Published online May 16, 2024. doi: 10.12998/wjcc.v12.i14.2359
Revised: February 18, 2024
Accepted: April 2, 2024
Published online: May 16, 2024
Processing time: 118 Days and 3 Hours
Non-alcoholic fatty liver disease (NAFLD) and alcohol-related liver disease (Ar
To investigate the correlation between fatty liver and mental disorders, thus ne
Data on NAFLD and ArLD were retrieved from the genome-wide association studies catalog, while information on mental disorders, including Alzheimer's disease, schizophrenia, anxiety disorder, attention deficit hyperactivity disorder (ADHD), bipolar disorder, major depressive disorder, multiple personality dis
After excluding weak instrumental variables, a causal relationship was identified between fatty liver disease and the occurrence and development of some psychia
Associations were found between genetic predisposition to fatty liver disease and an increased risk of a broad range of psychiatric disorders, namely bipolar disorder, OCD, and PTSD, highlighting the significance of preven
Core Tip: Non-alcoholic fatty liver disease and alcohol-related liver disease are the predominant forms of chronic liver diseases, with their incidence gradually increasing due to changing lifestyle habits. Observational studies have indicated a potential association between fatty liver and psychiatric disorders, necessitating Mendelian randomization studies to eluci
- Citation: Xu WM, Zhang HF, Feng YH, Li SJ, Xie BY. Genetically predicted fatty liver disease and risk of psychiatric disorders: A mendelian randomization study. World J Clin Cases 2024; 12(14): 2359-2369
- URL: https://www.wjgnet.com/2307-8960/full/v12/i14/2359.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i14.2359
As is extensively documented, non-alcoholic fatty liver disease (NAFLD) and alcohol-related liver disease (ArLD) have emerged as the most important causes of hepatic injury, and their incidence is steadily increasing due to changes in lifestyle habits[1]. The former is a chronic liver condition associated with obesity and metabolic syndrome, with a pre
While psychiatric disorders can be treated via various approaches, according to the World Health Organization (WHO), their incidence increases on a yearly basis[6]. Although the relationship between fatty liver and psychiatric disorders remains elusive, a higher prevalence of metabolic syndrome is observed in patients with psychiatric disorders[7]. Common psychiatric disorders, such as bipolar disorder, depressive disorder, or schizophrenia, may be associated with metabolic syndrome or substance abuse[8]. Elwing et al[9] observed a higher prevalence of major depression as well as anxiety disorders in NAFLD patients. At the same time, the incidence of ArLD is closely associated with excessive and chronic alcohol consumption, with addictive misuse of alcohol considered a highly prevalent psychiatric disorder[10]. For instance, anxiety is a common comorbidity in patients with alcohol abuse, causing significant discomfort and cognitive impairment[11].
Mendelian randomization (MR) is an approach for investigating causality between exposures and outcomes of interest[12] that utilizes single nucleotide polymorphisms (SNPs) as unconfounded proxies for exposures, thereby circumventing residual confounders and reverse causality commonly present in conventional observational studies[13]. The MR design represents a crucial strategy for causal inference without randomized clinical trials (RCTs), given that genetic variants are randomly assorted during meiosis, mimicking an RCT[14]. There is a non-negligible relationship between fatty liver and mental disorders, necessitating a comprehensive understanding of their shared characteristics to facilitate the develop
In this study, all data were derived from the Genetic Alliance's publicly available compilation of statistical data from genome-wide association studies (GWAS). All original studies underwent a thorough ethical review process and ob
Summary statistics on fatty liver disease and psychiatric disorders were collected from published GWAS to explore the causal effect of fatty liver disease on the risk of psychiatric disorders using two-sample MR.
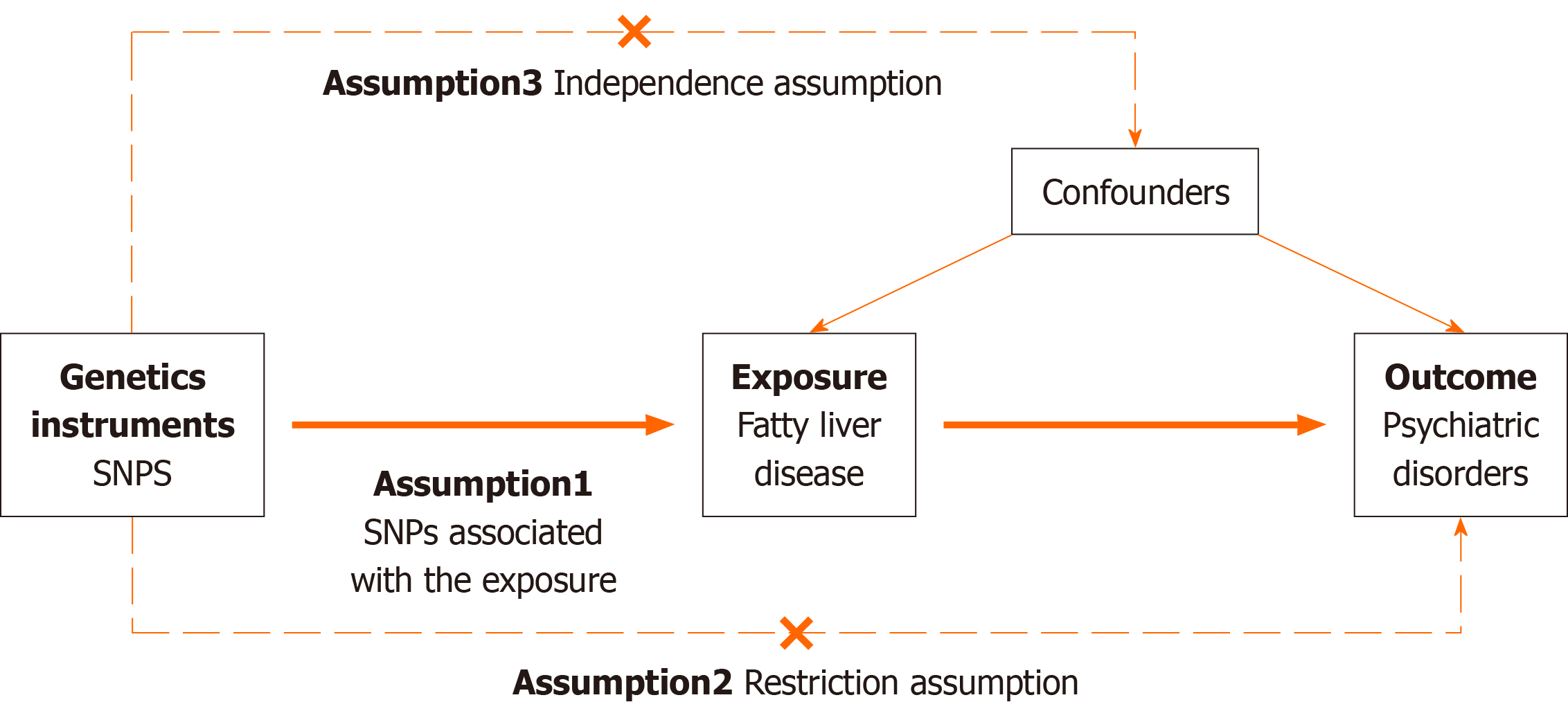
The MR Approach was constructed based on three primary assumptions: (1) Genetic variants as instrumental variables (IVs) should be significantly associated with the risk factor of interest; (2) the genetic variants used should not be asso
Summary statistics on fatty liver disease were downloaded from published GWAS. The GWAS Catalog database is pub
| Ref. | Outcome | PMID | Sample size | |
| Cases | Controls | |||
| Bellenguez et al[17], 2022 | Alzheimer’s disease | 35379992 | 39106 | 401577 |
| Watson et al[18], 2019 | Anorexia nervosa | 31308545 | 16992 | 55525 |
| Schoeler et al[19], 2023 | Anxiety disorder | 37106081 | 282802 | |
| Demontis et al[20], 2023 | Attention deficit hyperactivity disorder | 36702997 | 38691 | 186843 |
| Stahl et al[21], 2019 | Bipolar disorder | 31043756 | 20352 | 31358 |
| Howard et al[22], 2019 | Major depressive disorder | 30718901 | 246363 | 561190 |
| Cross-Disorder Group of the Psychiatric Genomics Consortium[23], 2019 | Multiple disorders | 31835028 | 232964 | 494162 |
| IOCDF-GC, OCGAS[24], 2018 | Obsessive-compulsive disorder | 28761083 | 2688 | 7037 |
| Nievergelt et al[25], 2019 | Post-traumatic stress disorder | 31594949 | 30000 | 170000 |
| Trubetskoy et al[26], 2022 | Schizophrenia | 35396580 | 76775 | 243649 |
The genetic instruments for each exposure trait or disease were meticulously chosen from the corresponding GWASs, surpassing the threshold of genome-wide significance (P < 5 × 108). Independent SNPs were defined by R2 < 0.001 and clump window > 10 kb without linkage disequilibrium (LD) were proposed as instrumental variables. LD among SNPs for each risk factor was calculated based on 1000 genomes LD reference panel European population[27] using the PLINK clumping approach (PLINK: a tool set for whole-genome association and population-based linkage analyses)[28].
The variance in fatty liver disease explained by the IVs was calculated, and weak IVs bias was analyzed using F-statistics. R2 was calculated based on the effect estimates (β) and allele frequencies (EAF) of each single SNP using the following formula: R2 = 2× EAF (1-EAF) × β2[29]. The F value was further calculated according to the formula F = R2 × (N-2)/(1-R2)[30]. F value > 10 was considered a strong genetic IV; otherwise, the SNP was discarded.
An assessment of heterogeneity across SNPs was conducted using Cochran's Q statistics. The primary analytical method used to examine causal associations was the random-effect inverse-variance-weighted model[31]. The MR-Egger method was used to determine the pleiotropic effects of the instrumental SNPs[32].
F values for each SNP were individually calculated, and SNPs with values greater than 10 were retained, suggesting a low risk of bias due to weak IVs. Consequently, all SNPs in this study had F-statistics larger than 10 (Table 2).
| Exposure/Outcome | SNPs | R2 | F-statistic |
| Alcohol-related liver disease | |||
| Alzheimer’s disease | / | / | / |
| Anorexia nervosa, anxiety disorder, attention deficit hyperactivity disorder, bipolar disorder, major depressive disorder, multiple disorders, obsessive-compulsive disorder, post-traumatic stress disorder, schizophrenia | rs738408 | 0.11 | 57352.08 |
| Non-alcoholic fatty liver disease | |||
| Alzheimer’s disease | rs28601761, rs3747207, rs73001065 | 0.04 | 35089.81 |
| Anorexia nervosa, anxiety disorder, attention deficit hyperactivity disorder, bipolar disorder, major depressive disorder, multiple disorders, obsessive-compulsive disorder, post-traumatic stress disorder, schizophrenia | rs3747207, rs429358 | 0.02 | 26942.59 |
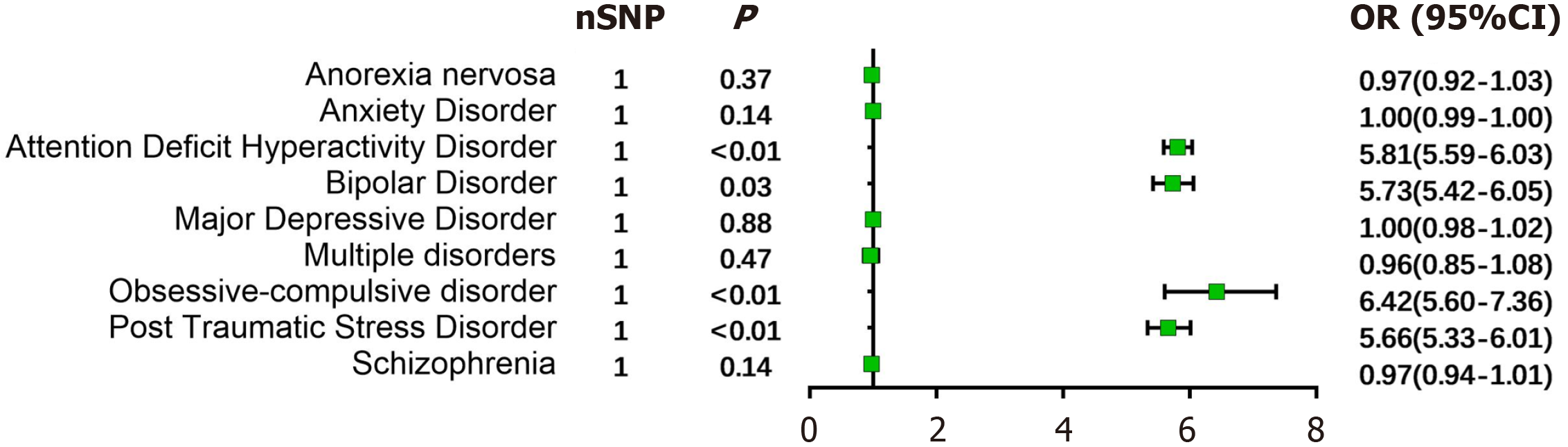
ArLD: In the ArLD population, one SNP (rs738408) was retained following screening and filtering. The findings indicated that ArLD significantly elevates the likelihood of developing attention deficit hyperactivity disorder (ADHD) (OR: 5.81, 95%CI: 5.59-6.03, P < 0.01), bipolar disorder (BD) (OR: 5.73, 95%CI: 5.42-6.05, P = 0.03), obsessive-compulsive disorder (OCD) (OR: 6.42, 95%CI: 5.60-7.36, P < 0.01), and post-traumatic stress disorder (PTSD) (OR: 5.66, 95%CI: 5.33-6.01, P < 0.01). In contrast, there was no evidence suggesting that ArLD increased the risk of anorexia nervosa (OR: 0.97, P = 0.37), anxiety disorder (OR: 1.00, P = 0.14), major depressive disorder (MDD) (OR: 1.00, P = 0.88), multiple personality disorders (OR: 0.96, P = 0.47), and schizophrenia (OR: 0.97, P = 0.14) (Figure 2). Considering that only one SNP was included, tests for heterogeneity and pleiotropy could not be performed.
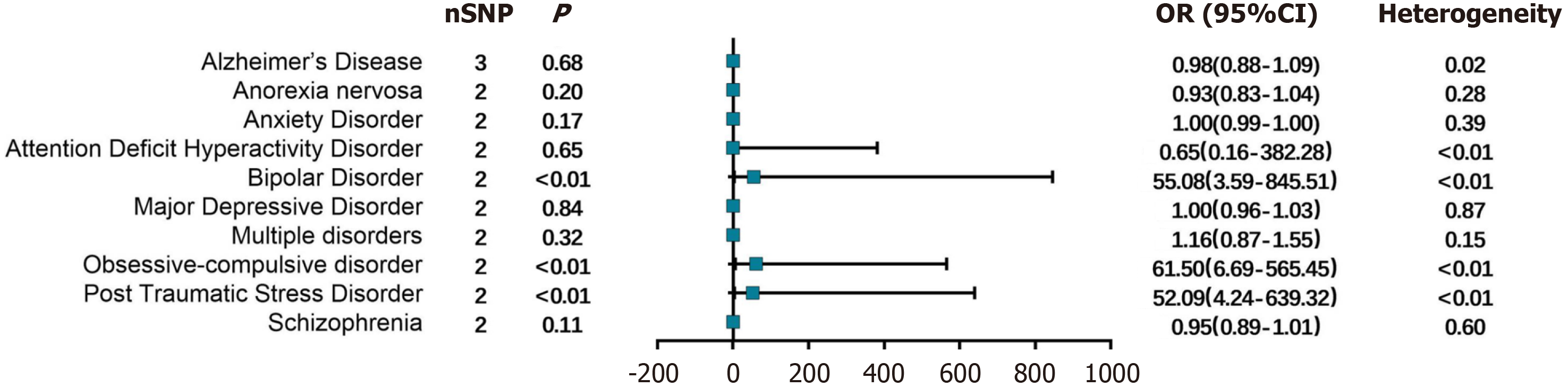
NAFLD: In the NAFLD population, 3 SNPs (rs28601761, rs3747207, and rs73001065) were included in the analysis of their association with Alzheimer's disease. At the same time, 2 SNPs (rs3747207 and rs429358) were analyzed for their asso
As fatty liver disease has emerged as a leading cause of chronic liver disease worldwide, and with a deeper understand
According to the WHO, 5.1% of the global burden of disease can be attributable to alcohol misuse, ranking it as the seventh leading risk factor globally[33]. Of note, persistent alcohol use disorder is strongly correlated with progressive hepatic damage and increased mortality[34]. Among patients diagnosed with fatty liver disease, non-interrupted con
Alcohol dependence and BD frequently coexist, with chronic alcoholism being associated with 24%-62% of BD cases worldwide[42]. Prolonged alcohol consumption can exert detrimental effects on both the immune system and nervous system while also increasing the frequency and severity of emotional episodes in individuals with BD[43]. Abnormalities in the N-methyl-d-asperate (NMDA) receptor play a critical role in the occurrence and development of BD. Conversely, ethanol acts on the brain to inhibit NMDA receptors, thereby elevating the risk of BD[44]. However, the risks of fatty liver appear to outweigh the risks associated with alcohol abuse, and patients with NAFLD seem to be at a higher risk com
NAFLD is closely linked to metabolic disorder syndrome that is characterized by obesity, insulin resistance, and hyperlipidemia[50]. Animal experiments have validated that metabolic syndrome can lead to astrogliosis and micro
The combination of fatty liver and persistent inflammation is associated with neuroinflammation, disruptions in neu
This is the first study to explore the causal relationship between psychiatric disorders and fatty liver disease using a two-sample MR analysis with pooled GWAS-level statistics, thereby minimizing potential confounding and reverse cau
In summary, this MR study provides genetic evidence supporting a causal relationship between fatty liver disease and psychiatric disorders. Our results collectively suggest that ArLD is a risk factor for ADHD, bipolar disorder, OCD, and PTSD. Moreover, our findings highlight a correlation between the presence of NAFLD and a higher risk of bipolar disorder, OCD, and PTSD. Thus, patients with fatty liver disease should be more vigilant to prevent the onset of mental disorders.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Khan MM, India S-Editor: Liu JH L-Editor: A P-Editor: Xu ZH
| 1. | Odriozola A, Santos-Laso A, Del Barrio M, Cabezas J, Iruzubieta P, Arias-Loste MT, Rivas C, Duque JCR, Antón Á, Fábrega E, Crespo J. Fatty Liver Disease, Metabolism and Alcohol Interplay: A Comprehensive Review. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 2. | Cotter TG, Rinella M. Nonalcoholic Fatty Liver Disease 2020: The State of the Disease. Gastroenterology. 2020;158:1851-1864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 842] [Article Influence: 168.4] [Reference Citation Analysis (2)] |
| 3. | Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62:S47-S64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1516] [Cited by in RCA: 2140] [Article Influence: 214.0] [Reference Citation Analysis (0)] |
| 4. | Liu SY, Tsai IT, Hsu YC. Alcohol-Related Liver Disease: Basic Mechanisms and Clinical Perspectives. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 5. | Saiman Y, Duarte-Rojo A, Rinella ME. Fatty Liver Disease: Diagnosis and Stratification. Annu Rev Med. 2022;73:529-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 6. | Jahani R, Khaledyan D, Jahani A, Jamshidi E, Kamalinejad M, Khoramjouy M, Faizi M. Evaluation and comparison of the antidepressant-like activity of Artemisia dracunculus and Stachys lavandulifolia ethanolic extracts: an in vivo study. Res Pharm Sci. 2019;14:544-553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 7. | Soto-Angona Ó, Anmella G, Valdés-Florido MJ, De Uribe-Viloria N, Carvalho AF, Penninx BWJH, Berk M. Non-alcoholic fatty liver disease (NAFLD) as a neglected metabolic companion of psychiatric disorders: common pathways and future approaches. BMC Med. 2020;18:261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 8. | Penninx BWJH, Lange SMM. Metabolic syndrome in psychiatric patients: overview, mechanisms, and implications. Dialogues Clin Neurosci. 2018;20:63-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 347] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 9. | Elwing JE, Lustman PJ, Wang HL, Clouse RE. Depression, anxiety, and nonalcoholic steatohepatitis. Psychosom Med. 2006;68:563-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 10. | Chivero ET, Sil S, Kumar M, Buch S. Substance use, microbiome and psychiatric disorders. Pharmacol Biochem Behav. 2022;219:173432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 11. | Anker JJ, Kushner MG. Co-Occurring Alcohol Use Disorder and Anxiety: Bridging Psychiatric, Psychological, and Neurobiological Perspectives. Alcohol Res. 2019;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 12. | Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1354] [Cited by in RCA: 2607] [Article Influence: 372.4] [Reference Citation Analysis (0)] |
| 13. | Smith GD, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;4:e352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 438] [Cited by in RCA: 398] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 14. | Pingault JB, O'Reilly PF, Schoeler T, Ploubidis GB, Rijsdijk F, Dudbridge F. Using genetic data to strengthen causal inference in observational research. Nat Rev Genet. 2018;19:566-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 304] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 15. | Jiang L, Zheng Z, Fang H, Yang J. A generalized linear mixed model association tool for biobank-scale data. Nat Genet. 2021;53:1616-1621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 350] [Article Influence: 87.5] [Reference Citation Analysis (0)] |
| 16. | Ghodsian N, Abner E, Emdin CA, Gobeil É, Taba N, Haas ME, Perrot N, Manikpurage HD, Gagnon É, Bourgault J, St-Amand A, Couture C, Mitchell PL, Bossé Y, Mathieu P, Vohl MC, Tchernof A, Thériault S, Khera AV, Esko T, Arsenault BJ. Electronic health record-based genome-wide meta-analysis provides insights on the genetic architecture of non-alcoholic fatty liver disease. Cell Rep Med. 2021;2:100437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 106] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 17. | Bellenguez C, Küçükali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, Naj AC, Campos-Martin R, Grenier-Boley B, Andrade V, Holmans PA, Boland A, Damotte V, van der Lee SJ, Costa MR, Kuulasmaa T, Yang Q, de Rojas I, Bis JC, Yaqub A, Prokic I, Chapuis J, Ahmad S, Giedraitis V, Aarsland D, Garcia-Gonzalez P, Abdelnour C, Alarcón-Martín E, Alcolea D, Alegret M, Alvarez I, Álvarez V, Armstrong NJ, Tsolaki A, Antúnez C, Appollonio I, Arcaro M, Archetti S, Pastor AA, Arosio B, Athanasiu L, Bailly H, Banaj N, Baquero M, Barral S, Beiser A, Pastor AB, Below JE, Benchek P, Benussi L, Berr C, Besse C, Bessi V, Binetti G, Bizarro A, Blesa R, Boada M, Boerwinkle E, Borroni B, Boschi S, Bossù P, Bråthen G, Bressler J, Bresner C, Brodaty H, Brookes KJ, Brusco LI, Buiza-Rueda D, Bûrger K, Burholt V, Bush WS, Calero M, Cantwell LB, Chene G, Chung J, Cuccaro ML, Carracedo Á, Cecchetti R, Cervera-Carles L, Charbonnier C, Chen HH, Chillotti C, Ciccone S, Claassen JAHR, Clark C, Conti E, Corma-Gómez A, Costantini E, Custodero C, Daian D, Dalmasso MC, Daniele A, Dardiotis E, Dartigues JF, de Deyn PP, de Paiva Lopes K, de Witte LD, Debette S, Deckert J, Del Ser T, Denning N, DeStefano A, Dichgans M, Diehl-Schmid J, Diez-Fairen M, Rossi PD, Djurovic S, Duron E, Düzel E, Dufouil C, Eiriksdottir G, Engelborghs S, Escott-Price V, Espinosa A, Ewers M, Faber KM, Fabrizio T, Nielsen SF, Fardo DW, Farotti L, Fenoglio C, Fernández-Fuertes M, Ferrari R, Ferreira CB, Ferri E, Fin B, Fischer P, Fladby T, Fließbach K, Fongang B, Fornage M, Fortea J, Foroud TM, Fostinelli S, Fox NC, Franco-Macías E, Bullido MJ, Frank-García A, Froelich L, Fulton-Howard B, Galimberti D, García-Alberca JM, García-González P, Garcia-Madrona S, Garcia-Ribas G, Ghidoni R, Giegling I, Giorgio G, Goate AM, Goldhardt O, Gomez-Fonseca D, González-Pérez A, Graff C, Grande G, Green E, Grimmer T, Grünblatt E, Grunin M, Gudnason V, Guetta-Baranes T, Haapasalo A, Hadjigeorgiou G, Haines JL, Hamilton-Nelson KL, Hampel H, Hanon O, Hardy J, Hartmann AM, Hausner L, Harwood J, Heilmann-Heimbach S, Helisalmi S, Heneka MT, Hernández I, Herrmann MJ, Hoffmann P, Holmes C, Holstege H, Vilas RH, Hulsman M, Humphrey J, Biessels GJ, Jian X, Johansson C, Jun GR, Kastumata Y, Kauwe J, Kehoe PG, Kilander L, Ståhlbom AK, Kivipelto M, Koivisto A, Kornhuber J, Kosmidis MH, Kukull WA, Kuksa PP, Kunkle BW, Kuzma AB, Lage C, Laukka EJ, Launer L, Lauria A, Lee CY, Lehtisalo J, Lerch O, Lleó A, Longstreth W Jr, Lopez O, de Munain AL, Love S, Löwemark M, Luckcuck L, Lunetta KL, Ma Y, Macías J, MacLeod CA, Maier W, Mangialasche F, Spallazzi M, Marquié M, Marshall R, Martin ER, Montes AM, Rodríguez CM, Masullo C, Mayeux R, Mead S, Mecocci P, Medina M, Meggy A, Mehrabian S, Mendoza S, Menéndez-González M, Mir P, Moebus S, Mol M, Molina-Porcel L, Montrreal L, Morelli L, Moreno F, Morgan K, Mosley T, Nöthen MM, Muchnik C, Mukherjee S, Nacmias B, Ngandu T, Nicolas G, Nordestgaard BG, Olaso R, Orellana A, Orsini M, Ortega G, Padovani A, Paolo C, Papenberg G, Parnetti L, Pasquier F, Pastor P, Peloso G, Pérez-Cordón A, Pérez-Tur J, Pericard P, Peters O, Pijnenburg YAL, Pineda JA, Piñol-Ripoll G, Pisanu C, Polak T, Popp J, Posthuma D, Priller J, Puerta R, Quenez O, Quintela I, Thomassen JQ, Rábano A, Rainero I, Rajabli F, Ramakers I, Real LM, Reinders MJT, Reitz C, Reyes-Dumeyer D, Ridge P, Riedel-Heller S, Riederer P, Roberto N, Rodriguez-Rodriguez E, Rongve A, Allende IR, Rosende-Roca M, Royo JL, Rubino E, Rujescu D, Sáez ME, Sakka P, Saltvedt I, Sanabria Á, Sánchez-Arjona MB, Sanchez-Garcia F, Juan PS, Sánchez-Valle R, Sando SB, Sarnowski C, Satizabal CL, Scamosci M, Scarmeas N, Scarpini E, Scheltens P, Scherbaum N, Scherer M, Schmid M, Schneider A, Schott JM, Selbæk G, Seripa D, Serrano M, Sha J, Shadrin AA, Skrobot O, Slifer S, Snijders GJL, Soininen H, Solfrizzi V, Solomon A, Song Y, Sorbi S, Sotolongo-Grau O, Spalletta G, Spottke A, Squassina A, Stordal E, Tartan JP, Tárraga L, Tesí N, Thalamuthu A, Thomas T, Tosto G, Traykov L, Tremolizzo L, Tybjærg-Hansen A, Uitterlinden A, Ullgren A, Ulstein I, Valero S, Valladares O, Broeckhoven CV, Vance J, Vardarajan BN, van der Lugt A, Dongen JV, van Rooij J, van Swieten J, Vandenberghe R, Verhey F, Vidal JS, Vogelgsang J, Vyhnalek M, Wagner M, Wallon D, Wang LS, Wang R, Weinhold L, Wiltfang J, Windle G, Woods B, Yannakoulia M, Zare H, Zhao Y, Zhang X, Zhu C, Zulaica M; EADB; GR@ACE; DEGESCO; EADI; GERAD; Demgene; FinnGen; ADGC; CHARGE, Farrer LA, Psaty BM, Ghanbari M, Raj T, Sachdev P, Mather K, Jessen F, Ikram MA, de Mendonça A, Hort J, Tsolaki M, Pericak-Vance MA, Amouyel P, Williams J, Frikke-Schmidt R, Clarimon J, Deleuze JF, Rossi G, Seshadri S, Andreassen OA, Ingelsson M, Hiltunen M, Sleegers K, Schellenberg GD, van Duijn CM, Sims R, van der Flier WM, Ruiz A, Ramirez A, Lambert JC. New insights into the genetic etiology of Alzheimer's disease and related dementias. Nat Genet. 2022;54:412-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 665] [Cited by in RCA: 1339] [Article Influence: 446.3] [Reference Citation Analysis (0)] |
| 18. | Watson HJ, Yilmaz Z, Thornton LM, Hübel C, Coleman JRI, Gaspar HA, Bryois J, Hinney A, Leppä VM, Mattheisen M, Medland SE, Ripke S, Yao S, Giusti-Rodríguez P; Anorexia Nervosa Genetics Initiative, Hanscombe KB, Purves KL; Eating Disorders Working Group of the Psychiatric Genomics Consortium, Adan RAH, Alfredsson L, Ando T, Andreassen OA, Baker JH, Berrettini WH, Boehm I, Boni C, Perica VB, Buehren K, Burghardt R, Cassina M, Cichon S, Clementi M, Cone RD, Courtet P, Crow S, Crowley JJ, Danner UN, Davis OSP, de Zwaan M, Dedoussis G, Degortes D, DeSocio JE, Dick DM, Dikeos D, Dina C, Dmitrzak-Weglarz M, Docampo E, Duncan LE, Egberts K, Ehrlich S, Escaramís G, Esko T, Estivill X, Farmer A, Favaro A, Fernández-Aranda F, Fichter MM, Fischer K, Föcker M, Foretova L, Forstner AJ, Forzan M, Franklin CS, Gallinger S, Giegling I, Giuranna J, Gonidakis F, Gorwood P, Mayora MG, Guillaume S, Guo Y, Hakonarson H, Hatzikotoulas K, Hauser J, Hebebrand J, Helder SG, Herms S, Herpertz-Dahlmann B, Herzog W, Huckins LM, Hudson JI, Imgart H, Inoko H, Janout V, Jiménez-Murcia S, Julià A, Kalsi G, Kaminská D, Kaprio J, Karhunen L, Karwautz A, Kas MJH, Kennedy JL, Keski-Rahkonen A, Kiezebrink K, Kim YR, Klareskog L, Klump KL, Knudsen GPS, La Via MC, Le Hellard S, Levitan RD, Li D, Lilenfeld L, Lin BD, Lissowska J, Luykx J, Magistretti PJ, Maj M, Mannik K, Marsal S, Marshall CR, Mattingsdal M, McDevitt S, McGuffin P, Metspalu A, Meulenbelt I, Micali N, Mitchell K, Monteleone AM, Monteleone P, Munn-Chernoff MA, Nacmias B, Navratilova M, Ntalla I, O'Toole JK, Ophoff RA, Padyukov L, Palotie A, Pantel J, Papezova H, Pinto D, Rabionet R, Raevuori A, Ramoz N, Reichborn-Kjennerud T, Ricca V, Ripatti S, Ritschel F, Roberts M, Rotondo A, Rujescu D, Rybakowski F, Santonastaso P, Scherag A, Scherer SW, Schmidt U, Schork NJ, Schosser A, Seitz J, Slachtova L, Slagboom PE, Slof-Op 't Landt MCT, Slopien A, Sorbi S, Świątkowska B, Szatkiewicz JP, Tachmazidou I, Tenconi E, Tortorella A, Tozzi F, Treasure J, Tsitsika A, Tyszkiewicz-Nwafor M, Tziouvas K, van Elburg AA, van Furth EF, Wagner G, Walton E, Widen E, Zeggini E, Zerwas S, Zipfel S, Bergen AW, Boden JM, Brandt H, Crawford S, Halmi KA, Horwood LJ, Johnson C, Kaplan AS, Kaye WH, Mitchell JE, Olsen CM, Pearson JF, Pedersen NL, Strober M, Werge T, Whiteman DC, Woodside DB, Stuber GD, Gordon S, Grove J, Henders AK, Juréus A, Kirk KM, Larsen JT, Parker R, Petersen L, Jordan J, Kennedy M, Montgomery GW, Wade TD, Birgegård A, Lichtenstein P, Norring C, Landén M, Martin NG, Mortensen PB, Sullivan PF, Breen G, Bulik CM. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat Genet. 2019;51:1207-1214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 705] [Cited by in RCA: 670] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 19. | Schoeler T, Speed D, Porcu E, Pirastu N, Pingault JB, Kutalik Z. Participation bias in the UK Biobank distorts genetic associations and downstream analyses. Nat Hum Behav. 2023;7:1216-1227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 174] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 20. | Demontis D, Walters GB, Athanasiadis G, Walters R, Therrien K, Nielsen TT, Farajzadeh L, Voloudakis G, Bendl J, Zeng B, Zhang W, Grove J, Als TD, Duan J, Satterstrom FK, Bybjerg-Grauholm J, Bækved-Hansen M, Gudmundsson OO, Magnusson SH, Baldursson G, Davidsdottir K, Haraldsdottir GS, Agerbo E, Hoffman GE, Dalsgaard S, Martin J, Ribasés M, Boomsma DI, Soler Artigas M, Roth Mota N, Howrigan D, Medland SE, Zayats T, Rajagopal VM; ADHD Working Group of the Psychiatric Genomics Consortium; iPSYCH-Broad Consortium, Nordentoft M, Mors O, Hougaard DM, Mortensen PB, Daly MJ, Faraone SV, Stefansson H, Roussos P, Franke B, Werge T, Neale BM, Stefansson K, Børglum AD. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat Genet. 2023;55:198-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 320] [Article Influence: 160.0] [Reference Citation Analysis (0)] |
| 21. | Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, Mattheisen M, Wang Y, Coleman JRI, Gaspar HA, de Leeuw CA, Steinberg S, Pavlides JMW, Trzaskowski M, Byrne EM, Pers TH, Holmans PA, Richards AL, Abbott L, Agerbo E, Akil H, Albani D, Alliey-Rodriguez N, Als TD, Anjorin A, Antilla V, Awasthi S, Badner JA, Bækvad-Hansen M, Barchas JD, Bass N, Bauer M, Belliveau R, Bergen SE, Pedersen CB, Bøen E, Boks MP, Boocock J, Budde M, Bunney W, Burmeister M, Bybjerg-Grauholm J, Byerley W, Casas M, Cerrato F, Cervantes P, Chambert K, Charney AW, Chen D, Churchhouse C, Clarke TK, Coryell W, Craig DW, Cruceanu C, Curtis D, Czerski PM, Dale AM, de Jong S, Degenhardt F, Del-Favero J, DePaulo JR, Djurovic S, Dobbyn AL, Dumont A, Elvsåshagen T, Escott-Price V, Fan CC, Fischer SB, Flickinger M, Foroud TM, Forty L, Frank J, Fraser C, Freimer NB, Frisén L, Gade K, Gage D, Garnham J, Giambartolomei C, Pedersen MG, Goldstein J, Gordon SD, Gordon-Smith K, Green EK, Green MJ, Greenwood TA, Grove J, Guan W, Guzman-Parra J, Hamshere ML, Hautzinger M, Heilbronner U, Herms S, Hipolito M, Hoffmann P, Holland D, Huckins L, Jamain S, Johnson JS, Juréus A, Kandaswamy R, Karlsson R, Kennedy JL, Kittel-Schneider S, Knowles JA, Kogevinas M, Koller AC, Kupka R, Lavebratt C, Lawrence J, Lawson WB, Leber M, Lee PH, Levy SE, Li JZ, Liu C, Lucae S, Maaser A, MacIntyre DJ, Mahon PB, Maier W, Martinsson L, McCarroll S, McGuffin P, McInnis MG, McKay JD, Medeiros H, Medland SE, Meng F, Milani L, Montgomery GW, Morris DW, Mühleisen TW, Mullins N, Nguyen H, Nievergelt CM, Adolfsson AN, Nwulia EA, O'Donovan C, Loohuis LMO, Ori APS, Oruc L, Ösby U, Perlis RH, Perry A, Pfennig A, Potash JB, Purcell SM, Regeer EJ, Reif A, Reinbold CS, Rice JP, Rivas F, Rivera M, Roussos P, Ruderfer DM, Ryu E, Sánchez-Mora C, Schatzberg AF, Scheftner WA, Schork NJ, Shannon Weickert C, Shehktman T, Shilling PD, Sigurdsson E, Slaney C, Smeland OB, Sobell JL, Søholm Hansen C, Spijker AT, St Clair D, Steffens M, Strauss JS, Streit F, Strohmaier J, Szelinger S, Thompson RC, Thorgeirsson TE, Treutlein J, Vedder H, Wang W, Watson SJ, Weickert TW, Witt SH, Xi S, Xu W, Young AH, Zandi P, Zhang P, Zöllner S; eQTLGen Consortium; BIOS Consortium, Adolfsson R, Agartz I, Alda M, Backlund L, Baune BT, Bellivier F, Berrettini WH, Biernacka JM, Blackwood DHR, Boehnke M, Børglum AD, Corvin A, Craddock N, Daly MJ, Dannlowski U, Esko T, Etain B, Frye M, Fullerton JM, Gershon ES, Gill M, Goes F, Grigoroiu-Serbanescu M, Hauser J, Hougaard DM, Hultman CM, Jones I, Jones LA, Kahn RS, Kirov G, Landén M, Leboyer M, Lewis CM, Li QS, Lissowska J, Martin NG, Mayoral F, McElroy SL, McIntosh AM, McMahon FJ, Melle I, Metspalu A, Mitchell PB, Morken G, Mors O, Mortensen PB, Müller-Myhsok B, Myers RM, Neale BM, Nimgaonkar V, Nordentoft M, Nöthen MM, O'Donovan MC, Oedegaard KJ, Owen MJ, Paciga SA, Pato C, Pato MT, Posthuma D, Ramos-Quiroga JA, Ribasés M, Rietschel M, Rouleau GA, Schalling M, Schofield PR, Schulze TG, Serretti A, Smoller JW, Stefansson H, Stefansson K, Stordal E, Sullivan PF, Turecki G, Vaaler AE, Vieta E, Vincent JB, Werge T, Nurnberger JI, Wray NR, Di Florio A, Edenberg HJ, Cichon S, Ophoff RA, Scott LJ, Andreassen OA, Kelsoe J, Sklar P; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium. Genome-wide association study identifies 30 Loci associated with bipolar disorder. Nat Genet. 2019;51:793-803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1295] [Cited by in RCA: 1026] [Article Influence: 171.0] [Reference Citation Analysis (0)] |
| 22. | Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, Coleman JRI, Hagenaars SP, Ward J, Wigmore EM, Alloza C, Shen X, Barbu MC, Xu EY, Whalley HC, Marioni RE, Porteous DJ, Davies G, Deary IJ, Hemani G, Berger K, Teismann H, Rawal R, Arolt V, Baune BT, Dannlowski U, Domschke K, Tian C, Hinds DA; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Trzaskowski M, Byrne EM, Ripke S, Smith DJ, Sullivan PF, Wray NR, Breen G, Lewis CM, McIntosh AM. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1774] [Cited by in RCA: 1628] [Article Influence: 271.3] [Reference Citation Analysis (0)] |
| 23. | Cross-Disorder Group of the Psychiatric Genomics Consortium. Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell. 2019;179:1469-1482.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 921] [Cited by in RCA: 865] [Article Influence: 144.2] [Reference Citation Analysis (0)] |
| 24. | International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS). Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol Psychiatry. 2018;23:1181-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 380] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 25. | Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen CY, Choi KW, Coleman JRI, Dalvie S, Duncan LE, Gelernter J, Levey DF, Logue MW, Polimanti R, Provost AC, Ratanatharathorn A, Stein MB, Torres K, Aiello AE, Almli LM, Amstadter AB, Andersen SB, Andreassen OA, Arbisi PA, Ashley-Koch AE, Austin SB, Avdibegovic E, Babić D, Bækvad-Hansen M, Baker DG, Beckham JC, Bierut LJ, Bisson JI, Boks MP, Bolger EA, Børglum AD, Bradley B, Brashear M, Breen G, Bryant RA, Bustamante AC, Bybjerg-Grauholm J, Calabrese JR, Caldas-de-Almeida JM, Dale AM, Daly MJ, Daskalakis NP, Deckert J, Delahanty DL, Dennis MF, Disner SG, Domschke K, Dzubur-Kulenovic A, Erbes CR, Evans A, Farrer LA, Feeny NC, Flory JD, Forbes D, Franz CE, Galea S, Garrett ME, Gelaye B, Geuze E, Gillespie C, Uka AG, Gordon SD, Guffanti G, Hammamieh R, Harnal S, Hauser MA, Heath AC, Hemmings SMJ, Hougaard DM, Jakovljevic M, Jett M, Johnson EO, Jones I, Jovanovic T, Qin XJ, Junglen AG, Karstoft KI, Kaufman ML, Kessler RC, Khan A, Kimbrel NA, King AP, Koen N, Kranzler HR, Kremen WS, Lawford BR, Lebois LAM, Lewis CE, Linnstaedt SD, Lori A, Lugonja B, Luykx JJ, Lyons MJ, Maples-Keller J, Marmar C, Martin AR, Martin NG, Maurer D, Mavissakalian MR, McFarlane A, McGlinchey RE, McLaughlin KA, McLean SA, McLeay S, Mehta D, Milberg WP, Miller MW, Morey RA, Morris CP, Mors O, Mortensen PB, Neale BM, Nelson EC, Nordentoft M, Norman SB, O'Donnell M, Orcutt HK, Panizzon MS, Peters ES, Peterson AL, Peverill M, Pietrzak RH, Polusny MA, Rice JP, Ripke S, Risbrough VB, Roberts AL, Rothbaum AO, Rothbaum BO, Roy-Byrne P, Ruggiero K, Rung A, Rutten BPF, Saccone NL, Sanchez SE, Schijven D, Seedat S, Seligowski AV, Seng JS, Sheerin CM, Silove D, Smith AK, Smoller JW, Sponheim SR, Stein DJ, Stevens JS, Sumner JA, Teicher MH, Thompson WK, Trapido E, Uddin M, Ursano RJ, van den Heuvel LL, Van Hooff M, Vermetten E, Vinkers CH, Voisey J, Wang Y, Wang Z, Werge T, Williams MA, Williamson DE, Winternitz S, Wolf C, Wolf EJ, Wolff JD, Yehuda R, Young RM, Young KA, Zhao H, Zoellner LA, Liberzon I, Ressler KJ, Haas M, Koenen KC. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019;10:4558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 421] [Cited by in RCA: 402] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 26. | Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, Bryois J, Chen CY, Dennison CA, Hall LS, Lam M, Watanabe K, Frei O, Ge T, Harwood JC, Koopmans F, Magnusson S, Richards AL, Sidorenko J, Wu Y, Zeng J, Grove J, Kim M, Li Z, Voloudakis G, Zhang W, Adams M, Agartz I, Atkinson EG, Agerbo E, Al Eissa M, Albus M, Alexander M, Alizadeh BZ, Alptekin K, Als TD, Amin F, Arolt V, Arrojo M, Athanasiu L, Azevedo MH, Bacanu SA, Bass NJ, Begemann M, Belliveau RA, Bene J, Benyamin B, Bergen SE, Blasi G, Bobes J, Bonassi S, Braun A, Bressan RA, Bromet EJ, Bruggeman R, Buckley PF, Buckner RL, Bybjerg-Grauholm J, Cahn W, Cairns MJ, Calkins ME, Carr VJ, Castle D, Catts SV, Chambert KD, Chan RCK, Chaumette B, Cheng W, Cheung EFC, Chong SA, Cohen D, Consoli A, Cordeiro Q, Costas J, Curtis C, Davidson M, Davis KL, de Haan L, Degenhardt F, DeLisi LE, Demontis D, Dickerson F, Dikeos D, Dinan T, Djurovic S, Duan J, Ducci G, Dudbridge F, Eriksson JG, Fañanás L, Faraone SV, Fiorentino A, Forstner A, Frank J, Freimer NB, Fromer M, Frustaci A, Gadelha A, Genovese G, Gershon ES, Giannitelli M, Giegling I, Giusti-Rodríguez P, Godard S, Goldstein JI, González Peñas J, González-Pinto A, Gopal S, Gratten J, Green MF, Greenwood TA, Guillin O, Gülöksüz S, Gur RE, Gur RC, Gutiérrez B, Hahn E, Hakonarson H, Haroutunian V, Hartmann AM, Harvey C, Hayward C, Henskens FA, Herms S, Hoffmann P, Howrigan DP, Ikeda M, Iyegbe C, Joa I, Julià A, Kähler AK, Kam-Thong T, Kamatani Y, Karachanak-Yankova S, Kebir O, Keller MC, Kelly BJ, Khrunin A, Kim SW, Klovins J, Kondratiev N, Konte B, Kraft J, Kubo M, Kučinskas V, Kučinskiene ZA, Kusumawardhani A, Kuzelova-Ptackova H, Landi S, Lazzeroni LC, Lee PH, Legge SE, Lehrer DS, Lencer R, Lerer B, Li M, Lieberman J, Light GA, Limborska S, Liu CM, Lönnqvist J, Loughland CM, Lubinski J, Luykx JJ, Lynham A, Macek M Jr, Mackinnon A, Magnusson PKE, Maher BS, Maier W, Malaspina D, Mallet J, Marder SR, Marsal S, Martin AR, Martorell L, Mattheisen M, McCarley RW, McDonald C, McGrath JJ, Medeiros H, Meier S, Melegh B, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mitjans M, Molden E, Molina E, Molto MD, Mondelli V, Moreno C, Morley CP, Muntané G, Murphy KC, Myin-Germeys I, Nenadić I, Nestadt G, Nikitina-Zake L, Noto C, Nuechterlein KH, O'Brien NL, O'Neill FA, Oh SY, Olincy A, Ota VK, Pantelis C, Papadimitriou GN, Parellada M, Paunio T, Pellegrino R, Periyasamy S, Perkins DO, Pfuhlmann B, Pietiläinen O, Pimm J, Porteous D, Powell J, Quattrone D, Quested D, Radant AD, Rampino A, Rapaport MH, Rautanen A, Reichenberg A, Roe C, Roffman JL, Roth J, Rothermundt M, Rutten BPF, Saker-Delye S, Salomaa V, Sanjuan J, Santoro ML, Savitz A, Schall U, Scott RJ, Seidman LJ, Sharp SI, Shi J, Siever LJ, Sigurdsson E, Sim K, Skarabis N, Slominsky P, So HC, Sobell JL, Söderman E, Stain HJ, Steen NE, Steixner-Kumar AA, Stögmann E, Stone WS, Straub RE, Streit F, Strengman E, Stroup TS, Subramaniam M, Sugar CA, Suvisaari J, Svrakic DM, Swerdlow NR, Szatkiewicz JP, Ta TMT, Takahashi A, Terao C, Thibaut F, Toncheva D, Tooney PA, Torretta S, Tosato S, Tura GB, Turetsky BI, Üçok A, Vaaler A, van Amelsvoort T, van Winkel R, Veijola J, Waddington J, Walter H, Waterreus A, Webb BT, Weiser M, Williams NM, Witt SH, Wormley BK, Wu JQ, Xu Z, Yolken R, Zai CC, Zhou W, Zhu F, Zimprich F, Atbaşoğlu EC, Ayub M, Benner C, Bertolino A, Black DW, Bray NJ, Breen G, Buccola NG, Byerley WF, Chen WJ, Cloninger CR, Crespo-Facorro B, Donohoe G, Freedman R, Galletly C, Gandal MJ, Gennarelli M, Hougaard DM, Hwu HG, Jablensky AV, McCarroll SA, Moran JL, Mors O, Mortensen PB, Müller-Myhsok B, Neil AL, Nordentoft M, Pato MT, Petryshen TL, Pirinen M, Pulver AE, Schulze TG, Silverman JM, Smoller JW, Stahl EA, Tsuang DW, Vilella E, Wang SH, Xu S; Indonesia Schizophrenia Consortium; PsychENCODE; Psychosis Endophenotypes International Consortium; SynGO Consortium, Adolfsson R, Arango C, Baune BT, Belangero SI, Børglum AD, Braff D, Bramon E, Buxbaum JD, Campion D, Cervilla JA, Cichon S, Collier DA, Corvin A, Curtis D, Forti MD, Domenici E, Ehrenreich H, Escott-Price V, Esko T, Fanous AH, Gareeva A, Gawlik M, Gejman PV, Gill M, Glatt SJ, Golimbet V, Hong KS, Hultman CM, Hyman SE, Iwata N, Jönsson EG, Kahn RS, Kennedy JL, Khusnutdinova E, Kirov G, Knowles JA, Krebs MO, Laurent-Levinson C, Lee J, Lencz T, Levinson DF, Li QS, Liu J, Malhotra AK, Malhotra D, McIntosh A, McQuillin A, Menezes PR, Morgan VA, Morris DW, Mowry BJ, Murray RM, Nimgaonkar V, Nöthen MM, Ophoff RA, Paciga SA, Palotie A, Pato CN, Qin S, Rietschel M, Riley BP, Rivera M, Rujescu D, Saka MC, Sanders AR, Schwab SG, Serretti A, Sham PC, Shi Y, St Clair D, Stefánsson H, Stefansson K, Tsuang MT, van Os J, Vawter MP, Weinberger DR, Werge T, Wildenauer DB, Yu X, Yue W, Holmans PA, Pocklington AJ, Roussos P, Vassos E, Verhage M, Visscher PM, Yang J, Posthuma D, Andreassen OA, Kendler KS, Owen MJ, Wray NR, Daly MJ, Huang H, Neale BM, Sullivan PF, Ripke S, Walters JTR, O'Donovan MC; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1733] [Cited by in RCA: 1465] [Article Influence: 488.3] [Reference Citation Analysis (0)] |
| 27. | 1000 Genomes Project Consortium; Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6041] [Cited by in RCA: 5952] [Article Influence: 457.8] [Reference Citation Analysis (0)] |
| 28. | Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5338] [Cited by in RCA: 7510] [Article Influence: 751.0] [Reference Citation Analysis (0)] |
| 29. | Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, Hottenga JJ, Fischer K, Esko T, Surakka I, Broer L, Nyholt DR, Mateo Leach I, Salo P, Hägg S, Matthews MK, Palmen J, Norata GD, O'Reilly PF, Saleheen D, Amin N, Balmforth AJ, Beekman M, de Boer RA, Böhringer S, Braund PS, Burton PR, de Craen AJ, Denniff M, Dong Y, Douroudis K, Dubinina E, Eriksson JG, Garlaschelli K, Guo D, Hartikainen AL, Henders AK, Houwing-Duistermaat JJ, Kananen L, Karssen LC, Kettunen J, Klopp N, Lagou V, van Leeuwen EM, Madden PA, Mägi R, Magnusson PK, Männistö S, McCarthy MI, Medland SE, Mihailov E, Montgomery GW, Oostra BA, Palotie A, Peters A, Pollard H, Pouta A, Prokopenko I, Ripatti S, Salomaa V, Suchiman HE, Valdes AM, Verweij N, Viñuela A, Wang X, Wichmann HE, Widen E, Willemsen G, Wright MJ, Xia K, Xiao X, van Veldhuisen DJ, Catapano AL, Tobin MD, Hall AS, Blakemore AI, van Gilst WH, Zhu H; CARDIoGRAM consortium, Erdmann J, Reilly MP, Kathiresan S, Schunkert H, Talmud PJ, Pedersen NL, Perola M, Ouwehand W, Kaprio J, Martin NG, van Duijn CM, Hovatta I, Gieger C, Metspalu A, Boomsma DI, Jarvelin MR, Slagboom PE, Thompson JR, Spector TD, van der Harst P, Samani NJ. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45:422-427, 427e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 648] [Cited by in RCA: 749] [Article Influence: 62.4] [Reference Citation Analysis (0)] |
| 30. | Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 1232] [Article Influence: 82.1] [Reference Citation Analysis (0)] |
| 31. | Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27:R195-R208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 794] [Cited by in RCA: 988] [Article Influence: 141.1] [Reference Citation Analysis (0)] |
| 32. | Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1177] [Cited by in RCA: 1845] [Article Influence: 205.0] [Reference Citation Analysis (0)] |
| 33. | Peacock A, Leung J, Larney S, Colledge S, Hickman M, Rehm J, Giovino GA, West R, Hall W, Griffiths P, Ali R, Gowing L, Marsden J, Ferrari AJ, Grebely J, Farrell M, Degenhardt L. Global statistics on alcohol, tobacco and illicit drug use: 2017 status report. Addiction. 2018;113:1905-1926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 664] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 34. | Fuster D, Samet JH. Alcohol Use in Patients with Chronic Liver Disease. N Engl J Med. 2018;379:1251-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 143] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 35. | Horrell J, Callaghan L, Dhanda A. Alcohol misuse in patients with alcohol-related liver disease: How can we do better? A narrative review of the literature. Alcohol Clin Exp Res. 2022;46:1364-1370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 36. | Weiss F, Tidona S, Carli M, Perugi G, Scarselli M. Triple Diagnosis of Attention-Deficit/Hyperactivity Disorder with Coexisting Bipolar and Alcohol Use Disorders: Clinical Aspects and Pharmacological Treatments. Curr Neuropharmacol. 2023;21:1467-1476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 37. | Elmquist J, Shorey RC, Anderson S, Stuart GL. Are Borderline Personality Symptoms Associated With Compulsive Sexual Behaviors Among Women in Treatment for Substance Use Disorders? An Exploratory Study. J Clin Psychol. 2016;72:1077-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 38. | Ferreira GM, Lee RSC, Piquet-Pessôa M, de Menezes GB, Moreira-de-Oliveira ME, Albertella L, Yücel M, Dos Santos Cruz M, Dos Santos-Ribeiro S, Fontenelle LF. Habitual vs affective motivations in obsessive-compulsive disorder and alcohol use disorder. CNS Spectr. 2021;26:243-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Burchi E, Makris N, Lee MR, Pallanti S, Hollander E. Compulsivity in Alcohol Use Disorder and Obsessive Compulsive Disorder: Implications for Neuromodulation. Front Behav Neurosci. 2019;13:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 40. | Hawn SE, Cusack SE, Amstadter AB. A Systematic Review of the Self-Medication Hypothesis in the Context of Posttraumatic Stress Disorder and Comorbid Problematic Alcohol Use. J Trauma Stress. 2020;33:699-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 41. | Shirley MC, Sirocco KY. Introduction to special section: ADHD, impulsivity, and alcohol abuse. Exp Clin Psychopharmacol. 2014;22:97-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Grunze H, Soyka M. The pharmacotherapeutic management of comorbid bipolar disorder and alcohol use disorder. Expert Opin Pharmacother. 2022;23:1181-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Rakofsky JJ, Dunlop BW. Do alcohol use disorders destabilize the course of bipolar disorder? J Affect Disord. 2013;145:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 44. | Chitty KM, Lagopoulos J, Kaur M, Hickie IB, Hermens DF. The N-methyl-D-aspartate receptor as a neurobiological intersection between bipolar disorder and alcohol use: a longitudinal mismatch negativity study. Int J Neuropsychopharmacol. 2015;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 45. | Kenneson A, Funderburk JS. Patatin-like phospholipase domain-containing protein 3 (PNPLA3): A potential role in the association between liver disease and bipolar disorder. J Affect Disord. 2017;209:93-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 46. | Fuller BE, Rodriguez VL, Linke A, Sikirica M, Dirani R, Hauser P. Prevalence of liver disease in veterans with bipolar disorder or schizophrenia. Gen Hosp Psychiatry. 2011;33:232-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 47. | Zarrinpar A, Gupta S, Maurya MR, Subramaniam S, Loomba R. Serum microRNAs explain discordance of non-alcoholic fatty liver disease in monozygotic and dizygotic twins: a prospective study. Gut. 2016;65:1546-1554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 48. | Zhang M, Sun W, Zhou M, Tang Y. MicroRNA-27a regulates hepatic lipid metabolism and alleviates NAFLD via repressing FAS and SCD1. Sci Rep. 2017;7:14493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 49. | Alural B, Genc S, Haggarty SJ. Diagnostic and therapeutic potential of microRNAs in neuropsychiatric disorders: Past, present, and future. Prog Neuropsychopharmacol Biol Psychiatry. 2017;73:87-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 50. | Huang DQ, El-Serag HB, Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2021;18:223-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 840] [Cited by in RCA: 1217] [Article Influence: 304.3] [Reference Citation Analysis (0)] |
| 51. | Alves Freire Ribeiro AC, Batista TH, Trujillo Rojas VC, Giusti-Paiva A, Cardoso Vilela F. Metabolic syndrome accentuates post-traumatic stress disorder-like symptoms and glial activation. Behav Brain Res. 2020;384:112557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 52. | Isomura K, Brander G, Chang Z, Kuja-Halkola R, Rück C, Hellner C, Lichtenstein P, Larsson H, Mataix-Cols D, Fernández de la Cruz L. Metabolic and Cardiovascular Complications in Obsessive-Compulsive Disorder: A Total Population, Sibling Comparison Study With Long-Term Follow-up. Biol Psychiatry. 2018;84:324-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 53. | Tutunchi H, Naeini F, Saghafi-Asl M, Farrin N, Monshikarimi A, Ostadrahimi A. Effects of oleoylethanolamide supplementation on atherogenic indices and hematological parameters in patients with nonalcoholic fatty liver disease: A clinical trial. Health Promot Perspect. 2020;10:373-382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 54. | Hori H, Kim Y. Inflammation and post-traumatic stress disorder. Psychiatry Clin Neurosci. 2019;73:143-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 226] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 55. | Attwells S, Setiawan E, Wilson AA, Rusjan PM, Mizrahi R, Miler L, Xu C, Richter MA, Kahn A, Kish SJ, Houle S, Ravindran L, Meyer JH. Inflammation in the Neurocircuitry of Obsessive-Compulsive Disorder. JAMA Psychiatry. 2017;74:833-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 136] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 56. | Mazza MG, De Lorenzo R, Conte C, Poletti S, Vai B, Bollettini I, Melloni EMT, Furlan R, Ciceri F, Rovere-Querini P; COVID-19 BioB Outpatient Clinic Study group, Benedetti F. Anxiety and depression in COVID-19 survivors: Role of inflammatory and clinical predictors. Brain Behav Immun. 2020;89:594-600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 967] [Cited by in RCA: 1015] [Article Influence: 203.0] [Reference Citation Analysis (0)] |
| 57. | Shao Q, Wu Y, Ji J, Xu T, Yu Q, Ma C, Liao X, Cheng F, Wang X. Interaction Mechanisms Between Major Depressive Disorder and Non-alcoholic Fatty Liver Disease. Front Psychiatry. 2021;12:711835. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |