Published online Oct 26, 2023. doi: 10.12998/wjcc.v11.i30.7294
Peer-review started: August 8, 2023
First decision: August 24, 2023
Revised: September 1, 2023
Accepted: September 27, 2023
Article in press: September 27, 2023
Published online: October 26, 2023
Processing time: 78 Days and 5.9 Hours
With the widespread use of antimicrobial drugs, bacterial resistance has become a significant problem, posing a serious threat to public health. The prevalence of clinical infection strains in hospitals and their drug sensitivities are key to the appropriate use of antibiotics in clinical practice.
To identify prevalent bacteria and their antibiotic resistance profiles in a hospital setting, thereby guiding effective antibiotic usage by clinicians.
Specimens from across the institution were collected by the microbiology la
A total of 12062 bacterial strains of key monitoring significance were detected. Staphylococcus aureus demonstrated widespread resistance to penicillin, but none of the strains were resistant to vancomycin or linezolid. Moreover, 219 strains of methicillin-resistant coagulase-negative staphylococci and 110 strains of me
Our hospital’s overall antibiotic resistance rate was relatively stable from 2017 to 2022. The detection rates of key monitoring strains are reported quarterly and their resistance dynamics are monitored and communicated to the entire hospital, which can guide clinical antibiotic selection.
Core Tip: This study provides essential insights into the distribution and drug resistance of key clinical bacteria in a hospital setting. The findings help clinicians to understand the prevalence of these strains and their resistance to antibiotics, which can guide effective treatment decisions. This contributes to the broader efforts to manage and combat antibiotic resistance, a significant global health challenge.
- Citation: Li ZY, Yang D, Hao CH. Six-year analysis of key monitoring for bacterial strain distribution and antibiotic sensitivity in a hospital. World J Clin Cases 2023; 11(30): 7294-7301
- URL: https://www.wjgnet.com/2307-8960/full/v11/i30/7294.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i30.7294
The widespread application of antimicrobial drugs has brought bacterial resistance to the forefront as a substantial public health threat[1]. To enhance the scientific management of these drugs, our hospital has initiated tracking tests in the microbiology laboratory to determine the distribution of key monitoring bacteria within its premises. This measure is in line with the State Council’s “National Action Plan to Contain Antibacterial Resistance (2016-2020)”, the National Health Commission’s “Clinical Application Management Measures of Antibacterial Drugs”, and the “Notice on Further Strengthening the Clinical Application Management of Antibacterial Drugs to Contain Antibacterial Resistance”. The hospital has selected eight bacteria associated with hospital-acquired infections for key monitoring and summarized the changes in the antibiotic sensitivity of various multidrug-resistant strains. The intention is to aid clinicians in comprehending the prevalence of strains causing clinical infection within the hospital and serve as a reference for the judicious use of antibiotics.
A total of 12062 clinical isolates were collected from the hospital during the period 2017-2022 from blood, sputum, urine, wound secretion, drainage fluid, cerebrospinal fluid, and pus samples.
The VITEK 2 compact fully automatic analyzer was used to identify the bacteria in the collected samples and determine their antibiotic sensitivity. The analysis and interpretation of the results complied with the Clinical and Laboratory Standards Institute 2017 guidelines. The WHONET5.6 software was used to statistically analyze the results.
According to the “Guidelines for Prevention and Control of Multidrug-Resistant Bacterial Hospital Infections (Trial)” issued by the Ministry of Health in 2011[2], multidrug-resistant bacteria are those that demonstrate resistance to three or more types of clinically used antimicrobial drugs.
The quality control strains used in this study were Staphylococcus aureus (S. aureus) (ATCC6538); Pseudomonas aeruginosa (P. aeruginosa) (ATCC27853); Escherichia coli (E. coli) (ATCC25922); Acinetobacter baumannii (A. baumannii) (ATCC19606); Staphylococcus epidermidis (S. epidermidis) (ATCC12228); Klebsiella pneumoniae (K. pneumoniae) (ATCC700603); Enterococcus faecium (E. faecium) (ATCC29212); Enterococcus faecalis (E. faecalis) (ATCC51558).
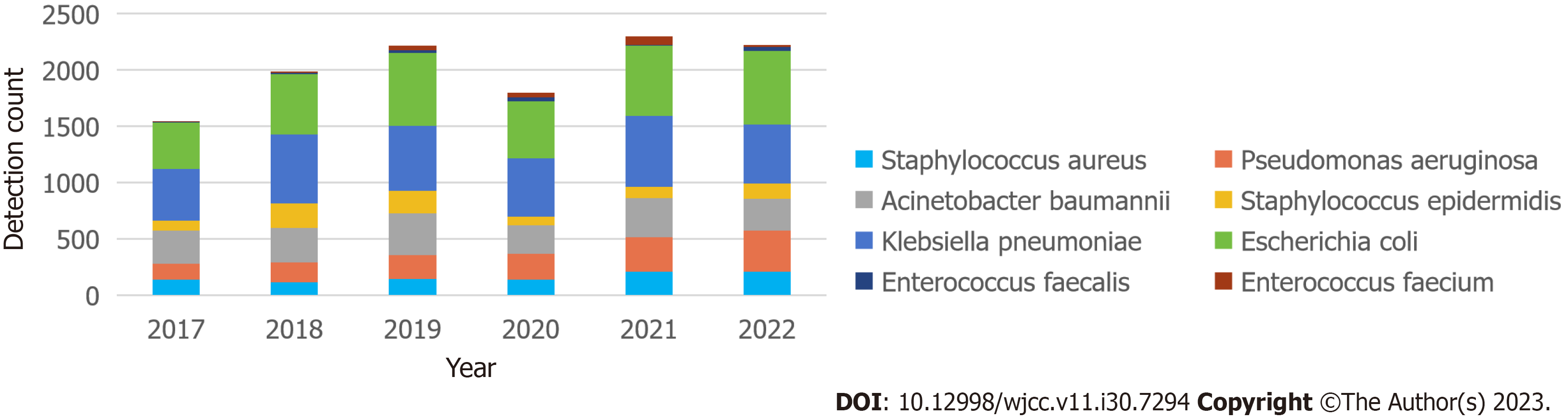
Between 2017 and 2022, the principal pathogens linked to hospital-acquired infections were: S. aureus, P. aeruginosa, A. baumannii, S. epidermidis, K. pneumoniae, E. coli, E. faecalis, and E. faecium. A rising trend in the detection rate of these eight key pathogens was observed from 2017 to 2019. However, in 2020, a significant reduction was noted in the overall detection rate, which could be attributed to decreased hospital visits owing to the impact of the pandemic. In 2021 and 2022, the rate returned to nearly the 2019 level. Of these, the three most commonly detected strains were E. coli (3384, 28%), K. pneumoniae (3326, 27.6%), and A. baumannii (1861, 15.4%). Following these strains, in descending order, were P. aeruginosa, S. aureus, S. epidermidis, E. faecium, and E. faecalis (see Figure 1). The incidence of multidrug-resistant bacteria was 20%-27% (see Table 1).
| Year | Count of multi-drug resistant bacteria | Count of key surveillance strains | Percentage of multi-drug resistant bacteria |
| 2017 | 438 | 1547 | 28.31% |
| 2018 | 447 | 1980 | 22.58% |
| 2019 | 459 | 2215 | 20.72% |
| 2020 | 455 | 1797 | 25.32% |
| 2021 | 542 | 2298 | 23.59% |
| 2022 | 526 | 2225 | 23.64% |
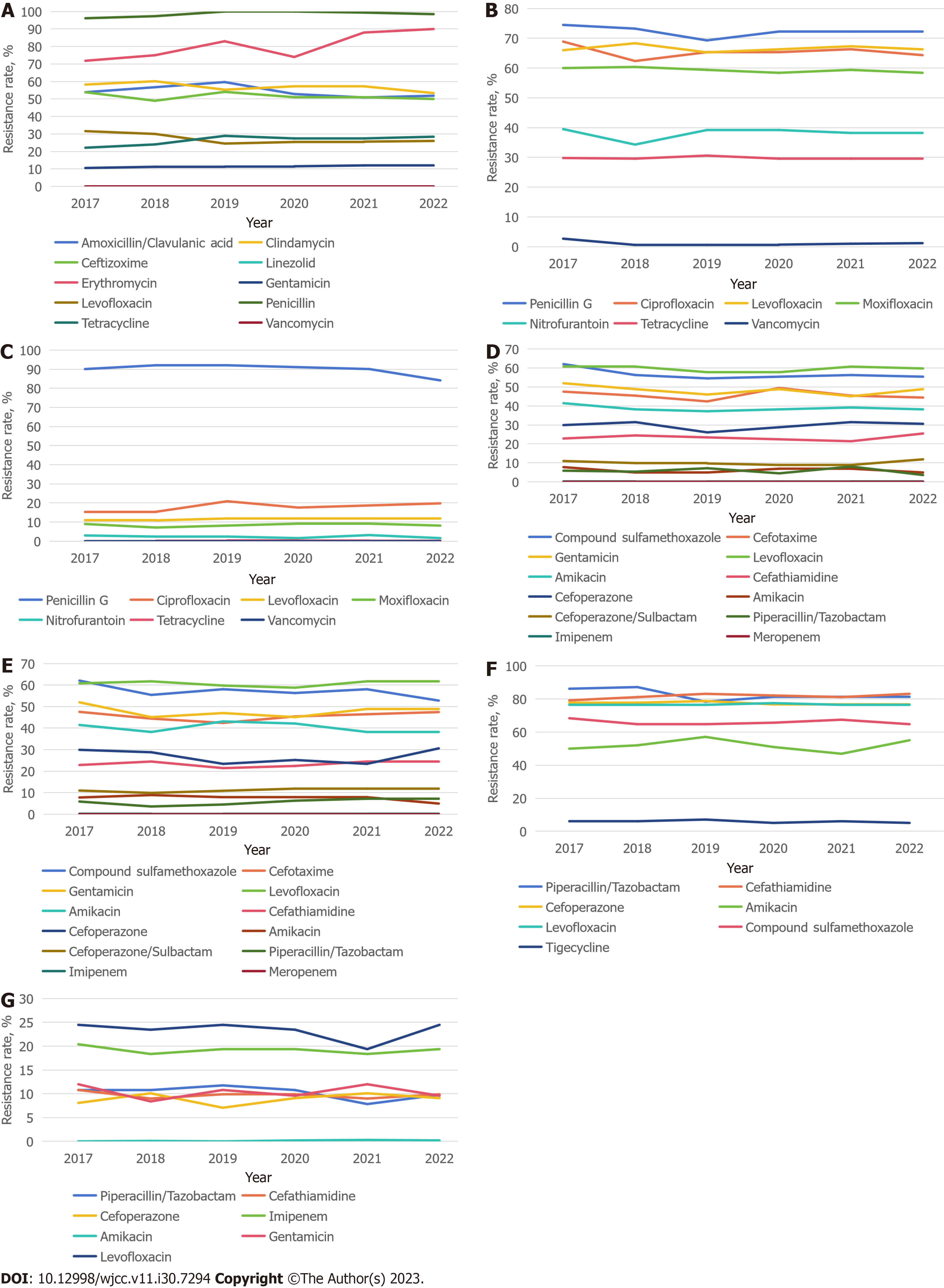
Staphylococcus: A total of 944 S. aureus strains and 812 S. epidermidis strains were identified. Of these, there were 219 methicillin-resistant coagulase-negative staphylococci (MRCNS) and 110 methicillin-resistant S. aureus (MRSA). S. aureus exhibited a high penicillin resistance rate of nearly 100%. Over the 6 years, the erythromycin resistance rate was high and increased gradually from 71.8% in 2017 to 90% in 2022. Concurrently, golden staphylococcus demonstrated significant clindamycin resistance, reaching 64.0% in 2022. Gentamicin remained relatively effective against S. aureus, with a stable resistance rate of approximately 11% over the period. Notably, no S. aureus strains resistant to vancomycin or linezolid were detected (refer to Figure 2A).
Enterococcus: A total of 191 E. faecium and 114 E. faecalis strains were identified in this study, both of which exhibited resistance to penicillin G. E. faecium showed resistance to the third-generation quinolones ciprofloxacin and levofloxacin, with resistance rates of 60.38%-68.9% and 66.28%-70.38%, respectively. Resistance to the fourth-generation fluoroquinolone moxifloxacin was slightly less, at approximately 60%. E. faecium demonstrated lower resistance to nitrofurantoin and tetracycline. Importantly, a few vancomycin-resistant E. faecium were detected from 2017 to 2022, although the resistance rate decreased gradually from 2.7% in 2017 and stabilized at approximately 1% (refer to Figure 2B).
In contrast, E. faecalis displayed a lower overall detection rate and demonstrated less resistance. The resistance rate to penicillin G exceeded 90%, the resistance rate to gentamicin remained between 15% and 20%, and the resistance rates to third- and fourth-generation quinolones were significantly lower than those of E. faecium. No vancomycin-resistant E. faecalis were detected (refer to Figure 2C).
Enterobacteriales:E. coli and K. pneumoniae were the top two pathogens monitored and accounted for 28.1% and 27.6% of the total strains detected, respectively. The overall resistance rate to the compound sulfamethoxazole exceeded 50% for both E. coli and K. pneumoniae, with no increasing trend over the 6 years. Both pathogens showed a high resistance rate to cefotaxime, which fluctuated around 50%. However, the pathogens demonstrated high sensitivity to cefotaxime, a third-generation cephalosporin, with a sensitivity rate of approximately 78% for E. coli and 76% for K. pneumoniae. Both pathogens displayed slightly higher sensitivity to fourth-generation cephalosporins. Cefoperazone/sulbactam, a key antibiotic combination, was effective against these two predominant strains of Enterobacteriales. Their resistance to the drug remained at approximately 9%. The pathogens depicted even lower resistance to piperacillin/tazobactam, which hovered in the range of 5%-8%. Notably, no E. coli or K. pneumoniae resistant to meropenem were detected, and their resistance rate to imipenem was a mere 0.2% (refer to Figures 2D and E).
Nonfermenting bacteria:A. baumannii and P. aeruginosa were the third and fourth most frequently detected bacterial strains in this study. The monitoring of A. baumannii revealed a high resistance rate to piperacillin/tazobactam of almost 90%. The pathogen’s resistance rates to cefotaxime, cefoperazone, and levofloxacin hovered at approximately 75%. The resistance rate to tigecycline was relatively low and stabilized at approximately 6% in recent years (refer to Figure 2F).
When compared with A. baumannii, P. aeruginosa exhibited lower antimicrobial resistance in susceptibility tests. Over the past 8 years, its resistance to imipenem fluctuated between 18.4% and 21.4%. Resistance to cephalosporins, such as cefotaxime and cefoperazone, was maintained at < 10% in the past 6 years. P. aeruginosa consistently demonstrated the lowest resistance to amikacin, maintaining a rate of 0.2% over the past 3 years (refer to Figure 2G).
From 2017 to 2022, the distribution and drug sensitivity of eight key pathogens, namely, S. aureus, P. aeruginosa, A. baumannii, S. epidermidis, K. pneumoniae, E. coli, E. faecalis, and E. faecium, were continuously monitored and analyzed. The empirical use of penicillin has culminated in widespread drug resistance in the detected S. aureus strains[3]. Although the detection rate of MRSA is lower than that in similar studies conducted previously[4,5], the overall resistance rate of S. aureus agrees with national bacterial resistance monitoring data[6,7]. Hospitals need to pay attention to the detection of MRCNS. Research on hospital-acquired Methicillin-resistant non-staphylococci indicates that infections are concentrated in the lower respiratory tract and surgical sites. Clinical departments should strengthen disinfection and prevention measures. Patients with two consecutive MRCNS detections should be placed under preventive isolation to mitigate the spread of hospital-acquired infections[8]. Coagulase-negative staphylococci possess fewer virulence properties than S. aureus, which results in different disease spectrums. Hence, host susceptibility becomes significantly more critical[9].
Of the eight key surveillance strains, the total number of E. faecium and E. faecalis identified in our hospital is relatively low. A few vancomycin-resistant enterococci (VRE) have been detected over the past 6 years. For VRE, experts suggest selecting the corresponding antibiotic based on the results of the strain’s drug sensitivity test and combining antibiotics with different mechanisms of action to avoid resistance[10-12].
The unique cell wall structure of gram-negative bacteria makes them more likely to acquire drug resistance than gram-positive bacteria[13,14]. E. coli and K. pneumoniae are the top two key surveillance strains. Our hospital’s K. pneumoniae drug resistance situation is serious, which mirrors the conclusions of drug resistance analysis conducted by other tertiary hospitals[15]. The resistance rates to gentamicin, cefotaxime, and the compound sulfamethoxazole exceed 50%. Cefoperazone/sulbactam and piperacillin/tazobactam are viable alternatives to treat infections caused by drug-resistant bacteria, but their adverse reactions warrant the special attention of clinical doctors and pharmacists[16,17].
The increase in the antibiotic resistance rate of A. baumannii isolates has altered the epidemiology of hospital-acquired severe infections. A systematic review has identified that carbapenem-resistant and multidrug-resistant A. baumannii account for 65% and 59%, respectively, of all hospital-acquired infections among patients admitted to the intensive care unit in Southeast Asia[18]. Of the key surveillance strains in our hospital, A. baumannii also exhibited a relatively high overall resistance rate. Clinically, strategies to handle the primary drug resistance of isolates are limited, and the main alternative is to combine polymyxin with tigecycline, a tetracycline derivative[19]. Our hospital has detected a few tigecycline-resistant strains, which warrant clinicians’ attention.
The global burden of antibiotic resistance is high in clinical isolates of P. aeruginosa[20]. Nonetheless, national monitoring data should be interpreted with caution because local resistance rates vary greatly and are closely related to local antibiotic usage patterns and patient types. The resistance rate of P. aeruginosa detected in our hospital is relatively low. An observational study found that the drug resistance risk was the lowest for cefoperazone and the highest for imipenem in patients treated with a single antipseudomonal agent[21]. This finding is consistent with the conclusions drawn from the drug resistance rate detected in our hospital.
The present study, while providing valuable insights into bacterial strain distribution and antibiotic sensitivity over a six-year period in a single hospital, is subject to several limitations. First, the geographic scope of the research is limited to one hospital, potentially constraining the generalizability of the findings. Second, the methods employed, including the use of the VITEK 2 compact analyzer for bacterial identification and WHONET5.6 software for statistical analysis, may have inherent limitations in terms of accuracy and analytical capabilities. Third, although the study spans six years, bacterial resistance patterns can evolve rapidly, rendering the data less applicable in the future. Fourth, the absence of a control group for comparative analysis poses a limitation in the interpretative strength of the results. Additionally, the possibility of human error or subjectivity in interpreting cultures and tests cannot be ruled out. Financial constraints may have also influenced the comprehensiveness of the study, and any gaps or omissions in the data further limit the scope of our conclusions.
Collection of samples from the entire hospital and summarizing the detection volume and drug sensitivity results of key strains can serve as a reference for the clinical use of antibiotics. Over the past 6 years, the antibiotic resistance rate in our hospital has remained relatively stable, with no large-scale drug-resistant strain outbreak. The findings of hospital-acquired infection management and the rational use of antibiotics are the result of collaboration among clinical departments, microbiology laboratories, pharmacy departments, hospital-acquired infection management departments, and medical affairs. The ultimate goal is to reduce patient infection risk, prevent antibiotic abuse, and curb superbug production from multiple perspectives, such as infection prevention and diagnosis, rational use of antibiotics, and long-term monitoring.
This research focuses on the critical public health issue of bacterial resistance due to prevalent antimicrobial drug use. The study’s objective is to understand the prevalent bacterial strains and their resistance patterns within a hospital setting. Advanced analytical tools and software were employed for comprehensive analysis. This initiative aims to guide clinicians in effective antibiotic selection, addressing the growing concern of antimicrobial resistance.
The motivation behind this research is the urgent need to address the escalating problem of bacterial resistance in hospitals. By understanding the prevalent strains and their antibiotic resistance profiles, this study provides essential insights to clinicians. The findings assist in making informed decisions on antibiotic usage, contributing significantly to managing antimicrobial resistance effectively.
The main goal of this research is to identify prevalent bacteria in hospitals and assess their antibiotic resistance profiles. The research focuses on key strains and evaluates their resistance to common antibiotics, aiming to guide clinical antibiotic selection. The establishment of a regular reporting system is also a crucial objective, ensuring the continuous monitoring of resistance data to aid clinical decisions.
The study involves collecting specimens from varied sources within the hospital, utilizing the VITEK 2 compact analyzer for bacterial identification and antibiotic sensitivity testing. WHONET5.6 software facilitated effective data analysis and interpretation. The methodologies ensured accurate and reliable findings, providing critical insights for appropriate antibiotic selection in clinical practice.
The study identified 12062 bacterial strains, revealing diverse resistance patterns among them. Notable findings include Staphylococcus aureus’s resistance to penicillin and Pseudomonas aeruginosa’s minimal resistance to certain antibiotics. Regular reporting and monitoring of these strains and their resistance dynamics are conducted to continually inform and guide clinical antibiotic selection within the hospital.
The findings of this study disclose significant insights into bacterial resistance within a hospital. It outlines specific resistance patterns of various bacteria, emphasizing the importance of regular reporting and monitoring. These conclusions are pivotal in guiding clinicians in optimizing antibiotic selection and contribute to the broader goal of managing antimicrobial resistance effectively.
This study underscores the need for continuous surveillance of antibiotic resistance and encourages further research into the mechanisms behind resistance development. Investigating the effects of interventions and focusing on collaborative efforts to implement effective infection control measures are pivotal. Long-term studies and public awareness campaigns are essential for developing guidelines and promoting responsible antibiotic usage, aiming to curb the spread of bacterial resistance.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nunes R, United States; Singh S, India S-Editor: Wang JJ L-Editor: A P-Editor: Xu ZH
| 1. | Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3674] [Cited by in RCA: 3325] [Article Influence: 221.7] [Reference Citation Analysis (0)] |
| 2. | Cosgrove SE, Carmeli Y. The impact of antimicrobial resistance on health and economic outcomes. Clin Infect Dis. 2003;36:1433-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 360] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 3. | Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629-641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2027] [Cited by in RCA: 1822] [Article Influence: 113.9] [Reference Citation Analysis (0)] |
| 4. | Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4167] [Cited by in RCA: 4277] [Article Influence: 158.4] [Reference Citation Analysis (0)] |
| 5. | Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK; Active Bacterial Core surveillance (ABCs) MRSA Investigators. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763-1771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2491] [Cited by in RCA: 2472] [Article Influence: 137.3] [Reference Citation Analysis (0)] |
| 6. | Livermore DM. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med. 2012;27:128-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 246] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 7. | Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3433] [Cited by in RCA: 3632] [Article Influence: 227.0] [Reference Citation Analysis (0)] |
| 8. | Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3023] [Cited by in RCA: 3123] [Article Influence: 148.7] [Reference Citation Analysis (0)] |
| 9. | Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27:870-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1011] [Cited by in RCA: 995] [Article Influence: 90.5] [Reference Citation Analysis (0)] |
| 10. | Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 959] [Cited by in RCA: 1135] [Article Influence: 87.3] [Reference Citation Analysis (0)] |
| 11. | Jaspan HB, Brothers AW, Campbell AJ, McGuire JK, Browd SR, Manley TJ, Pak D, Weissman SJ. Multidrug-resistant Enterococcus faecium meningitis in a toddler: characterization of the organism and successful treatment with intraventricular daptomycin and intravenous tigecycline. Pediatr Infect Dis J. 2010;29:379-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Schutt AC, Bohm NM. Multidrug-resistant Enterococcus faecium endocarditis treated with combination tigecycline and high-dose daptomycin. Ann Pharmacother. 2009;43:2108-2112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol. 2011;2:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 517] [Cited by in RCA: 585] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 14. | Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1392] [Cited by in RCA: 1560] [Article Influence: 91.8] [Reference Citation Analysis (0)] |
| 15. | Falagas ME, Karageorgopoulos DE. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among Gram-negative bacilli: need for international harmonization in terminology. Clin Infect Dis. 2008;46:1121-2; author reply 1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 187] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 16. | Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J Jr; Infectious Diseases Society of America. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:155-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1079] [Cited by in RCA: 1094] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 17. | Tamma PD, Cosgrove SE, Maragakis LL. Combination therapy for treatment of infections with gram-negative bacteria. Clin Microbiol Rev. 2012;25:450-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 578] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 18. | Teerawattanapong N, Panich P, Kulpokin D, Na Ranong S, Kongpakwattana K, Saksinanon A, Goh BH, Lee LH, Apisarnthanarak A, Chaiyakunapruk N. A Systematic Review of the Burden of Multidrug-Resistant Healthcare-Associated Infections Among Intensive Care Unit Patients in Southeast Asia: The Rise of Multidrug-Resistant Acinetobacter baumannii. Infect Control Hosp Epidemiol. 2018;39:525-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 19. | Karageorgopoulos DE, Kelesidis T, Kelesidis I, Falagas ME. Tigecycline for the treatment of multidrug-resistant (including carbapenem-resistant) Acinetobacter infections: a review of the scientific evidence. J Antimicrob Chemother. 2008;62:45-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 139] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 20. | Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013;62:499-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 315] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 21. | Livermore DM, Warner M, Mushtaq S, Doumith M, Zhang J, Woodford N. What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int J Antimicrob Agents. 2011;37:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 260] [Article Influence: 18.6] [Reference Citation Analysis (0)] |