbiota-related genome-wide association study (GWAS) sum
We identified several gut microbiota taxa exhibiting putative causal relationships with dementia or its subtypes, potentially serving as risk or protective factors for the disease. In addition, reverse MR analysis indicated that the relative abundance of several gut microbiota taxa might be influenced by dementia or its subtypes. An exhaustive sensitivity analysis confirmed the absence of heterogeneity and horizontal pleiotropy. After applying correction for multiple testing, we observed that the order Bacillales (odds ratio: 0.830, 95% confidence interval: 0.740-0.932, P = 0.00155, Padjust = 0.0311) exhibited a strong association with Alzheimer’s disease-related dementia.
The results suggest that gut microbiota is causally associated with dementia. Our findings provide novel insights into the pathophysiology of dementia and have important implications for its treatment and prevention.
Core Tip: We identified several gut microbiota taxa as being associated with the risk of or protection against dementia, including its subtypes. These gut microbiota also exhibited potential as therapeutic targets for the disease. Conversely, reverse Mendelian randomization analysis indicated that dementia or its subtypes also influenced the abundance of several gut bacteria, implying that these taxa may serve as biomarkers for assessing the occurrence and progression of dementia. This study offers novel insights into the mechanisms underlying the role of the gut microbiome in this disorder.
- Citation: Ren ZL, Zhou HH, Chen CP, He H, Wang DL, Liu Z. Causal relationships between gut microbiota and dementia: A two-sample, bidirectional, Mendelian randomization study. World J Clin Cases 2024; 12(16): 2780-2788
- URL: https://www.wjgnet.com/2307-8960/full/v12/i16/2780.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i16.2780
Dementia refers to any disease that results in a significant decline in an individual’s cognitive ability, impacting their language, attention, orientation, judgment, and planning skills. Dementia can have numerous causes, with neurodegenerative and cerebrovascular diseases being predominant[1,2]. The global prevalence of dementia is projected to increase approximately 2.7-fold by 2050 compared with 2019 due to population growth and aging[3]. Alzheimer’s disease (AD) is a neurodegenerative disorder characterized by progressive cognitive decline and behavioral impairment and is the most common cause of dementia. The etiology of AD remains uncertain, but it is hypothesized to be associated with genetic factors, protein aggregation, and neuroinflammation[4]. Vascular dementia is the second most common form of dementia, accounting for approximately 15% of the total number of cases. It is attributed to conditions such as cerebral infarction and chronic small vessel disease in the brain[5]. Numerous other types of dementia have also been identified, including frontotemporal dementia, Lewy body dementia, and dementia associated with Parkinson’s disease, among others[6,7].
Gut microbiota have a significant impact on the well-being of their host throughout life. They play a role in the regulation of brain function and behavior through the gut–brain axis and their dysbiosis is associated with the patho
Mendelian randomization (MR) studies exploit associations between genetic variants and modifiable exposures to evaluate the possible causal relationships between said exposures and outcomes while minimizing bias stemming from confounding and reverse causation. Two-sample, bidirectional MR analysis utilizes exposure and outcome data from two independent samples to estimate causal relationships and assists in elucidating the directionality of complex causal associations[15]. STROBE-MR provides guidelines for reporting MR studies[16].
In this work, we sought to identify causal relationships between gut microbiota and different forms and subtypes of dementia via a comprehensive two-sample MR analysis using datasets from the MiBioGen and FinnGen consortia. Employing bidirectional MR, we investigated the potential causal impact of gut microbiota on dementia risk, as well as the influence of dementia on gut microbiota abundance.
The exposure data utilized in this study were obtained from the publicly accessible IEU OpenGWAS project published by the MiBioGen consortium[17]. To investigate the influence of host genetics on gut microbiota composition, gut microbiota-associated genome-wide association study (GWAS) data for multiple cohorts, comprising 14306 participants from several European countries, were collected and analyzed. A total of 196 taxa were included, encompassing 9 phyla, 16 classes, 20 orders, 32 families, and 119 genera.
The GWAS data utilized as the outcome dataset in this study were obtained from the FinnGen R10 cohort[18]. Additionally, we obtained GWAS summary data for three types of dementia, namely, Alzheimer’s dementia, vascular dementia, and dementia in other diseases classified elsewhere. The diagnosis of dementia in FinnGen was based on version 10 of the International Classification of Diseases. Cases of different types of dementia were considered as narrower endpoints of dementia based on a strict definition. The specific details of the exposure and outcome variables analyzed in this MR study are presented in Table 1.
| Trait | Data source | Consortium | No. of samples | No. of cases | No. of controls |
| Gut microbiota | A total of 196 taxa | MiBioGen | 14306 | / | / |
| F5_ALZHDEMENT | Dementia in AD | FinnGen (R10) | 394705 | 6145 | 388560 |
| F5_VASCDEM | Vascular dementia | FinnGen (R10) | 395741 | 2717 | 393024 |
| F5_DEMINOTH | Dementia in other diseases classified elsewhere | FinnGen (R10) | 389828 | 1268 | 388560 |
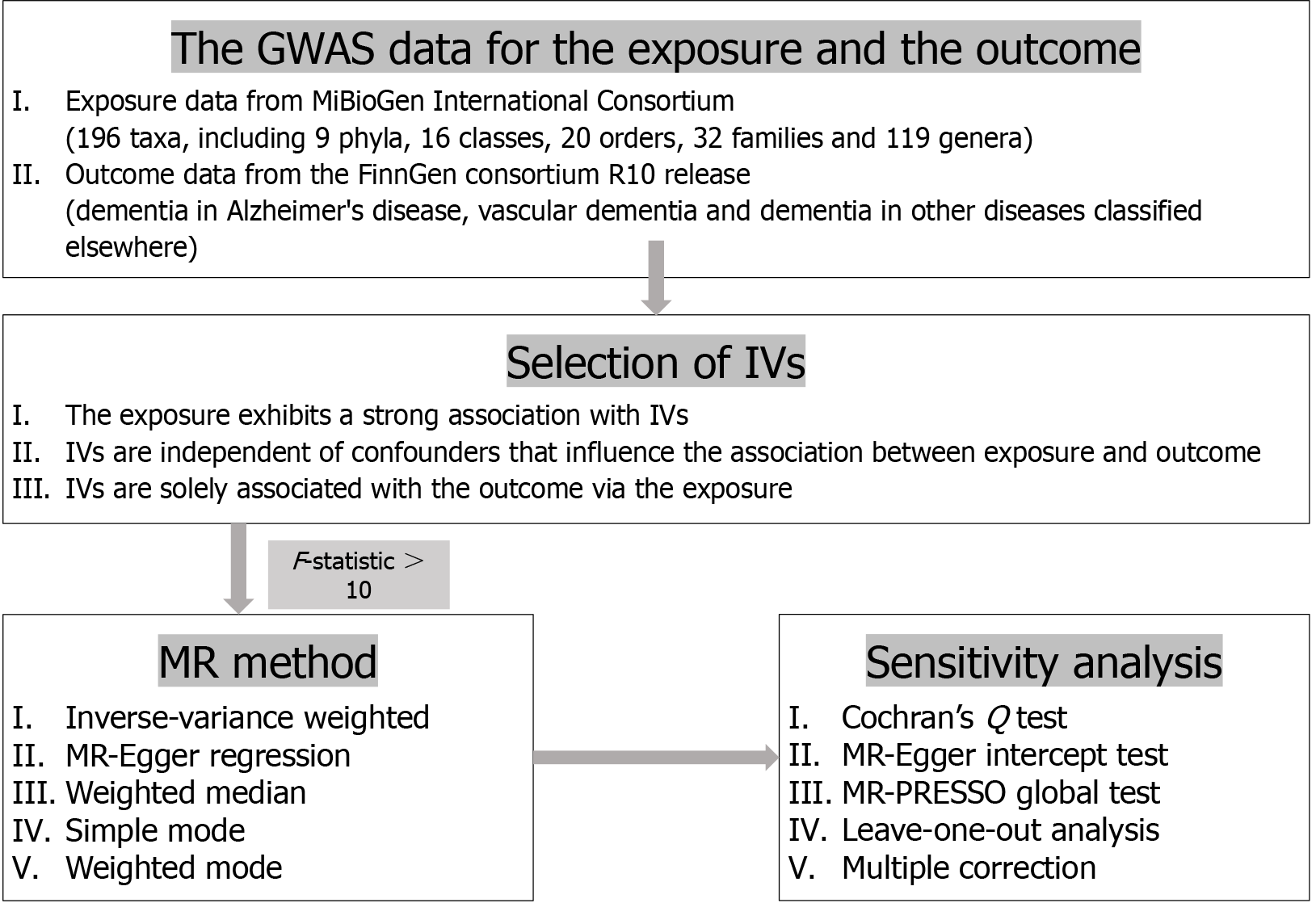
Instrumental variables (IVs) selection was based on the three fundamental assumptions of MR: There must be a significant association between a single nucleotide polymorphism (SNP) and exposure, maintaining the independence of the SNP; confounding variables must be eliminated to ensure that the IV does not influence the causal link between the exposure and the outcome; and the outcome is not influenced by the SNP. In the MR analysis, the gut microbiota was considered as the exposure factor, while dementia and its subtypes served as the outcomes. SNPs that were strongly associated with the gut microbiota were utilized as IVs. Because of the limited number of IVs obtained under the stringent threshold (P < 5e−8) when considering the gut microbiota as the exposure factor, a more inclusive threshold (P < 1e−5) was subsequently used to increase the number of IVs and ensure more reliable results. When the gut microbiota was considered as the outcome factor, the strict threshold (P < 5e−8) was used when there were a sufficient number of IVs for conducting a reverse MR analysis. In cases where IV availability was limited, alternative thresholds (P < 5e−6 or P < 1e−5) were selected. Additionally, SNP pruning was performed within a 10000 kb window with a threshold of r2 < 0.001 to address the issue of linkage disequilibrium and ensure the independence of each IV. The selected SNPs were entered into the PhenoScanner database to identify those associated with confounding factors[19,20]. An F-statistic threshold greater than 10 was indicative of the absence of bias resulting from a weak IV[21]. The F-statistic was calculated using the formula F = (beta/se)2[22].
A two-sample, bidirectional MR analysis was conducted to investigate the causal relationship between gut microbiota abundance and dementia or its subtypes. Five commonly used MR methods, namely, inverse-variance weighted (IVW), Egger regression, weighted median, simple mode, and weighted mode, were utilized for features that encompassed multiple IVs[23]. For features containing only one IV, associations were reported as odds ratios (ORs) and tested using the Wald ratio method[24]. Studies have reported that the IVW method demonstrates slightly higher statistical power than other methods in specific scenarios[25]. Thus, the IVW method was employed to determine whether a result was positive overall. A P-value < 0.05 was considered significant. An OR greater than 1 indicated that the exposure was a risk factor for the outcome, while an OR smaller than 1 indicated that the exposure was a protective factor for the outcome.
Cochran’s Q-statistic and the MR-Egger intercept test were employed to assess the heterogeneity among IVs[26]. A P-value greater than 0.05 for the Q-statistic was considered as indicating non-heterogeneity. The MR-Egger intercept test and the MR-PRESSO global test were used to evaluate the horizontal pleiotropy among the IVs[27,28]. No horizontal pleiotropy was inferred if the P-values in the MR-Egger intercept analysis or the MR-PRESSO global test exceeded 0.05. The MR-PRESSO outlier test was used for removing outlier SNPs and estimating the corrected results, while the MR-PRESSO distortion test was employed to evaluate differences between the pre-correction and post-correction outcomes. Additionally, to detect possible heterogeneity among the SNPs, a sequential leave-one-out analysis was conducted, excluding each instrumental SNP. When performing the MR analysis by sequentially removing each SNP, it was found that the overall results were only minimally affected, indicating a limited direct impact of SNP heterogeneity.
Multiple testing significance thresholds were established at each taxonomic level (phylum, class, order, family, and genus) using the Bonferroni method[29]. These thresholds were defined as P < 0.05/n, where n represents the number of independent gut microbiota taxa in the corresponding taxonomic level. Subsequently, the observed P-values were compared to this modified threshold to determine statistical significance.
Statistical analyses were conducted using R version 4.2.3. MR analyses were performed using the TwosampleMR and MR-PRESSO R packages[30,31]. Graphs were generated using the forestploter and ggplot2 R packages. A flow chart of the overall study design is presented in Figure 1.
In the MR analysis, we identified a total of 2036 SNPs (IVs) that were associated with gut microbiota at different taxonomical levels. Of these SNPs, 103 were associated with 9 phyla, 185 were associated with 16 classes, 224 were associated with 20 orders, 352 were associated with 32 families, and 1220 were associated with 119 genera. All these IVs demonstrated a stronger association with the exposure variable (P < 1e-5) than with the outcome variable (P > 5e-5), with all the respective F-statistics being greater than 10. Details of the SNPs are presented in Supplementary Table 1.
In the reverse MR analysis, we evaluated the impact of dementia and its subtypes as exposures on the abundance of 196 gut microbiota taxa. We identified 11 SNPs from the set of IVs (P < 5e-8) identified when examining Alzheimer’s dementia as an exposure factor; 18 SNPs were chosen from the set of IVs (P < 5e-6) identified when considering vascular dementia as an exposure factor; and 8 SNPs were chosen from the set of IVs (P < 5e-6) identified when examining dementia in other diseases classified elsewhere as an exposure factor. All the respective F-statistics were greater than 10. Details of the SNPs are presented in Supplementary Table 2.
All selected SNPs were examined for associations with potential confounding factors using the online tool PhenoScanner. No evidence was found for the existence of correlations between the selected IVs and other exposure factors that might influence the outcomes; accordingly, no SNPs were excluded from this study.
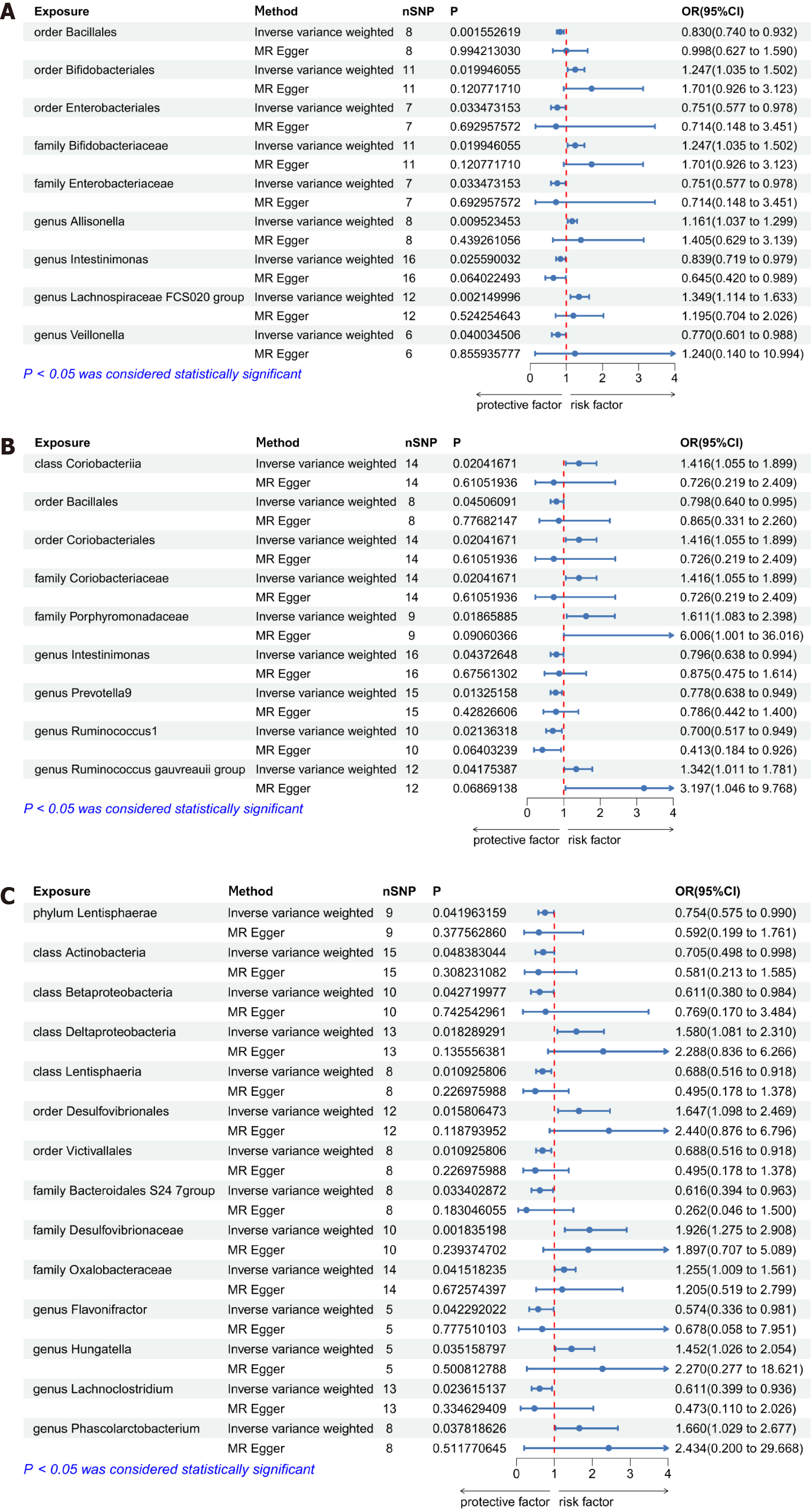
Causal relationships between gut microbiota and AD-related dementia: We conducted a bidirectional MR analysis to evaluate whether there was a causal relationship between the abundance of 196 gut microbiota taxa and dementia in AD. The results of the IVW analysis demonstrated that 9 gut microbiota taxa exerted an influence on dementia in AD (Figure 2A). Four of the 9 taxa were associated with risk factors for dementia in AD, including the order Bifidobacteriales, the family Bifidobacteriaceae, and the genera Allisonella and Lachnospiraceae FCS020 group. Meanwhile, 5 gut microbiota taxa - the orders Bacillales and Enterobacteriales, the family Enterobacteriaceae, and the genera Intestinimonas and Veillonella - were found to be associated with protective factors, with the order Bacillales exhibiting statistical significance in this regard (P = 0.0001, OR = 0.78, 95% confidence interval = 0.69-0.88) after correcting for multiple testing using the Bonferroni method. In the reverse MR analysis, IVW results revealed that the abundance of 9 gut microbiota taxa was influenced by dementia in AD (Supplementary Figure 1). Among individuals diagnosed with AD, the abundance of 6 taxa of gut microbiota was increased, including the class Erysipelotrichia, the order Erysipelotrichales, the families Erysipelotrichaceae and Family XIII, and the genera Eubacterium coprostanoligenes group and Odoribacter.
The causal relationship between gut microbiota and vascular dementia: We also undertook a bidirectional MR analysis to assess the causal associations between the abundance of 196 gut microbiota taxa and vascular dementia. IVW analysis results suggested that a total of 9 taxa exerted influence on vascular dementia (Figure 2B). Five of these were associated with risk factors for vascular dementia, including the class Coriobacteriia, the order Coriobacteriales, the families Coriobacteriaceae and Porphyromonadaceae, and the genus Ruminococcus gauvreauii group. Additionally, 4 gut microbiota taxa were associated with protective factors, including the order Bacillales and the genera Intestinimonas, Prevotella9, and Ruminococcus1. In the reverse MR analysis, the results of inverse variance weighting revealed that the abundance of 8 taxa of gut microbiota was also influenced by vascular dementia (Supplementary Figure 2). The abundance of 5 of these 8 taxa was increased, including the class Erysipelotrichia, the order Erysipelotrichales, the family Erysipelotrichaceae, and the genera Parasutterella and Veillonella, while that of 3 taxa (the family Bacteroidaceae and the genera Bacteroides and Eubacterium rectale group) was decreased.
The causal relationship between gut microbiota and dementia in other diseases classified elsewhere: A bidirectional MR analysis was conducted to evaluate the causal associations between the abundance of 196 gut microbiota and dementia in other diseases classified elsewhere. The results of the IVW analysis suggested that 14 gut microbiota taxa exerted an influence on dementia in other diseases classified elsewhere (Figure 2C). Among these 14 taxa, 6 were associated with risk factors for dementia in other diseases classified elsewhere, including the class Deltaproteobacteria, the order Desulfovibrionales, the families Desulfovibrionaceae and Oxalobacteraceae, and the genera Hungatella and Phascolarctobacterium. Meanwhile, 8 gut microbiota taxa were found to be associated with protective factors, including the phylum Lentisphaerae; the classes Actinobacteria, Betaproteobacteria, and Lentisphaeria; the order Victivallales; the family Bacteroidales S247 group; and the genera Flavonifractor and Lachnoclostridium. In the reverse MR analysis, no significant effect of dementia in other diseases classified elsewhere on the abundance of gut microbiota was observed with any of the five MR methods employed in this study.
In the two-sample, bidirectional MR analysis, the Q-statistics of both the IVW and MR-Egger analyses indicated that there was no heterogeneity within the selected IVs (P > 0.05). No significant directional horizontal pleiotropy was detected in the remaining results according to the MR-Egger intercept test and the MR-PRESSO global test (P > 0.05). Additionally, the leave-one-out analysis demonstrated the robustness of the MR results, as the exclusion of any individual IV did not affect the overall findings. Details of the heterogeneity and horizontal pleiotropy analyses are presented in Supplementary Table 3.
Although there is some evidence linking gut microbiota to dementia, specifically Alzheimer’s and vascular dementias, establishing a definitive causal relationship is difficult due to the complex molecular mechanisms involved. In this study, we explored the potential causal link between gut microbiota and dementia using a bidirectional, two-sample MR analysis. The results indicated that several taxa of gut microbiota may have causal roles in modulating the risk of dementia, either providing protection or increasing susceptibility. We found some evidence suggesting that dementia influenced the abundance of several gut microbiota taxa, indicative of reciprocal interactions between the gut microbiome and dementia. Investigating the causal connections between specific gut bacteria and dementia in both directions yielded valuable insights into the mechanisms that drive the interactions between the microbiota and the brain, laying the foundation for microbiome-based interventions in preventing and managing this condition.
The results of this study suggested that there is a strong association between the protective effect of the order Bacillales and the development of dementia in AD, with suggestive associations also found for vascular dementia. Although no study to date has reported on a relationship between Bacillales and dementias such as AD and vascular dementia, one MR-based study found a negative correlation between Bacillales and systemic lupus erythematosus[32]. This suggests that members of the order Bacillales may contribute to the inhibition of autoimmune inflammation, thus potentially exerting a protective effect against dementia. Our findings further indicated that the order Enterobacteriales and the family Enterobacteriaceae are associated with a protective effect against AD-related dementia in the context of genetic variation, whereas the order Bifidobacteriales and the family Bifidobacteriaceae are associated with an increased risk for this condition. Some evidence supports that the abundance of both Enterobacteriales and Enterobacteriaceae is increased in patients with AD[33,34]. In contrast, it has been reported that 5xFAD mice, widely employed as a model of AD, exhibit a decline in Bifidobacteriales abundance in comparison to wild-type mice[35]. However, specific dietary supplements were found to reverse the disturbance in gut microbiota and metabolic disorders in AD model mice by enhancing the proportions of Bifidobacterium[36]. These results indicate that gut microbiota may serve dual roles in disease pathogenesis; nevertheless, shifts in their abundance do not directly equate to their influence on disease initiation and progression.
Three gut microbiota taxa - the class Coriobacteriia, the order Coriobacteriales, and the family Coriobacteriaceae - were found to exert influence on vascular dementia, functioning as risk factors. Although no direct link between vascular dementia and these three taxa has been established to date, research has shown that changes in their abundance contribute to increased systemic inflammation[37]. Dementia in other diseases classified elsewhere includes Lewy body dementia and Parkinson’s disease-related dementia, among others. The gut microbiota taxa identified in this study may contribute to the exploration of the mechanisms underlying the role of gut bacteria in rare types of dementia.
In the reverse MR analysis, the abundance of the order Erysipelotrichales, the class Erysipelotrichia, and the family Erysipelotrichaceae was positively influenced by both Alzheimer’s and vascular dementias. There is evidence to support that patients with AD exhibit an increase in the abundance of gut microbiota taxa such as Erysipelatoclostridiaceae, Erysipelotrichales, Patescibacteria, Saccharimonadales, and Saccharimonadia[38]. Parts of other gut microbiota are also supported by the literature.
This study had multiple strengths. Our methodology involved using MR to address common confounders seen in observational studies. Additionally, we selected strong IVs based on the three core assumptions of MR. We also performed sensitivity tests, including Cochran's Q test, the MR-Egger intercept test, the MR-PRESSO global test, and a leave-one-out analysis, to assess the robustness of the study findings. Furthermore, Bonferroni correction was used to control for possible false positives. Importantly, our research design allowed us to undertake a pioneering exploration of the causal relationships between gut microbiota and multiple subtypes of dementia. Our objective was to compare both shared and distinct mechanisms underlying the associations between different subtypes of dementia and gut microbiota.
While multiple causal relationships between gut microorganisms and dementia were identified and their robustness confirmed, the present study also had several limitations. First, we primarily included European patients in the GWAS summary data. The demographic composition of the analyzed datasets could impact the generalizability of the study findings. Second, in cases where only a few IVs met the stringent threshold (P < 5e-8), a relatively relaxed threshold (P < 1e-5 or P < 5e-6) was employed for the selection process. Third, among the outcome data, patients with outcomes other than Alzheimer’s dementia and vascular dementia were diagnosed based solely on meeting the criteria for dementia, without specifying the disease. Multiple types of diseases can lead to dementia, each with diverse and intricate underlying mechanisms.
In brief, the results of this study suggested that gut microbiota are causally associated with dementia. These findings provide innovative insights into the pathophysiology of the disease and may thus have important implications for its treatment and prevention. Gut microbiota also exhibit potential as therapeutic targets for dementia, irrespective of whether they act as risk or protective factors. Conversely, dementia and its subtypes were found to influence the abundance of several gut microbes. Accordingly, these bacteria may serve as biomarkers for assessing the occurrence and progression of dementia.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s classification
Scientific Quality: Grade B
Novelty: Grade A
Creativity or Innovation: Grade A
Scientific Significance: Grade B
P-Reviewer: Hakim GD, Turkey S-Editor: Che XX L-Editor: A P-Editor: Zhao S
| 1. | Arvanitakis Z, Bennett DA. What Is Dementia? JAMA. 2019;322:1728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 2. | Gale SA, Acar D, Daffner KR. Dementia. Am J Med. 2018;131:1161-1169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 390] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 3. | GBD 2019 Dementia Forecasting Collaborators. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: an analysis for the Global Burden of Disease Study 2019. Lancet Public Health. 2022;7:e105-e125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2320] [Cited by in RCA: 2089] [Article Influence: 696.3] [Reference Citation Analysis (1)] |
| 4. | Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chételat G, Teunissen CE, Cummings J, van der Flier WM. Alzheimer's disease. Lancet. 2021;397:1577-1590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1527] [Cited by in RCA: 2570] [Article Influence: 642.5] [Reference Citation Analysis (0)] |
| 5. | O'Brien JT, Thomas A. Vascular dementia. Lancet. 2015;386:1698-1706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 723] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 6. | Walker Z, Possin KL, Boeve BF, Aarsland D. Lewy body dementias. Lancet. 2015;386:1683-1697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 376] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 7. | Bang J, Spina S, Miller BL. Frontotemporal dementia. Lancet. 2015;386:1672-1682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 575] [Cited by in RCA: 669] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 8. | Cryan JF, O'Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, Codagnone MG, Cussotto S, Fulling C, Golubeva AV, Guzzetta KE, Jaggar M, Long-Smith CM, Lyte JM, Martin JA, Molinero-Perez A, Moloney G, Morelli E, Morillas E, O'Connor R, Cruz-Pereira JS, Peterson VL, Rea K, Ritz NL, Sherwin E, Spichak S, Teichman EM, van de Wouw M, Ventura-Silva AP, Wallace-Fitzsimons SE, Hyland N, Clarke G, Dinan TG. The Microbiota-Gut-Brain Axis. Physiol Rev. 2019;99:1877-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1156] [Cited by in RCA: 2729] [Article Influence: 454.8] [Reference Citation Analysis (2)] |
| 9. | Tremlett H, Bauer KC, Appel-Cresswell S, Finlay BB, Waubant E. The gut microbiome in human neurological disease: A review. Ann Neurol. 2017;81:369-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 362] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 10. | Chen C, Liao J, Xia Y, Liu X, Jones R, Haran J, McCormick B, Sampson TR, Alam A, Ye K. Gut microbiota regulate Alzheimer's disease pathologies and cognitive disorders via PUFA-associated neuroinflammation. Gut. 2022;71:2233-2252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 290] [Cited by in RCA: 258] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 11. | Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, Johnson SC, Carlsson CM, Asthana S, Zetterberg H, Blennow K, Bendlin BB, Rey FE. Gut microbiome alterations in Alzheimer's disease. Sci Rep. 2017;7:13537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1248] [Cited by in RCA: 1370] [Article Influence: 171.3] [Reference Citation Analysis (0)] |
| 12. | Zhuang ZQ, Shen LL, Li WW, Fu X, Zeng F, Gui L, Lü Y, Cai M, Zhu C, Tan YL, Zheng P, Li HY, Zhu J, Zhou HD, Bu XL, Wang YJ. Gut Microbiota is Altered in Patients with Alzheimer's Disease. J Alzheimers Dis. 2018;63:1337-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 589] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 13. | Li S, Shao Y, Li K, HuangFu C, Wang W, Liu Z, Cai Z, Zhao B. Vascular Cognitive Impairment and the Gut Microbiota. J Alzheimers Dis. 2018;63:1209-1222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 14. | Nishiwaki H, Ueyama J, Kashihara K, Ito M, Hamaguchi T, Maeda T, Tsuboi Y, Katsuno M, Hirayama M, Ohno K. Gut microbiota in dementia with Lewy bodies. NPJ Parkinsons Dis. 2022;8:169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 15. | Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization. JAMA. 2017;318:1925-1926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 2125] [Article Influence: 265.6] [Reference Citation Analysis (0)] |
| 16. | Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, VanderWeele TJ, Higgins JPT, Timpson NJ, Dimou N, Langenberg C, Golub RM, Loder EW, Gallo V, Tybjaerg-Hansen A, Davey Smith G, Egger M, Richards JB. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. JAMA. 2021;326:1614-1621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 828] [Cited by in RCA: 1990] [Article Influence: 497.5] [Reference Citation Analysis (0)] |
| 17. | Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, Le Roy CI, Raygoza Garay JA, Finnicum CT, Liu X, Zhernakova DV, Bonder MJ, Hansen TH, Frost F, Rühlemann MC, Turpin W, Moon JY, Kim HN, Lüll K, Barkan E, Shah SA, Fornage M, Szopinska-Tokov J, Wallen ZD, Borisevich D, Agreus L, Andreasson A, Bang C, Bedrani L, Bell JT, Bisgaard H, Boehnke M, Boomsma DI, Burk RD, Claringbould A, Croitoru K, Davies GE, van Duijn CM, Duijts L, Falony G, Fu J, van der Graaf A, Hansen T, Homuth G, Hughes DA, Ijzerman RG, Jackson MA, Jaddoe VWV, Joossens M, Jørgensen T, Keszthelyi D, Knight R, Laakso M, Laudes M, Launer LJ, Lieb W, Lusis AJ, Masclee AAM, Moll HA, Mujagic Z, Qibin Q, Rothschild D, Shin H, Sørensen SJ, Steves CJ, Thorsen J, Timpson NJ, Tito RY, Vieira-Silva S, Völker U, Völzke H, Võsa U, Wade KH, Walter S, Watanabe K, Weiss S, Weiss FU, Weissbrod O, Westra HJ, Willemsen G, Payami H, Jonkers DMAE, Arias Vasquez A, de Geus EJC, Meyer KA, Stokholm J, Segal E, Org E, Wijmenga C, Kim HL, Kaplan RC, Spector TD, Uitterlinden AG, Rivadeneira F, Franke A, Lerch MM, Franke L, Sanna S, D'Amato M, Pedersen O, Paterson AD, Kraaij R, Raes J, Zhernakova A. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53:156-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 1022] [Article Influence: 255.5] [Reference Citation Analysis (0)] |
| 18. | Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, Loukola A, Lahtela E, Mattsson H, Laiho P, Della Briotta Parolo P, Lehisto AA, Kanai M, Mars N, Rämö J, Kiiskinen T, Heyne HO, Veerapen K, Rüeger S, Lemmelä S, Zhou W, Ruotsalainen S, Pärn K, Hiekkalinna T, Koskelainen S, Paajanen T, Llorens V, Gracia-Tabuenca J, Siirtola H, Reis K, Elnahas AG, Sun B, Foley CN, Aalto-Setälä K, Alasoo K, Arvas M, Auro K, Biswas S, Bizaki-Vallaskangas A, Carpen O, Chen CY, Dada OA, Ding Z, Ehm MG, Eklund K, Färkkilä M, Finucane H, Ganna A, Ghazal A, Graham RR, Green EM, Hakanen A, Hautalahti M, Hedman ÅK, Hiltunen M, Hinttala R, Hovatta I, Hu X, Huertas-Vazquez A, Huilaja L, Hunkapiller J, Jacob H, Jensen JN, Joensuu H, John S, Julkunen V, Jung M, Junttila J, Kaarniranta K, Kähönen M, Kajanne R, Kallio L, Kälviäinen R, Kaprio J; FinnGen, Kerimov N, Kettunen J, Kilpeläinen E, Kilpi T, Klinger K, Kosma VM, Kuopio T, Kurra V, Laisk T, Laukkanen J, Lawless N, Liu A, Longerich S, Mägi R, Mäkelä J, Mäkitie A, Malarstig A, Mannermaa A, Maranville J, Matakidou A, Meretoja T, Mozaffari SV, Niemi MEK, Niemi M, Niiranen T, O Donnell CJ, Obeidat ME, Okafo G, Ollila HM, Palomäki A, Palotie T, Partanen J, Paul DS, Pelkonen M, Pendergrass RK, Petrovski S, Pitkäranta A, Platt A, Pulford D, Punkka E, Pussinen P, Raghavan N, Rahimov F, Rajpal D, Renaud NA, Riley-Gillis B, Rodosthenous R, Saarentaus E, Salminen A, Salminen E, Salomaa V, Schleutker J, Serpi R, Shen HY, Siegel R, Silander K, Siltanen S, Soini S, Soininen H, Sul JH, Tachmazidou I, Tasanen K, Tienari P, Toppila-Salmi S, Tukiainen T, Tuomi T, Turunen JA, Ulirsch JC, Vaura F, Virolainen P, Waring J, Waterworth D, Yang R, Nelis M, Reigo A, Metspalu A, Milani L, Esko T, Fox C, Havulinna AS, Perola M, Ripatti S, Jalanko A, Laitinen T, Mäkelä TP, Plenge R, McCarthy M, Runz H, Daly MJ, Palotie A. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508-518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1241] [Cited by in RCA: 1957] [Article Influence: 978.5] [Reference Citation Analysis (0)] |
| 19. | Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, Young R, Butterworth AS. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207-3209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 541] [Cited by in RCA: 1045] [Article Influence: 116.1] [Reference Citation Analysis (0)] |
| 20. | Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, Butterworth AS, Staley JR. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851-4853. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 1419] [Article Influence: 283.8] [Reference Citation Analysis (0)] |
| 21. | Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 1195] [Article Influence: 79.7] [Reference Citation Analysis (0)] |
| 22. | Li H, Zhang X, Wang Z, Feng S, Zhang G. Can Intelligence Affect Alcohol-, Smoking-, and Physical Activity-Related Behaviors? A Mendelian Randomization Study. J Intell. 2023;11:29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 23. | Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1330] [Cited by in RCA: 3719] [Article Influence: 309.9] [Reference Citation Analysis (1)] |
| 24. | Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26:2333-2355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 368] [Cited by in RCA: 1063] [Article Influence: 106.3] [Reference Citation Analysis (0)] |
| 25. | Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4015] [Cited by in RCA: 5629] [Article Influence: 625.4] [Reference Citation Analysis (0)] |
| 26. | Bowden J, Del Greco M F, Minelli C, Zhao Q, Lawlor DA, Sheehan NA, Thompson J, Davey Smith G. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int J Epidemiol. 2019;48:728-742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 596] [Article Influence: 99.3] [Reference Citation Analysis (1)] |
| 27. | Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2275] [Cited by in RCA: 6144] [Article Influence: 614.4] [Reference Citation Analysis (0)] |
| 28. | Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693-698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2495] [Cited by in RCA: 5276] [Article Influence: 753.7] [Reference Citation Analysis (0)] |
| 29. | Sedgwick P. Multiple hypothesis testing and Bonferroni's correction. BMJ. 2014;349:g6284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 145] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 30. | Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4747] [Cited by in RCA: 4742] [Article Influence: 677.4] [Reference Citation Analysis (0)] |
| 31. | Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 411] [Cited by in RCA: 1388] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 32. | Xiang K, Wang P, Xu Z, Hu YQ, He YS, Chen Y, Feng YT, Yin KJ, Huang JX, Wang J, Wu ZD, Yang XK, Wang DG, Ye DQ, Pan HF. Causal Effects of Gut Microbiome on Systemic Lupus Erythematosus: A Two-Sample Mendelian Randomization Study. Front Immunol. 2021;12:667097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 136] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 33. | Liu P, Wu L, Peng G, Han Y, Tang R, Ge J, Zhang L, Jia L, Yue S, Zhou K, Li L, Luo B, Wang B. Altered microbiomes distinguish Alzheimer's disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav Immun. 2019;80:633-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 420] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 34. | Hou M, Xu G, Ran M, Luo W, Wang H. APOE-ε4 Carrier Status and Gut Microbiota Dysbiosis in Patients With Alzheimer Disease. Front Neurosci. 2021;15:619051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 35. | Kameno K, Hasegawa Y, Hayashi K, Takemoto Y, Uchikawa H, Mukasa A, Kim-Mitsuyama S. Loss of body weight in old 5xFAD mice and the alteration of gut microbiota composition. Exp Gerontol. 2022;166:111885. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 36. | Zhang J, Hao J, Liu R, Wu T, Sui W, Zhang M. Hawthorn flavonoid ameliorates cognitive deficit in mice with Alzheimer's disease by increasing the levels of Bifidobacteriales in gut microbiota and docosapentaenoic acid in serum metabolites. Food Funct. 2022;13:12371-12382. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 37. | Yusufu I, Ding K, Smith K, Wankhade UD, Sahay B, Patterson GT, Pacholczyk R, Adusumilli S, Hamrick MW, Hill WD, Isales CM, Fulzele S. A Tryptophan-Deficient Diet Induces Gut Microbiota Dysbiosis and Increases Systemic Inflammation in Aged Mice. Int J Mol Sci. 2021;22:5005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 38. | Zhu Z, Ma X, Wu J, Xiao Z, Wu W, Ding S, Zheng L, Liang X, Luo J, Ding D, Zhao Q. Altered Gut Microbiota and Its Clinical Relevance in Mild Cognitive Impairment and Alzheimer's Disease: Shanghai Aging Study and Shanghai Memory Study. Nutrients. 2022;14:3959. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |