Published online Oct 6, 2023. doi: 10.12998/wjcc.v11.i28.6895
Peer-review started: June 19, 2023
First decision: August 16, 2023
Revised: August 23, 2023
Accepted: September 14, 2023
Article in press: September 14, 2023
Published online: October 6, 2023
Processing time: 97 Days and 20.9 Hours
Quantitative fluorescent polymerase chain reaction (QF-PCR) is a rapid prenatal diagnostic method for abnormalities on chromosomes 21, 18, and 13 and sex chromosomal aneuploidy. However, the value of QF-PCR in diagnosing chromosomal structural abnormalities is limited. In this article, we report a confusing QF-PCR finding in a pregnant woman who underwent amniocentesis.
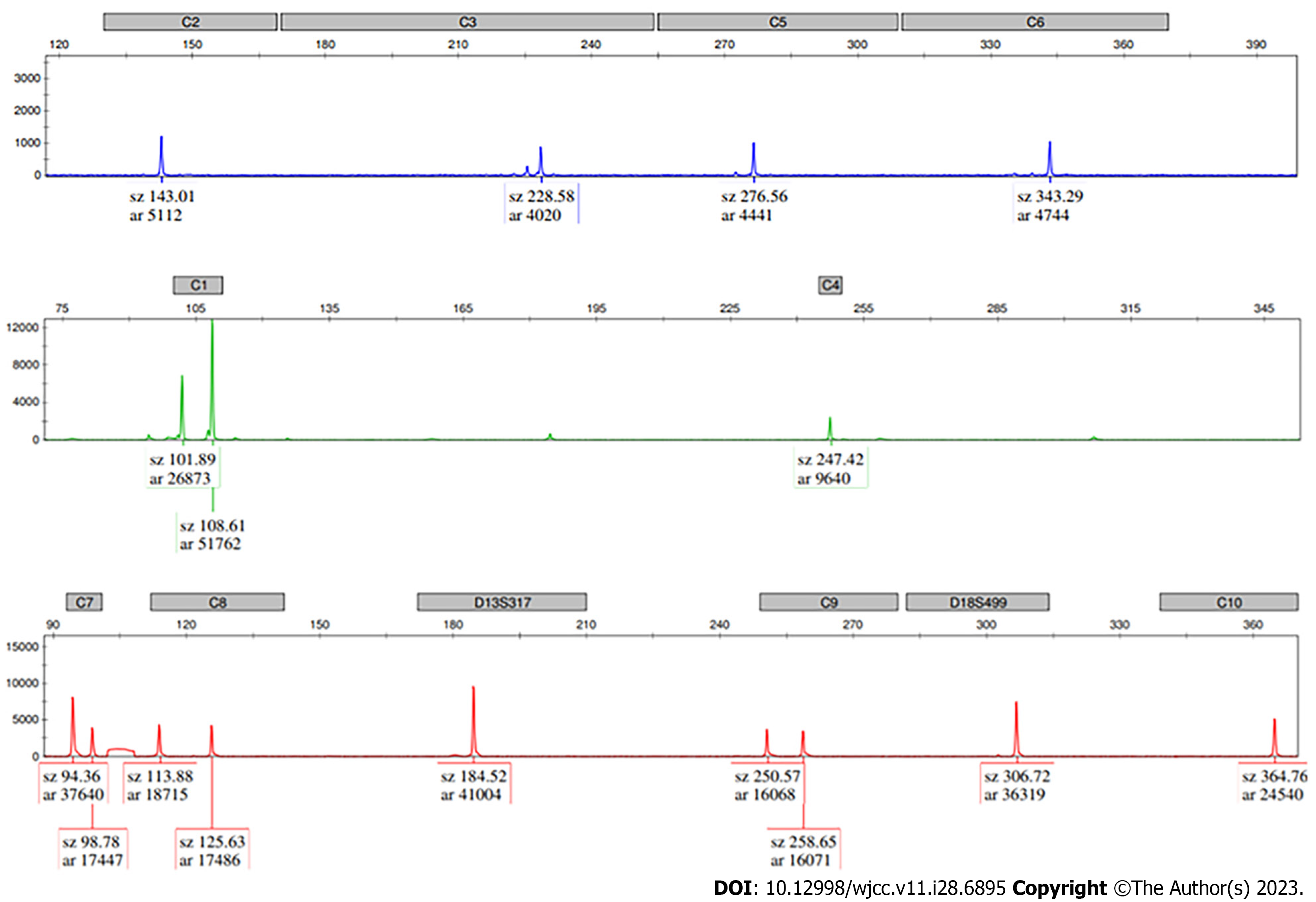
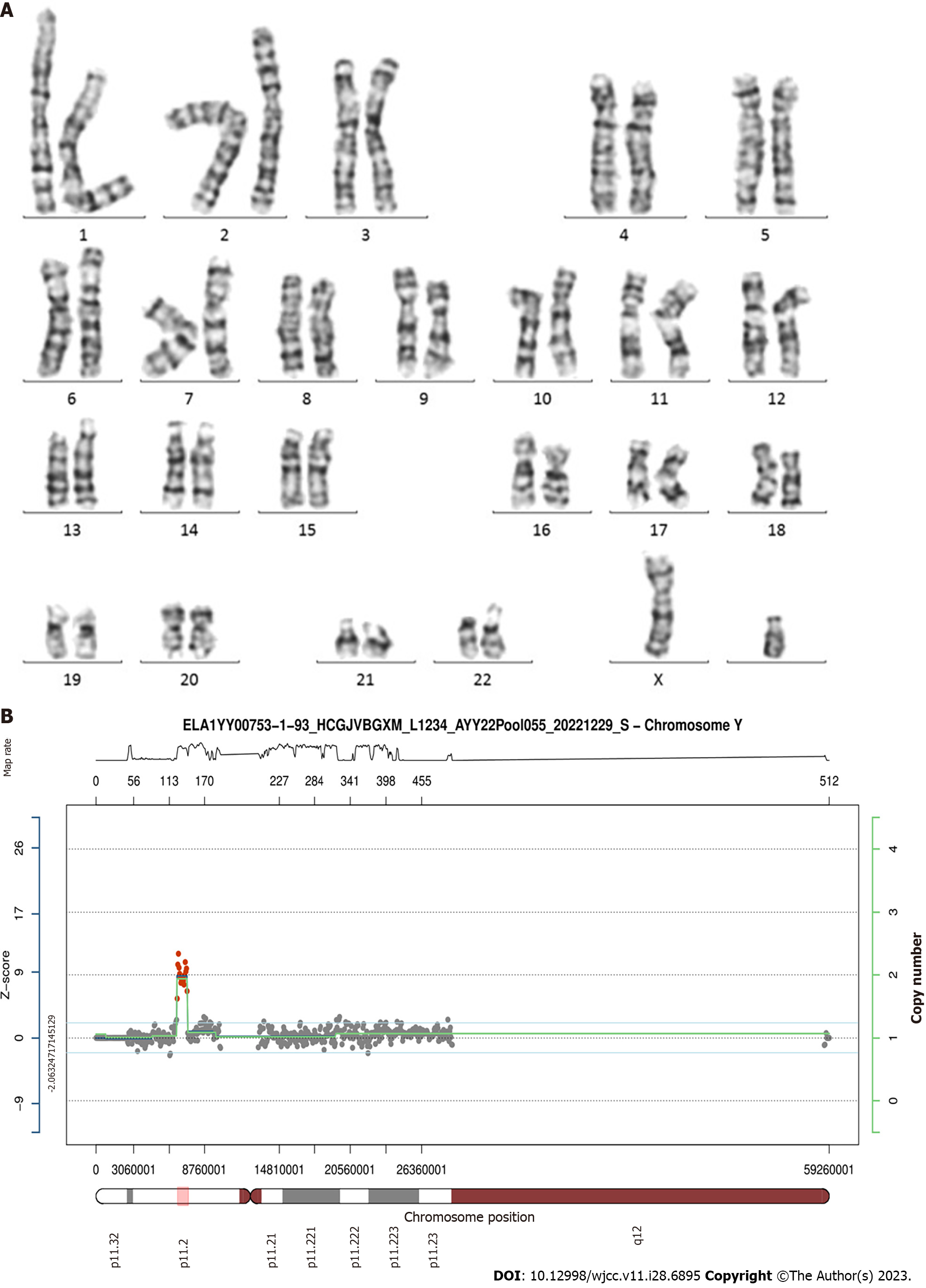
The short tandem repeat marker AMXY (Xp22.2/Yp11.2) located on the sex chromosome exhibited a trisomic biallelic pattern, indicating that the karyotype of the fetus might be 47,XYY. Chromosome analysis performed on cultured amniocytes showed a normal male karyotype of the fetus. Copy number variation sequencing confirmed a 500 kb duplication at Yp11.2-Yp11.2 (chrY:6610001_ 7110000) and a 250 kb duplication at Yp11.2-Yp11.2 (chrY:7110001_7360000).
In conclusion, the comprehensive application of different methods could achieve a higher detection rate and accuracy for the prenatal diagnosis of chromosomal disorders through chromosomal testing.
Core Tip: This case represented an interesting and clinically rare occurrence. The short tandem repeat marker AMXY (Xp22.2/Yp11.2) located on the sex chromosome exhibited a trisomic biallelic pattern in quantitative fluorescent polymerase chain reaction (QF-PCR), indicating the possible existence of two Y chromosomes. However, by analyzing the results of copy number variation sequencing and karyotyping, the fetus was confirmed to have only a partial duplication of the Y chromosome instead of the 47,XYY karyotype. The duplications identified do not include any genes annotated in the Online Mendelian Inheritance in Man database and will not cause the occurrence of birth defects. Therefore, when an abnormality is detected in the QF-PCR data mentioned above, additional methods should be used for comprehensive judgment to avoid misdiagnosis. The strength of the present report is that it provides valuable experience for prenatal diagnosis.
- Citation: Chen C, Tang T, Song QL, He YJ, Cai Y. Confusing finding of quantitative fluorescent polymerase chain reaction analysis in invasive prenatal genetic diagnosis: A case report. World J Clin Cases 2023; 11(28): 6895-6901
- URL: https://www.wjgnet.com/2307-8960/full/v11/i28/6895.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i28.6895
Aneuploidy is one of the most common chromosome abnormalities. Aneuploidy usually occurs in chromosomes 13, 18, and 21 and sex chromosomes. Quantitative fluorescent polymerase chain reaction (QF-PCR) is an accurate and rapid detection method for chromosomal aneuploidy. It can detect the number of abnormalities on chromosomes 13, 18, and 21 and the X and Y chromosomes using short tandem repeat (STR) markers. The advantages of QF-PCR include a short detection time, high throughput, and not requiring cell culture[1]. Due to rapid testing, low cost, and high efficiency, QF-PCR has been widely used in prenatal diagnostic examinations for pregnant women worldwide.
However, the value of QF-PCR in diagnosing chromosomal structural abnormalities is limited. To facilitate the recognition of structural aberrations, karyotyping and copy number variation sequencing (CNV-Seq) have been applied. CNV-Seq offers a higher test resolution than traditional karyotyping and has been recommended as the first-line genetic test for prenatal diagnosis. Here, we report a confusing QF-PCR result from a pregnant woman’s amniotic fluid. We provide an explanation for this abnormal result.
A 29-year-old woman had undergone first-trimester serological Down syndrome screening. The results indicated that the fetus had a high risk of Down syndrome.
There were no remarkable abnormalities.
Unremarkable.
There was no history of previous pregnancy complications.
Unremarkable.
The pregnant woman visited our center and underwent amniocentesis at 18 wk of gestation. QF-PCR results from amniotic cells excluded aneuploidies in chromosomes 21, 18, and 13. The peak area ratio of AMXY (Xp22.2/Yp11.2) was 0.569, indicating the possible existence of two Y chromosomes (Figure 1). According to these findings, the first impression of the karyotype of this fetus was 47,XYY. However, chromosome analysis performed on cultured amniocytes showed a normal male karyotype of the fetus. The results of karyotype analysis indicated that the number of chromosomes was 46, and the construction of the chromosomes was regular, which did not support the diagnosis of 47,XYY syndrome (Figure 2).
It was strange that the results of the two methods were inconsistent. Therefore, we added STR markers on the sex chromosomes for further detection. Among the additional STR markers, the peak area ratio of TAF9b (3p24.2/Xq21.1) was 2.15, indicating monosomy X. The peak height ratios of DXYS218 (Xq22.33/Yp11.32) and DXYS267 (Xq21.31/Yp11.31) were both 1.0, indicating monosomy Y. Consequently, the result did not meet the criteria for 47,XYY syndrome. Therefore, a duplication in the Yp11.2 region was considered. To further clarify this issue, CNV-Seq was conducted (Figure 2). CNV-Seq identified a 500 kb duplication at Yp11.2-Yp11.2 (chrY:6610001_7110000) and a 250 kb duplication at Yp11.2-Yp11.2 (chrY:7110001_7360000), which did not include any genes annotated in the Online Mendelian Inheritance in Man (OMIM) database. The STR marker AMXY (Xp22.2/Yp11.2) was located in the duplicated region at Yp11.2-Yp11.2.
No abnormalities were found in the obstetrics ultrasound examination.
The results of the laboratory examinations indicated only a partial duplication of the Y chromosome, instead of the 47,XYY karyotype.
No specific treatment was needed.
The pregnant woman was recommended to continue the pregnancy. Ultimately, she gave birth to a healthy child.
Prenatal diagnosis is an essential tool in providing genetic counseling for the prognosis and outcome of birth defects[2]. There are many methods for diagnosing chromosomal abnormalities, such as QF-PCR, karyotyping, CNV-Seq, and chromosomal microarray analysis. They each have their own advantages and shortcomings in prenatal diagnosis. By analyzing the polymorphism of STR genetic markers, QF-PCR can not only identify maternal contamination but also detect common chromosomal aneuploidy, triploidy, and uniparental disomy[1,3]. The advantages of QF-PCR include its shorter turnaround time and affordability. However, it cannot detect structural chromosome aberrations. Karyotyping, which analyzes cells extracted from the amniotic fluid, is the gold standard for diagnosing fetal chromosomal abnormalities. It can detect numerical and structural anomalies (resolution > 5 Mb) for 23 pairs of chromosomes. On the other hand, there are many shortcomings of karyotyping, such as its cell culture requirements and high manpower and time costs[4,5]. The CNV-Seq technique is a new testing method based on low-depth whole-genome sequencing. It has been extensively used in the detection of chromosomal aberrations such as aneuploidy, microdeletion, and microduplication (resolution > 100 kb)[6]. However, CNV-Seq is complex, and the cost is high. Therefore, the comprehensive application of these methods is particularly important for different patients.
One of the most common male sex chromosome abnormalities is 47,XYY syndrome, which is a sex chromosome abnormality in males caused by nondisjunction during paternal meiosis II or postzygotic mitosis[7]. Patients with 47,XYY syndrome usually have normal gonadal function and the ability to produce sperm[7,8]. However, some patients may still develop testicular failure, which can lead to problems with fertility. Due to the lack of clinical symptoms during birth and the growth process, the diagnosis of 47,XYY is easy to miss. Therefore, it is of great importance to improve prenatal diagnostic techniques and draw pregnant women’s attention to prenatal examinations.
In this case, we observed that the bimodal area ratio of AMXY (Xp22.2/Yp11.2) on the sex chromosome was 0.519 in QF-PCR. This result indicated that the fetus might have had an extra Y chromosome. Then, additional STR markers were applied for further detection. The peak area ratio of TAF9b (3P24.2/Xq21.1) was 2.14. The peak height ratios of DXYS218 (Xq22.33/Yp11.32) and DXYS267 (Xq21.31/Yp11.31) were both 1.0. Based on these results, this patient did not meet the criteria for 47,XYY syndrome. The results of the karyotype analysis showed 46,XY. CNV-Seq revealed a 500 kb duplication at Yp11.2-Yp11.2 (chrY:6610001_7110000) and a 250 kb duplication at Yp11.2-Yp11.2 (chrY:7110001_7360000). The duplications identified do not include any genes annotated in the OMIM database and will not cause the occurrence of birth defects. Finally, the diagnosis in this case was not 47,XYY syndrome. Although the QF-PCR results were abnormal, the actual diagnosis was a duplication at the Yp11.2 region. In fact, this is a rare phenomenon, although it is possible. The nonrecombinant region of the Y chromosome contains many highly homologous repeat sequences, which may lead to structural abnormalities of chromosomes through deletions, duplications, and inversions[9].
QF-PCR is reliable for the prenatal diagnosis of numerical anomalies of chromosomes 13, 18, 21, X, and Y. However, QF-PCR cannot detect inversions or translocations. The deletion or duplication region outside the STR loci will also be missed[10,11]. Consequently, QF-PCR may overlook structural chromosomal abnormalities. When the QF-PCR analysis shows abnormal peak height, additional techniques for prenatal diagnosis should be applied. In this case, we reported a duplicated region located on chromosome Y. The STR marker AMXY (Xp22.2/Yp11.2) was located in the duplicated region at Yp11.2-Yp11.2. This is the first case of Yp11.2 duplication that we have found among more than 3000 patients who have undergone QF-PCR in our laboratory. Previous studies have reported the function of QF-PCR in detecting mosaicism, deletions, or duplications[11-13]. These cases had deletion or duplication regions within the STR marker range designed in the QF-PCR kit. This is consistent with the conclusion of our study.
Moreover, it is important to analyze the peak ratios of more QF-PCR STR markers with trisomic biallelic patterns of the AMXY (Xp22.2/Yp11.2) marker. This might suggest the presence of copy number variants instead of two existing Y chromosomes. In addition, it is necessary to further analyze uncertain cases by karyotyping or next-generation sequencing. However, the low resolution of karyotyping analysis (> 5 Mb) may affect the accuracy of the results[5]. In our case, the result displayed a normal karyotype of the fetus. To further identify the potential structural abnormality on the Y chromosome, CNV-Seq analysis was performed and identified 250 kb and 500 kb duplications at Yp11.2-Yp11.2. By doing this, we can minimize the number of misdiagnosed cases. Amy Inkster et al[13] found that fetal QF-PCR showed a biallelic trisomic pattern for the X chromosome microsatellite marker DXS1187, with an otherwise normal male amplification pattern for all other sex chromosome markers. Chromosomal microarray analysis results indicated a maternally inherited 304 kb triplication within chromosome Xq26.2. As reported in the previous literature, QF-PCR cannot determine the starting location or fragment length of the deletion or duplication region[12,13]. Therefore, QF-PCR cannot be used as a routine method for detecting chromosomal deletions or duplications. It can only be used as a supplementary method to discover deletions or duplications in the detection of 13/18/21/X/Y chromosome aneuploidy[11,14]. To compensate for the shortcomings of QF-PCR, karyotyping and CNV-seq have been used in prenatal diagnosis. In recent years, with the development of molecular biology, molecular diagnostic technology has provided an effective supplement to traditional karyotype analysis. Some scholars have found that the combination of CNV-seq and QF-PCR could effectively reduce the risk of misdiagnosis and may be considered a routine method for prenatal chromosomal abnormality diagnosis[1,15].
In the present case, we confirmed two duplication regions within chromosome Yp11.2 by CNV-Seq after the detection of a trisomic biallelic pattern of the AMXY (Xp22.2/Yp11.2) marker by QF-PCR. There are some strengths of the present paper. First, this is the first report of duplications at Yp11.2-Yp11.2 by QF-PCR analysis. Second, duplications of these chromosome segments do not seem to cause birth defects. Last but not least, it provides valuable experience in the comprehensive application of different prenatal diagnostic techniques. QF-PCR can help detect deletions or duplications within STR loci. When an abnormality is detected in the QF-PCR data mentioned above, another method should be used for comprehensive judgment to avoid misdiagnosis, such as karyotyping, CNV-seq, and microarray analysis.
In conclusion, the comprehensive application of different methods could achieve a higher detection rate and accuracy for the prenatal diagnosis of chromosomal disorders through chromosomal testing.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Genetics and heredity
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mhatre S, India S-Editor: Chen YL L-Editor: Wang TQ P-Editor: Yu HG
| 1. | Qiao J, Yuan J, Hu W, Li Q, Fang H, Xu Y, Dai Y. Combined diagnosis of QF-PCR and CNV-Seq in fetal chromosomal abnormalities: A new perspective on prenatal diagnosis. J Clin Lab Anal. 2022;36:e24311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 2. | Wagner R, Tse WH, Gosemann JH, Lacher M, Keijzer R. Prenatal maternal biomarkers for the early diagnosis of congenital malformations: A review. Pediatr Res. 2019;86:560-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Liu Y, Guo L, Chen H, Lu J, Hu J, Li X, Wang T, Li F, Yin A. Discrepancy of QF-PCR, CMA and karyotyping on a de novo case of mosaic isodicentric Y chromosomes. Mol Cytogenet. 2019;12:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Dai P, Zhu X, Pei Y, Chen P, Li J, Gao Z, Liang Y, Kong X. Evaluation of optical genome mapping for detecting chromosomal translocation in clinical cytogenetics. Mol Genet Genomic Med. 2022;10:e1936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 5. | Zhang Q, Wang Y, Zhou J, Zhou R, Liu A, Meng L, Ji X, Hu P, Xu Z. 11q13.3q13.4 deletion plus 9q21.13q21.33 duplication in an affected girl arising from a familial four-way balanced chromosomal translocation. Mol Genet Genomic Med. 2023;e2248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | She Q, Zhen L, Fu F, Lei TY, Li LS, Li R, Wang D, Zhang YL, Jing XY, Yi CX, Zhong HZ, Tan WH, Li FG, Liao C. [Prenatal genetic diagnosis of the fetuses with isolated corpus callosum abnormality]. Zhonghua Fu Chan Ke Za Zhi. 2022;57:671-677. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 7. | Berglund A, Stochholm K, Gravholt CH. Morbidity in 47,XYY syndrome: a nationwide epidemiological study of hospital diagnoses and medication use. Genet Med. 2020;22:1542-1551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Berglund A, Stochholm K, Gravholt CH. The epidemiology of sex chromosome abnormalities. Am J Med Genet C Semin Med Genet. 2020;184:202-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 9. | Lange J, Noordam MJ, van Daalen SK, Skaletsky H, Clark BA, Macville MV, Page DC, Repping S. Intrachromosomal homologous recombination between inverted amplicons on opposing Y-chromosome arms. Genomics. 2013;102:257-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Masoudzadeh N, Teimourian S. Comparison of quantitative fluorescent polymerase chain reaction and karyotype analysis for prenatal screening of chromosomal aneuploidies in 270 amniotic fluid samples. J Perinat Med. 2019;47:631-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Xu C, Peng J, Zhang Y, Liang S, Wang D. Detection of partial deletion and mosaicism using quantitative fluorescent polymerase chain reaction: Case reports and a review of the literature. J Clin Lab Anal. 2022;36:e24574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 12. | Le TNU, Nguyen VN, Doan TDA, Doan HNB, Le PTQ, Le TL, Ha TMT. An experience in prenatal diagnosis via QF-PCR of a female child with a 9.9 Mb pure deletion at 18p11.32-11.22. Nagoya J Med Sci. 2020;82:783-790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 13. | Inkster A, Thomas MA, Gamache NS, Chan M, Stenroos P, Chernos JE, Argiropoulos B. A Challenging Prenatal QF-PCR Rapid Aneuploidy Test Result Caused by a Maternally Inherited Triplication within Chromosome Xq26.2. Cytogenet Genome Res. 2018;156:5-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 14. | Tekcan A, Tural S, Elbistan M, Kara N, Guven D, Kocak I. The combined QF-PCR and cytogenetic approach in prenatal diagnosis. Mol Biol Rep. 2014;41:7431-7436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 15. | Chen L, Wang L, Tang F, Zeng Y, Yin D, Zhou C, Zhu H, Li L, Zhang L, Wang J. Copy number variation sequencing combined with quantitative fluorescence polymerase chain reaction in clinical application of pregnancy loss. J Assist Reprod Genet. 2021;38:2397-2404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |