Published online Oct 16, 2023. doi: 10.12998/wjcc.v11.i29.6974
Peer-review started: April 26, 2023
First decision: July 27, 2023
Revised: August 12, 2023
Accepted: September 4, 2023
Article in press: September 4, 2023
Published online: October 16, 2023
Processing time: 170 Days and 0.7 Hours
Time series analysis is a valuable tool in epidemiology that complements the classical epidemiological models in two different ways: Prediction and forecast. Prediction is related to explaining past and current data based on various internal and external influences that may or may not have a causative role. Forecasting is an exploration of the possible future values based on the predictive ability of the model and hypothesized future values of the external and/or internal influences. The time series analysis approach has the advantage of being easier to use (in the cases of more straightforward and linear models such as Auto-Regressive Integrated Moving Average). Still, it is limited in forecasting time, unlike the classical models such as Susceptible-Exposed-Infectious-Removed. Its applicability in forecasting comes from its better accuracy for short-term prediction. In its basic form, it does not assume much theoretical knowledge of the mechanisms of spreading and mutating pathogens or the reaction of people and regulatory structures (governments, companies, etc.). Instead, it estimates from the data directly. Its predictive ability allows testing hypotheses for different factors that positively or negatively contribute to the pandemic spread; be it school closures, emerging variants, etc. It can be used in mortality or hospital risk estimation from new cases, seroprevalence studies, assessing properties of emerging variants, and estimating excess mortality and its relationship with a pandemic.
Core tip: Time-series analysis allows us to do easily and, in less time, precise short-term forecasting in novel pandemics by estimating directly from data. These models do not need extensive knowledge of pandemic mechanisms and interactions between peoples, societal structures, and pathogens. Its secondary but equally important role is distinguishing factors contributing to the spread or slowing it down. Of course, the time series analysis approach cannot give a forecast for an end of a pandemic, nor the precise moment of its peak, but it is invaluable for fast response based on sound statistical methodology.
- Citation: Tomov L, Chervenkov L, Miteva DG, Batselova H, Velikova T. Applications of time series analysis in epidemiology: Literature review and our experience during COVID-19 pandemic. World J Clin Cases 2023; 11(29): 6974-6983
- URL: https://www.wjgnet.com/2307-8960/full/v11/i29/6974.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i29.6974
Time series analysis studies are consecutive collections of observations in time to predict or forecast behavior[1]. Pre
Since correlation cannot be estimated directly for nonstationary processes via the standard regression techniques, the so-called spurious correlations[2], the existing number of time series models such as Box-Jenkins оr Auto-Regressive Integrated Moving Average (ARIMA) models that can deal with nonstationarity. Why do we use them in epidemiology? First, these are linear models that are simpler and easier to use than the classical nonlinear epidemiological models such as Susceptible-Infectious-Removed (SIR), Susceptible-Exposed-Infectious-Removed (SEIR), etc., for which no closed-form exact analytical solutions exist and need numerical simulations or special techniques for approximation for the long-term behavior of the model[3].
However, the difficulty in predicting cases and deaths during the coronavirus disease 2019 (COVID-19) pandemic was also emphasized by Roda et al[4]. They raised concerns about utilizing the affirmed case information as nonidentifiability in model alignments[4].
Talking about the classical nonlinear epidemiological models for epidemics, SIR is able to propose a simple model based on the two-reaction mechanism. In this way, the conditions for epidemic development, the course of a simple closed epidemic, as well as the mitigation strategies could be explained[5]. SIR has been used successfully to estimate the number of cases and deaths in outbreaks such as influenza H1N1 (2009-2010) and Ebola (2014-2016) viruses, examining the early growth, including with modifications, such as SIR with reactive behavioral changes and SIR with inhomogeneous mixing.
The SEIR model was also used mostly for influenza epidemics. Zhan et al[6] additionally used the COVID-19 historical data of 367 Chinese cities to create the transmission mechanisms and contact topology utilizing a set of profile codes. Then the method was applied to South Korea, Italy, and Iran, to predict the infection peaks before the end of March 2020[7].
However, when comparing the SIR and the ARIMA models, it was shown that ARIMA outperformed the SIR in predicting the cases of COVID-19[8].
Abolmaali and Shirzaei[7] were among the first to compare multiple epidemiological methods that can be used during the COVID-19 pandemic to monitor and even prevent the spread of infection. The authors demonstrated that predictive models, such as SIR, SEIR, ARIMA, etc., have proven useful and effective in predicting the incidence of infections. However, it was shown that SIR could not provide helpful early prediction in cases of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, giving a significant error in estimating for countries such as Brazil, United States, India and Russia. Moreover, SIR was useful mainly for predictions in the short term. On the contrary, SEIR showed better results than SIR in the long term in forecasting COVID-19[9].
Regarding ARIMA, it was shown that this model requires further information for a more precise point-by-point forecast, but the spread of COVID-19 was forecasted ultimately. Additionally, ARIMA itself helps the data to remain stationary, resulting in modeling flexibility while capturing any changes at every stage. Last but not least, the ARIMA model managed to maintain minimum error in the forecasting. Based on all these outcomes, ARIMA outperforms all other compared models[7].
Many other research groups confirmed the effectiveness and usefulness of the ARIMA model for COVID-19 pandemic forecasting[10-12].
Additionally, the ARIMA model has three parameters: p, d and q, which define the order of its three components-autoregression, differencing and moving average. The autoregressive part captures the dependence on past values or the process dynamics-the variable is regressed on its previous or lagged values. The moving average part indicates that regression errors are not independent but are a linear combination of error terms from different earlier moments[13]. The differencing part is used to transform the process into a broad sense stationary process vs consecutive differencing of its values. The combinations of these three parts with proper orders depending on the process allow us to make linear regression (LR) that accounts for the dependence of error terms for nonstationarity and assess dependence with previous moments of time. The ARIMA model can be combined with external regressors or factors. Thus, it can be applied as regression with ARIMA errors for the process, made stationary by differencing its values. The use of LR for prediction is, therefore, straightforward.
One example is the model in our recent research[13]. We predicted some weekly deaths from COVID-19 in Bulgaria from new cases by age groups with different lags. We used the viral variants as external regressors.
Another example is the analysis of the incidence and death of AIDS and HIV in China[14]. Time series models such as ARIMA need historical data to be able to extrapolate future trends. They are beneficial for short-term forecast[15] and for prediction, even for complex dynamics, such as for COVID-19, with different policies to control the spread and different viral variants in various stages of the pandemic[16]. In addition, there is a practical advantage of time series analysis over theoretical epidemiological models, and no detailed knowledge of the causal mechanism is required; instead, the structure is inferred from real-life data. This allows fast development and deployment of models during an ongoing pandemic with limited knowledge of key parameters related to the influence of measures or the characteristics of newly emerging variants.
Based on the characteristics and effectiveness of ARIMA, we created the following objectives for our study: (1) To apply time series analysis for assessment of SARS-CoV-2 variants spread; (2) to assess the excess mortality from COVID-19 in Bulgaria; (3) to demonstrate the importance of time series analysis during the COVID-19 pandemic in radiology departments; and (4) to determine the SARS-CoV-2 seroprevalence with time series analysis.
Time series regression studies have been extensively used in environmental epidemiology, especially to assess short-term associations between exposures[15]. The most commonly studied factors are air pollution, weather, and pollen, but health outcomes like mortality or disease-specific hospital admissions could also be investigated. Typically, data are available at regular time intervals for both exposure and result (e.g., daily pollution levels and daily death counts). The goal is to investigate short-term relationships between them.
In line with this, since COVID-19 has expanded globally, this results in a continuous pandemic, imposing limitations and expenditures on many governments. Therefore, anticipating the number of new cases and fatalities throughout this period can be crucial in predicting future expenses and facilities[17]. Furthermore, it is necessary to analyze the data during the pandemic to expect one or other intervention strategies, mitigations, etc. In such a way, the pandemic could be monitored, controlled, and properly managed. Many studies demonstrated that among the epidemiological models, the ARIMA model showed the desired precision in predicting the number of cases and fatalities with minimum error[10,18].
Although some researchers hesitantly avoided ARIMA for analyzing COVID-19 epidemiological data, Alabdulrazzaq et al[19] demonstrated that the ARIMA technique showed accurate and valid forecasting; especially the ARIMA best-fit model for predicting the confirmed and recovered cases of COVID-19. Despite the many dynamic aspects based on the novelty of the virus and the nature of the disease, the actual values for most of the periods were within the model prediction of a 95% confidence interval. Pearson’s correlation showed high correlations between the forecast points and the actual recorded data (r = 0.996)[19]. This confirms why ARIMA is one of the best-suited models with satisfactory results and minimum error.
Different methods in time series analyses can be used, such as the hybrid machine learning approach (using multiple simple algorithms to complement and facilitate each other) to anticipate the number of infected people and mortality rate[20]; LR (based on using regression models enabling subject-matter interpretation of the data); Least Absolute Shrinkage and Selection Operator (a model that uses over regression methods for more accurate predictions); support vector machine (using optimal hyperplane in an N-dimensional space, separating the data points in different classes); exponential smoothing (forecasting univariate time series data) to determine the affected by the virus people and the deceased cases[21]; numerical modeling to assess the effect of the population age on the mortality rate[22]; numerical modeling methods such as polynomial regression (fitting of a nonlinear relationship between the value of something and the condition mean of other); Bayesian Edge (estimating probability influenced by the belief of the likelihood of a certain outcome) and long short-term memory (having the ability to learn long term sequences of observations) to estimate the prevalence of SARS-CoV-2 infection and to predict the scale of the pandemic along with the mortality rate[23]; a deep learning system for the prediction of the COVID-19 time series[24]; mathematical model about the spread of COVID-19[25]; a stochastic model considering comorbidities and age[26]; an SIQR model made stochastic, considering the uncertainty of infection progress[27]; a fractional-order dynamical system[28]; fractional calculus and natural decom
We describe an example of application of time series analysis in epidemiology: Analysis of overall mortality and its dependence on the number of registered cases of COVID-19. In other words, we used ARIMA and tried to establish which part of the COVID-19 deaths were unregistered and which part of the overall mortality was influenced by the pandemic. In our modeling, we did not try to define and determine which part of the mortality was excess-this needs serious theoretical modeling that includes seasonal climate variations, dependence on population growth models, identification of other causes of mortality and their distribution to determine expected mortality, etc. With time series analysis, we could identify the excess part directly from data, by testing how much of the mortality variance for 2020-2021 could be explained by new COVID-19 cases. We used weekly data for deaths from the National Statistical Institute[33] and from John Hopkins University for total cases[34].
We added to the model categorical variable to account for the changes of dominant variants in Bulgaria during the first 2 years of the pandemic wild type, alpha and delta. The coevolutionary arms race between the acquired immunity for the survival of infections and reinfections and the mutating virus did not allow capturing of a clean and powerful connection between mortality and the disease severity caused by the virus variants because mortality was the product of interaction between the two coevolving agents.
Since the immune system adapts to decrease mortality, the mortality from previous waves also suppressed further mortality via natural selection. Thus, any positive correlation between variants and mortality in time-series analysis models carries more information than it appears purely from the coefficients and their standard estimation errors. Even the slightest edge of the variants over the immune system and the process of natural selection should be treated as significant here. This was the reason to include not only weekly COVID-19 cases with lags of 0-7 and 7-14 d (L1 and L2) but also variants as factor variables with the same lags in our optimal model.
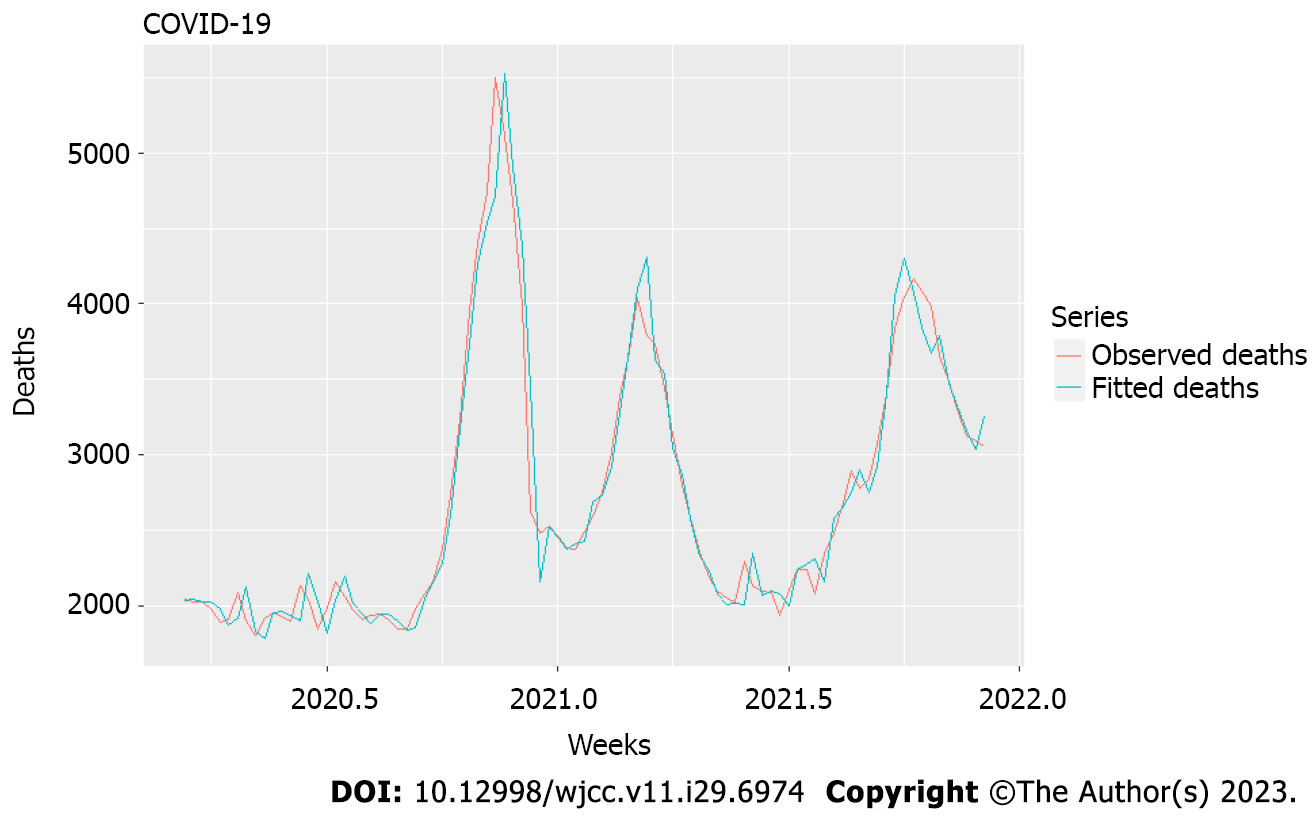
The model, described in Tables 1 and 2 and Figures 1 and 2, resulted from selecting different lags of the two chosen variables that produced the best fit (Figure 2). Also, the model was developed with the language R. It contained differencing of order I, as suggested by the ndiffs function via different unit root tests to achieve stationarity and meaningful correlations[35].
| Variables | Cases L1 | Cases L2 | Variants L1 | Variants L2 |
| Cases L1 | 1 | 0.69 | 0.02 | 0.03 |
| Cases L2 | 0.69 | 1 | 0.02 | 0.02 |
| Variants L1 | 0.02 | 0.02 | 1 | -0.02 |
| Variants L2 | 0.03 | 0.02 | -0.02 | 1 |
| Coefficient | Estimate | Standard error |
| Cases L1 | 0.0348 | 0.0103 |
| Cases L2 | 0.0394 | 0.0105 |
| Variants L1 | 106.4794 | 134.3759 |
| Variants L2 | 96.3354 | 134.5631 |
| MA1 | 0.2594 | 0.1100 |
| R2 | 0.953 | |
| RMSE | 188.83 | |
| Bias | -2.632 | |
| MAPE | 0.152 |
Factor variables such as variants L1 and L2 were not different. There was a significant influence on cases with lags L1 and L2 with a coefficient ratio to standard error over 3:1 (2:1 is required as a rule) and a sufficiently small correlation coefficient between their first differences of 0.69. Variants L1 and L2 had positive values but high standard errors of estimation.
Nonetheless, we considered their appearance significant after three different variants and the enormous increase of mortality over the 2 years of nearly 25% (on average) over the mean for 2015–2019 and with a high number of officially registered cases which was 11.5% of the overall population, with polymerase chain reaction (PCR) positivity on the average 12.14% (maximum 33.75%) indicating many more unregistered cases. Even this slight edge that we detected here indicated increasing severity with variants up to and including delta. Our model explains > 95% of the variation in deaths even though there was considerable variation in the mean age of new cases during these 2 years, and the exponentially increasing hospitalization (and therefore, mortality) risk with age[36]. This is evidence that most of the mortality increase in 2020 and 2021 was due to COVID-19. The model had shallow bias and mean absolute percentage error.
Conclusions from this model. Time-series analysis can serve as a first step in studying causal connections between an epidemic and excess mortality. Models such as ARIMA can show whether two or more nonstationary processes are moving together so that we can predict one behavior from another. Our models allowed us to catch when several different processes contributed with additional time lags to a resulting process, helping us uncover the link between two or more processes invisible to the naked eye. In this case, new weekly cases from the previous 2 wk, together with the changes in viral variants (factor variables), could explain 95% of the excess mortality. This is relevant as an answer to the often-appearing question of which part of the mortality during an epidemic is hidden from the official figures.
Moreover, during a pandemic with a high hospital burden, other patients have delayed treatment and are collateral victims of the pandemic. Time-series analysis can help quantify the excess mortality caused by a pandemic. It can help answer the question: Is the excess mortality due to closures or other mitigation measures, or due to the pandemic itself (although we did not try to answer this question in our study)? Closures could be added as a factor variable via the Oxford Stringency Index[37], and in a similar fashion, viral variants were added by us. Our research tried to check if variant evolution contributed to increased deaths. Still, it cannot be conclusively shown-the standard error of the relevant parameters, variants L1 and variants L2, is not small enough for that purpose (it should be at most half of the absolute value of the coefficient). This is possibly due to the only delta variant being significantly deadlier for that period[38].
However, it is essential to mention that virus variants change over time along with the clinical and epidemiological picture, as it was discussed recently by Miteva et al[39].
Medical imaging is crucial for initial diagnosis, staging, and follow-up. Therefore, the organization in departments of radiology is a significant topic because there has to be a separation of COVID-19 and possible COVID-19 patients from other patients in the hospital. Usually, it is done by arranging so-called COVID corridors in the hospital, when only COVID-19 patients are being scanned. This causes an interruption of regular hospital activity.
Usually, there is a delay in diagnosing patients with other diseases, which is a problem, especially with emergency patients. In the COVID corridor, the personnel in the radiology department are fully equipped to diagnose patients. However, the equipment was not always available, especially at the beginning of the pandemic. Moreover, it was costly which limited its use. Also, deep cleaning of the department is done after the end of the corridor, which causes even more delays in the other patients’ diagnoses and more expenses.
Forecasting the COVID-19 waves is crucial because the management of the radiology departments can be done according to it. A durable prediction model allows the departmental heads to organize the necessary COVID-19 corridors according to the expected wave. The duration and the exact hours of the corridors can be correctly adjusted, thus providing the required diagnosis of COVID-19 and non-COVID-19 patients with minimization of the delay of diagnosis for each group.
Also, the personnel shifts can be arranged according to the predicted model, providing enough X-ray technicians and radiologists. Furthermore, the necessary equipment will be provided in advance, reducing the needed time for changing clothes, and lowering expenses. The heads of the departments in which patients require diagnostic imaging can organize their work according to the COVID-19 corridor active hours, thus providing a safe and calm environment. Ambulatory patients can also arrange their examination according to the available hours.
In 2020, a genetic programming prediction model was introduced and further developed into a gene expression programming model. This model predicts the cases according to two parameters-confirmed cases and number of deaths[40]. Another prediction model used in India is ARIMA, which has value in predicting cases and shows the effect of unlocking after lockdown. The ARIMA model relies on the number of positive cases, the number of performed tests per day and the average positive percentage[41]. In the United Kingdom, weighted interval scoring was used for the prediction model, which used the data from the linear progression of 7-day cases[42]. In Chile, ARIMA (henceforth), exponential smoothing techniques, and Poisson models for time-dependent count data are used[43].
SARS-CoV-2 serology is used to identify previous infections, both in individuals and in populations. For this purpose, changes in antibody levels against SARS-CoV-2 are monitored to assess how they change over time and how long the protective immunity is preserved[44-46].
Many seroepidemiological studies are aimed at specific populations, such as health staff, police officers, and hospitalized patients with chronic diseases and COVID-19. Sometimes they use poorly and not well-validated laboratory methods[47] and mainly aim to study only the immunoglobulin G (IgG) response[48]. Therefore, various antibody responses began to be tested to improve the evaluation and diagnosis.
Many studies demonstrated that people with confirmed COVID-19 infection developed IgA, IgM and IgG against the S1 domain of the spike protein and nucleocapsid protein within 2 wk of symptoms[49,50]. Specific IgM antibodies are detected after 5-7 d of symptoms. After approximately 14 d, IgG begins to appear. IgA responses are detected almost simultaneously with IgM or earlier[51-53].
It was also found that the levels of antibodies correlated with the severity of the disease[54-57]. Previous research has found that with time, immunity to SARS-CoV-2 natural infection is short-lived and leads to a risk of reinfection[58-60].
А cohort study was conducted among health workers at the first SARS-CoV-2 epidemic peak in London[61]. They were tested weekly for symptoms, with RT-PCR and blood samples for 16-21 wk. Serological analysis was for IgG to the S1-domain of the spike protein and nucleocapsid protein. Asymptomatic or mild SARS-CoV-2 infection has been shown to elicit faster heterogeneous responses, and antibodies are cleared more quickly, which may affect the longevity of humoral immunity to SARS-CoV-2.
Another population-based study in Catalonia was conducted from February to November 2020. A multiplex serological test was used on 5000 participants from blood samples. Responses to 15 isotype-antigen combinations were monitored, and seroprevalence of 18.1% was found in adults and 15.3% in a simulation of the total population of Catalonia[62]. Based on the severity of the disease, immune profiles reveal that with increasing severity of infection, serum responses are more stable. The age and sex of the participants, overweight/obesity, compared with normal weight, and if they were or were not smokers when included in the study. There were no significant differences in seroprevalence between the two sexes. The seroprevalence among children was lower than in adults. Children were at lower risk of seropositivity than their parents in one family. Overweight/obese participants had higher antibody levels than those with normal weight. This is confusing because of suggesting that higher levels were adjusted for the severity of the disease[62].
A study on the seroprevalence of SARS-CoV-2 is currently being conducted in Barcelona from February 2021 to March 2022. The SeroCAP sentinel monitoring system is being used. IgG detection against SARS-CoV-2 spike protein will be performed monthly from blood samples collected from three hospitals. About 3000 samples are taken per month, and the prevalence will be assessed by age, sex and time in the three health zones in Barcelona. A complete analysis of the prevalence of SARS-CoV-2 infections will be performed, considering the demographic, social and economic factors. The correlation between seroprevalence confirmed cases of COVID-19. All measures applied so far will be studied[63].
A systematic review analyzed 47 studies from 23 countries[64]. In addition, other representative population-based studies at the national or regional level have been published[65-68]. The data showed that the SARS-CoV-2 seroprevalence in the general population varied from 0.37% to 22.1%. Biological, behavioral and social factors, including vaccine coverage, influence these percentages. We must acknowledge that the titers of protection against SARS-CoV-2 are currently unknown. However, virus-neutralizing antibodies are needed to protect and control the infection[69].
Mathematical modeling of pandemics is of vital importance for several reasons. One is to study the mechanisms of interactions between people, societal structures, and pathogens for past epidemics to develop knowledge that would help predict and control future ones. These are the classical epidemiological models, such as SIR and SEIR. Another reason is to enable us to manage a current epidemic by distinguishing productive from unproductive measurements and delivering precise short-term forecasts for the number of new cases, the number of new hospital admissions, expected deaths, etc.
This is the application of time series analysis, which relies on past values to predict future ones without extensive use of theoretical knowledge with all its uncertainties during an ongoing epidemic. It is used in mortality risk estimation, seroprevalence studies, reliable short-term forecasting for the healthcare system burden, and excess mortality estimation and analysis. It has a broad spectrum of linear and nonlinear, and single and multidimensional models. This allows one to choose ease of use vs capabilities according to different contexts where they are applied. Time series analysis supplements the classical epidemiological models in predictive and forecasting capabilities. It reinforces our decisions on how to act vs an epidemic with additional analytical approaches and results on which to step on.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: Bulgaria
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gupta L, Indonesia S-Editor: Fan JR L-Editor: Kerr C P-Editor: Fan JR
| 1. | Zeger SL, Irizarry R, Peng RD. On time series analysis of public health and biomedical data. Annu Rev Public Health. 2006;27:57-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 125] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 2. | Gu YL. Why do we Sometimes get Nonsense-Correlations between Time-Series?--A Study in Sampling and the Nature of Time-Series. J R Stat Soc. 1926;89:1. [RCA] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 692] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 3. | Barlow NS, Weinstein SJ. Accurate closed-form solution of the SIR epidemic model. Physica D. 2020;408:132540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 4. | Roda WC, Varughese MB, Han D, Li MY. Why is it difficult to accurately predict the COVID-19 epidemic? Infect Dis Model. 2020;5:271-281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 318] [Cited by in RCA: 262] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 5. | McBane G. SIR (Susceptible–Infectious–Removed) Model of Epidemiology as an Extended Example for Chemical Kinetics Students. J Chem Educ. 2021;98:2906-2911. [DOI] [Full Text] |
| 6. | Zhan C, Tse CK, Lai Z, Hao T, Su J. Prediction of COVID-19 spreading profiles in South Korea, Italy and Iran by data-driven coding. PLoS One. 2020;15:e0234763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 7. | Abolmaali S, Shirzaei S. A comparative study of SIR Model, Linear Regression, Logistic Function and ARIMA Model for forecasting COVID-19 cases. AIMS Public Health. 2021;8:598-613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Abuhasel KA, Khadr M, Alquraish MM. Analyzing and forecasting COVID-19 pandemic in the Kingdom of Saudi Arabia using ARIMA and SIR models. Comput Intell. 2022;38:770-783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Furtado P. Epidemiology SIR with Regression, Arima, and Prophet in Forecasting Covid-19. Eng Proc. 2021;5:52. [DOI] [Full Text] |
| 10. | Abolmaali S, Shirzaei S. Forecasting COVID-19 Number of Cases by Implementing ARIMA and SARIMA with Grid Search in the United States. 2021 Preprint. Available from: medRxiv:21258041. [DOI] [Full Text] |
| 11. | Roosa K, Lee Y, Luo R, Kirpich A, Rothenberg R, Hyman JM, Yan P, Chowell G. Real-time forecasts of the COVID-19 epidemic in China from February 5th to February 24th, 2020. Infect Dis Model. 2020;5:256-263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 354] [Cited by in RCA: 279] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 12. | Gupta R, Pal S. Trend Analysis and Forecasting of COVID-19 outbreak in India. 2020 Preprint. Available from: medRxiv:20044511. [DOI] [Full Text] |
| 13. | Tomov L, Tchorbadjieff A, Angelov S. Age-specific mortality risk from Covid-19 in Bulgaria, Computer Science and Education in Computer Science. 2021 Preprint. [DOI] [Full Text] |
| 14. | Xu B, Li J, Wang M. Epidemiological and time series analysis on the incidence and death of AIDS and HIV in China. BMC Public Health. 2020;20:1906. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 15. | Bhaskaran K, Gasparrini A, Hajat S, Smeeth L, Armstrong B. Time series regression studies in environmental epidemiology. Int J Epidemiol. 2013;42:1187-1195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 594] [Cited by in RCA: 771] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 16. | Claris S, Peter N. ARIMA model in predicting of COVID-19 epidemic for the southern Africa Region. Afr J Infect Dis. 2023;17:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 17. | Ayoobi N, Sharifrazi D, Alizadehsani R, Shoeibi A, Gorriz JM, Moosaei H, Khosravi A, Nahavandi S, Gholamzadeh Chofreh A, Goni FA, Klemeš JJ, Mosavi A. Time series forecasting of new cases and new deaths rate for COVID-19 using deep learning methods. Results Phys. 2021;27:104495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 18. | Rguibi MA, Moussa N, Madani A, Aaroud A, Zine-Dine K. Forecasting Covid-19 Transmission with ARIMA and LSTM Techniques in Morocco. SN Comput Sci. 2022;3:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Alabdulrazzaq H, Alenezi MN, Rawajfih Y, Alghannam BA, Al-Hassan AA, Al-Anzi FS. On the accuracy of ARIMA based prediction of COVID-19 spread. Results Phys. 2021;27:104509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 20. | Pinter G, Felde I, Mosavi A, Ghamisi P, Gloaguen R. COVID-19 Pandemic Prediction for Hungary; a Hybrid Machine Learning Approach. Mathematics. 2020;8:890. [RCA] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 101] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 21. | Burke RM, Midgley CM, Dratch A, Fenstersheib M, Haupt T, Holshue M, Ghinai I, Jarashow MC, Lo J, McPherson TD, Rudman S, Scott S, Hall AJ, Fry AM, Rolfes MA. Active Monitoring of Persons Exposed to Patients with Confirmed COVID-19 - United States, January-February 2020. MMWR Morb Mortal Wkly Rep. 2020;69:245-246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 274] [Cited by in RCA: 291] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 22. | Dowd JB, Andriano L, Brazel DM, Rotondi V, Block P, Ding X, Liu Y, Mills MC. Demographic science aids in understanding the spread and fatality rates of COVID-19. Proc Natl Acad Sci U S A. 2020;117:9696-9698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 648] [Cited by in RCA: 499] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 23. | Arun SS, Iyer GN. On the Analysis of COVID19 - Novel Corona Viral Disease Pandemic Spread Data Using Machine Learning Techniques. 2020 4th International Conference on Intelligent Computing and Control Systems (ICICCS); 2020: 1222-1227. Available from: https://ieeexplore.ieee.org/document/9121027. |
| 24. | Zeroual A, Harrou F, Dairi A, Sun Y. Deep learning methods for forecasting COVID-19 time-Series data: A Comparative study. Chaos Solitons Fractals. 2020;140:110121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 337] [Cited by in RCA: 146] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 25. | Babaei A, Ahmadi M, Jafari H, Liya A. A mathematical model to examine the effect of quarantine on the spread of coronavirus. Chaos Solitons Fractals. 2021;142:110418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Carannante M, D'Amato V, Iaccarino G. The Future Evolution of the Mortality Acceleration Due to the COVID-19: The Charlson Comorbidity Index in Stochastic Setting. Front Cardiovasc Med. 2022;9:938086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 27. | Wu LI, Feng Z. Homoclinic Bifurcation in an SIQR Model for Childhood Diseases. J Differ Equ. 2000;168:150-167. [DOI] [Full Text] |
| 28. | Singh H, Srivastava HM, Hammouch Z, Sooppy Nisar K. Numerical simulation and stability analysis for the fractional-order dynamics of COVID-19. Results Phys. 2021;20:103722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Gao W, Veeresha P, Prakasha DG, Baskonus HM. Novel Dynamic Structures of 2019-nCoV with Nonlocal Operator via Powerful Computational Technique. Biology (Basel). 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 30. | Boudaoui A, El Hadj Moussa Y, Hammouch Z, Ullah S. A fractional-order model describing the dynamics of the novel coronavirus (COVID-19) with nonsingular kernel. Chaos Solitons Fractals. 2021;146:110859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Tang B, Wang X, Li Q, Bragazzi NL, Tang S, Xiao Y, Wu J. Estimation of the Transmission Risk of the 2019-nCoV and Its Implication for Public Health Interventions. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 929] [Cited by in RCA: 701] [Article Influence: 140.2] [Reference Citation Analysis (0)] |
| 32. | Zamir M, Nadeem F, Abdeljawad T, Hammouch Z. Threshold condition and non pharmaceutical interventions's control strategies for elimination of COVID-19. Results Phys. 2021;20:103698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 33. | Republic of Bulgaria. Deaths in Bulgaria by weeks. [cited 20 August 2023]. Available from: https://www.nsi.bg/en/content/18121/basic-page/deaths-bulgaria-weeks. |
| 34. | Time series COVID-19. CSSEGISandData. [cited 20 August 2023]. Available from: https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv. |
| 35. | Search all 27018 R packages on CRAN and Bioconductor. RDocumentation. [cited 20 August 2023]. Available from: https://www.rdocumentation.org/packages/forecast/versions/8.16/topics/ndiffs. |
| 36. | Palmer S, Cunniffe N, Donnelly R. COVID-19 hospitalization rates rise exponentially with age, inversely proportional to thymic T-cell production. J R Soc Interface. 2021;18:20200982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 37. | OXFORD COVID-19 Government Response Stringency index. OCHA Services. [cited 20 August 2023]. Available from: https://data.humdata.org/dataset/oxford-covid-19-government-response-tracker?. |
| 38. | Tomov L, Batselova H, Velikova T. Estimating COVID Case Fatality Rate in Bulgaria for 2020–2021. In: Zlateva, T., Goleva, R. (eds) Computer Science and Education in Computer Science. CSECS 2022. Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering. Springer: Cham; 2022: 450. [DOI] [Full Text] |
| 39. | Miteva D, Kitanova M, Batselova H, Lazova S, Chervenkov L, Peshevska-Sekulovska M, Sekulovski M, Gulinac M, Vasilev GV, Tomov L, Velikova T. The End or a New Era of Development of SARS-CoV-2 Virus: Genetic Variants Responsible for Severe COVID-19 and Clinical Efficacy of the Most Commonly Used Vaccines in Clinical Practice. Vaccines (Basel). 2023;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 40. | Salgotra R, Gandomi M, Gandomi AH. Time Series Analysis and Forecast of the COVID-19 Pandemic in India using Genetic Programming. Chaos Solitons Fractals. 2020;138:109945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 41. | Singh S, Chowdhury C, Panja AK, Neogy S. Time Series Analysis of COVID-19 Data to Study the Effect of Lockdown and Unlock in India. J Inst Eng India Ser B. 2021;102:1275-1281. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 42. | Meakin S, Abbott S, Bosse N, Munday J, Gruson H, Hellewell J, Sherratt K; CMMID COVID-19 Working Group, Funk S. Comparative assessment of methods for short-term forecasts of COVID-19 hospital admissions in England at the local level. BMC Med. 2022;20:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Barría-Sandoval C, Ferreira G, Benz-Parra K, López-Flores P. Prediction of confirmed cases of and deaths caused by COVID-19 in Chile through time series techniques: A comparative study. PLoS One. 2021;16:e0245414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang ML, Jerome KR, Bloom JD, Greninger AL. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J Clin Microbiol. 2020;58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 457] [Cited by in RCA: 433] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 45. | Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings O, O'Byrne A, Kouphou N, Galao RP, Betancor G, Wilson HD, Signell AW, Winstone H, Kerridge C, Huettner I, Jimenez-Guardeño JM, Lista MJ, Temperton N, Snell LB, Bisnauthsing K, Moore A, Green A, Martinez L, Stokes B, Honey J, Izquierdo-Barras A, Arbane G, Patel A, Tan MKI, O'Connell L, O'Hara G, MacMahon E, Douthwaite S, Nebbia G, Batra R, Martinez-Nunez R, Shankar-Hari M, Edgeworth JD, Neil SJD, Malim MH, Doores KJ. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598-1607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1049] [Cited by in RCA: 935] [Article Influence: 187.0] [Reference Citation Analysis (0)] |
| 46. | Jones JM, Stone M, Sulaeman H, Fink RV, Dave H, Levy ME, Di Germanio C, Green V, Notari E, Saa P, Biggerstaff BJ, Strauss D, Kessler D, Vassallo R, Reik R, Rossmann S, Destree M, Nguyen KA, Sayers M, Lough C, Bougie DW, Ritter M, Latoni G, Weales B, Sime S, Gorlin J, Brown NE, Gould CV, Berney K, Benoit TJ, Miller MJ, Freeman D, Kartik D, Fry AM, Azziz-Baumgartner E, Hall AJ, MacNeil A, Gundlapalli AV, Basavaraju SV, Gerber SI, Patton ME, Custer B, Williamson P, Simmons G, Thornburg NJ, Kleinman S, Stramer SL, Opsomer J, Busch MP. Estimated US Infection- and Vaccine-Induced SARS-CoV-2 Seroprevalence Based on Blood Donations, July 2020-May 2021. JAMA. 2021;326:1400-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 162] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 47. | Chen X, Chen Z, Azman AS, Deng X, Sun R, Zhao Z, Zheng N, Chen X, Lu W, Zhuang T, Yang J, Viboud C, Ajelli M, Leung DT, Yu H. Serological evidence of human infection with SARS-CoV-2: a systematic review and meta-analysis. Lancet Glob Health. 2021;9:e598-e609. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 163] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 48. | Lai CC, Wang JH, Hsueh PR. Population-based seroprevalence surveys of anti-SARS-CoV-2 antibody: An up-to-date review. Int J Infect Dis. 2020;101:314-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 148] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 49. | Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, García-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26:1033-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1452] [Cited by in RCA: 1379] [Article Influence: 275.8] [Reference Citation Analysis (2)] |
| 50. | Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken CBEM, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg Infect Dis. 2020;26:1478-1488. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1123] [Cited by in RCA: 1120] [Article Influence: 224.0] [Reference Citation Analysis (0)] |
| 51. | Vogl T, Leviatan S, Segal E. SARS-CoV-2 antibody testing for estimating COVID-19 prevalence in the population. Cell Rep Med. 2021;2:100191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 52. | Sterlin D, Mathian A, Miyara M, Mohr A, Anna F, Claër L, Quentric P, Fadlallah J, Devilliers H, Ghillani P, Gunn C, Hockett R, Mudumba S, Guihot A, Luyt CE, Mayaux J, Beurton A, Fourati S, Bruel T, Schwartz O, Lacorte JM, Yssel H, Parizot C, Dorgham K, Charneau P, Amoura Z, Gorochov G. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 715] [Cited by in RCA: 769] [Article Influence: 192.3] [Reference Citation Analysis (0)] |
| 53. | Yu HQ, Sun BQ, Fang ZF, Zhao JC, Liu XY, Li YM, Sun XZ, Liang HF, Zhong B, Huang ZF, Zheng PY, Tian LF, Qu HQ, Liu DC, Wang EY, Xiao XJ, Li SY, Ye F, Guan L, Hu DS, Hakonarson H, Liu ZG, Zhong NS. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur Respir J. 2020;56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 246] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 54. | Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin Infect Dis. 2020;71:2027-2034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1590] [Cited by in RCA: 1840] [Article Influence: 368.0] [Reference Citation Analysis (0)] |
| 55. | Liu L, To KK, Chan KH, Wong YC, Zhou R, Kwan KY, Fong CH, Chen LL, Choi CY, Lu L, Tsang OT, Leung WS, To WK, Hung IF, Yuen KY, Chen Z. High neutralizing antibody titer in intensive care unit patients with COVID-19. Emerg Microbes Infect. 2020;9:1664-1670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 114] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 56. | van Tol S, Mögling R, Li W, Godeke GJ, Swart A, Bergmans B, Brandenburg A, Kremer K, Murk JL, van Beek J, Wintermans B, Reimerink J, Bosch BJ, Reusken C. Accurate serology for SARS-CoV-2 and common human coronaviruses using a multiplex approach. Emerg Microbes Infect. 2020;9:1965-1973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 57. | Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, Hu JL, Xu W, Zhang Y, Lv FJ, Su K, Zhang F, Gong J, Wu B, Liu XM, Li JJ, Qiu JF, Chen J, Huang AL. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1878] [Cited by in RCA: 1975] [Article Influence: 395.0] [Reference Citation Analysis (0)] |
| 58. | Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol. 2020;101:791-797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 259] [Article Influence: 51.8] [Reference Citation Analysis (1)] |
| 59. | Edridge AWD, Kaczorowska J, Hoste ACR, Bakker M, Klein M, Loens K, Jebbink MF, Matser A, Kinsella CM, Rueda P, Ieven M, Goossens H, Prins M, Sastre P, Deijs M, van der Hoek L. Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26:1691-1693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 530] [Article Influence: 106.0] [Reference Citation Analysis (0)] |
| 60. | Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, Nakao C, Rayaprolu V, Rawlings SA, Peters B, Krammer F, Simon V, Saphire EO, Smith DM, Weiskopf D, Sette A, Crotty S. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2187] [Cited by in RCA: 1935] [Article Influence: 483.8] [Reference Citation Analysis (0)] |
| 61. | Manisty C, Treibel TA, Jensen M, Semper A, Joy G, Gupta RK, Cutino-Moguel T, Andiapen M, Jones J, Taylor S, Otter A, Pade C, Gibbons J, Lee J, Bacon J, Thomas S, Moon C, Jones M, Williams D, Lambourne J, Fontana M, Altmann DM, Boyton R, Maini M, McKnight A, Chain B, Noursadeghi M, Moon JC. Time series analysis and mechanistic modelling of heterogeneity and sero-reversion in antibody responses to mild SARS-CoV-2 infection. EBioMedicine. 2021;65:103259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 62. | Karachaliou M, Moncunill G, Espinosa A, Castaño-Vinyals G, Jiménez A, Vidal M, Santano R, Barrios D, Puyol L, Carreras A, Mayer L, Rubio R, Cortés B, Pleguezuelos V, O'Callaghan-Gordo C, Fossati S, Rivas I, Casabonne D, Vrijheid M, Izquierdo L, Aguilar R, Basagaña X, Garcia-Aymerich J, de Cid R, Dobaño C, Kogevinas M. Infection induced SARS-CoV-2 seroprevalence and heterogeneity of antibody responses in a general population cohort study in Catalonia Spain. Sci Rep. 2021;11:21571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 63. | Sentís A, Torán P, Esperalba J, Agustí C, Ángel M, Fernández MG, Dopico E, Salvador-González B, González MV, Bordas A, Antón A, Violan C, Montoro-Fernández M, Aceiton J, Egea-Cortés L, Alonso L, Dacosta-Aguayo R, Calatayud L, Lejardi Y, Mendioroz J, Basora J, Reyes-Urueña J, Casabona J. Monitoring of SARS-CoV-2 seroprevalence among primary healthcare patients in the Barcelona Metropolitan Area: the SeroCAP sentinel network protocol. BMJ Open. 2022;12:e053237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 64. | Rostami A, Sepidarkish M, Leeflang MMG, Riahi SM, Nourollahpour Shiadeh M, Esfandyari S, Mokdad AH, Hotez PJ, Gasser RB. SARS-CoV-2 seroprevalence worldwide: a systematic review and meta-analysis. Clin Microbiol Infect. 2021;27:331-340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 262] [Cited by in RCA: 242] [Article Influence: 60.5] [Reference Citation Analysis (0)] |
| 65. | Stringhini S, Wisniak A, Piumatti G, Azman AS, Lauer SA, Baysson H, De Ridder D, Petrovic D, Schrempft S, Marcus K, Yerly S, Arm Vernez I, Keiser O, Hurst S, Posfay-Barbe KM, Trono D, Pittet D, Gétaz L, Chappuis F, Eckerle I, Vuilleumier N, Meyer B, Flahault A, Kaiser L, Guessous I. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet. 2020;396:313-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 849] [Cited by in RCA: 701] [Article Influence: 140.2] [Reference Citation Analysis (0)] |
| 66. | Stadlbauer D, Tan J, Jiang K, Hernandez MM, Fabre S, Amanat F, Teo C, Arunkumar GA, McMahon M, Capuano C, Twyman K, Jhang J, Nowak MD, Simon V, Sordillo EM, van Bakel H, Krammer F. Repeated cross-sectional sero-monitoring of SARS-CoV-2 in New York City. Nature. 2021;590:146-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 105] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 67. | Carlos B, Jordi S, Diana F. Seroprevalence against COVID-19 and follow-up of suspected cases in primary health care in Spain. 2020 Preprint. Available from: medRxiv:13.20130575. [DOI] [Full Text] |
| 68. | Castro Dopico X, Muschiol S, Christian M, Hanke L, Sheward DJ, Grinberg NF, Rorbach J, Bogdanovic G, Mcinerney GM, Allander T, Wallace C, Murrell B, Albert J, Karlsson Hedestam GB. Seropositivity in blood donors and pregnant women during the first year of SARS-CoV-2 transmission in Stockholm, Sweden. J Intern Med. 2021;290:666-676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 69. | Slabakova Y, Gerenska D, Ivanov N, Velikova T. Immune titers of protection against severe acute respiratory syndrome coronavirus 2: are we there yet? Explor Immunol. 2022;2:9-24. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |