Published online Sep 26, 2023. doi: 10.12998/wjcc.v11.i27.6383
Peer-review started: June 22, 2023
First decision: July 17, 2023
Revised: August 2, 2023
Accepted: August 23, 2023
Article in press: August 23, 2023
Published online: September 26, 2023
Processing time: 90 Days and 4.1 Hours
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer. With highly invasive biological characteristics and a lack of obvious clinical manifestations, HCC usually has a poor prognosis and ranks fourth in cancer mortality. The aetiology and exact molecular mechanism of primary HCC are still unclear.
To select the characteristic genes that are significantly associated with the prognosis of HCC patients and construct a prognosis model of this malignancy.
By comparing the gene expression levels of patients with different cancer grades of HCC, we screened out differentially expressed genes associated with tumour grade. By protein-protein interaction (PPI) network analysis, we obtained the top 2 PPI networks and hub genes from these differentially expressed genes. By using least absolute shrinkage and selection operator Cox regression, 13 prognostic genes were selected for feature extraction, and a prognostic risk model of HCC was established.
The model had significant prognostic ability in HCC. We also analysed the biological functions of these prognostic genes.
By comparing the gene profiles of patients with different stages of HCC, We have constructed a prognosis model consisting of 13 genes that have important prognostic value. This model has good application value and can be explained clinically.
Core Tip: By comparing the gene expression levels of hepatocellular carcinoma patients with different grades, we investigated the biological function of genes and selected 13 genes to construct a prognostic model. The results show that the model has effective predictive ability for liver cancer prognosis and appreciated clinical interpretation value.
- Citation: Zhang GX, Ding XS, Wang YL. Prognostic model of hepatocellular carcinoma based on cancer grade. World J Clin Cases 2023; 11(27): 6383-6397
- URL: https://www.wjgnet.com/2307-8960/full/v11/i27/6383.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i27.6383
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer and accounts for more than 90% of primary tumours of the liver. HCC is now the world’s fifth most widespread cancer type and the second leading cause of cancer death[1,2]. Currently, the morbidity of HCC worldwide is still rising owing to the absence of obvious signs in early stages and rapid tumour development[3]. Moreover, the five-year survival rate for HCC is only 18%, as more than 60% of HCC patients are identified in late stages and have a poor prognosis[4]. The recurrence-free survival and overall survival (OS) of patients with HCC are still largely unsatisfactory[5-7]. At present, serum alpha-fetoprotein (AFP), abdominal ultrasound, and triple-phase helical computed tomography are the main diagnostic methods for HCC[8-11]. Regrettably, the accuracy and specificity of these approaches are still unsatisfying, especially in early stages of disease[12]. To date, there are few effective biomarkers for HCC monitoring and early diagnosis. Thus, there is a pressing need to evaluate innovative treatment targets and early diagnostic markers to improve the prognosis of HCC patients.
In recent years, with the rapid development of high-throughput measurement technologies, bioinformatics analysis of expression profiles has been widely used to find potential targets for cancer and biomarkers for patient prognostic analysis. Numerous studies have demonstrated that biomarkers have promising diagnostic abilities in HCC, such as AFP-L3, GP73, DCP, GPC3, SCCA, and OPN[13,14]. Differential gene expression analysis between different clinical phenotypes is a common method to search for potential biomarkers of diseases. Weighted gene coexpression network analysis allows for construction of coexpressed gene modules by finding coordinately expressed gene modules and analysing the relationship between different modules and specific features.
The histological grade of HCC is one of the most predictive factors for postoperative recurrence and survival[15,16]. Survival rates for high-grade HCC are lower than those for low- and medium-grade HCC[17]. Image-based histological grade prediction will help to establish appropriate strategies for HCC management. The rapid development of genomics in recent years allows for exploring the genomic characteristics of cancer patients in depth. The genetic characteristics of different stages have been studied, and some progress has been made in other tumours, such as prostate cancer and breast cancer[18,19]. However, in the field of liver cancer, comparison of gene expression characteristics for different stages to mine characteristic genes is scarce. By comparing the different genetic characteristics expressed in different grades of liver cancer patients, we can understand the relevant information at the molecular level regarding the occurrence and development of liver cancer and understand the role of different genes in these processes. Such genes are associated with the prognosis of liver cancer and have potential as clinical therapeutic target genes. This has very important practical significance in present precision medicine.
In the present study, we sought to explore genomic characteristics between different grades of HCC. We selected prognostic genes associated with histologic grades and built a prognostic model of HCC.
We obtained RNA-seq data and associated clinical information for patients with HCC from The Cancer Genome Atlas (TCGA) database (http://xena.ucsc.edu/). Patients without clinical survival data were eliminated from the clinical information screening, and 366 patients were retained for subsequent analysis. There were 55 patients with G1 stage, 177 with G2 stage, 122 with G3 stage, and 12 with G4 stage. We combined G1 and G2 stages into a low-grade group of 232 patients and the G3 and G4 stages into a high-grade group of 134 patients. The sample size between the two groups met the requirements for comparison of differences. Subsequently, we downloaded the HCC dataset from the International Cancer Genome Consortium (ICGC) database (https://dcc.icgc.org/) as the validation dataset.
The patients with liver cancer were divided into two groups: Grade I and grade II were defined as low grade, and grade III and grade IV were defined as high grade. Differentially expressed genes (DEGs) between the high- and low-grade groups were obtained using the R package “limma” by setting the filtering criteria as false discovery rate < 0.050 and
Gene set enrichment analysis was conducted to explore the biological pathways and processes in which DEGs might be involved. The DEGs were subjected to Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and REACTOM analyses via the “ClusterProfiler” package in R (4.2.1) for gene set enrichment analysis (GSEA). The c5.all.v7.2.symbols.gmt, c2.cp.kegg.v7.4.symbols.gmt, and c2.cp.reactome.v7.4.symbols.gmt curated reference gene sets from the MSigDB file were chosen for GSEA. The enriched pathways were sorted using the normalized enrichment score and the corrected P value. Function or pathway terms with an adjusted P value < 0.05 and false discovery rate < 0.25 were considered statistically significantly enriched.
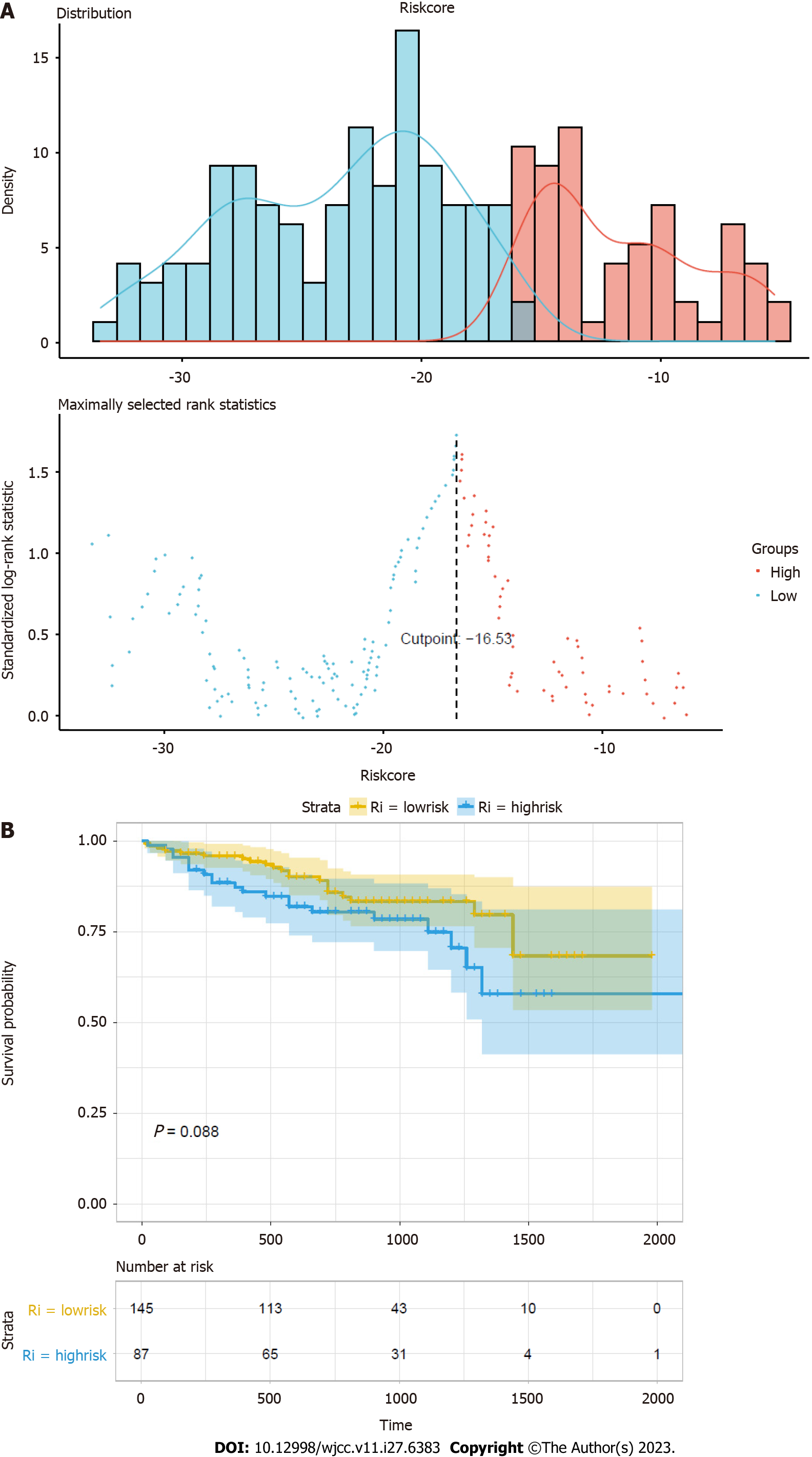
The expression matrix of the 382 DEGs was extracted by differential expression analysis. Then, genes associated with the prognosis of HCC were found using univariate Cox regression analysis (P < 0.05). To further screen grade-related genes and build a prognostic model, multivariate Cox regression analysis and least absolute shrinkage and selection operator (LASSO) regression analysis were conducted. We calculated the risk score as Coef gene × Expr gene, where Coef gene denotes the correlation between genes and patient survival in HCC, and the expression level of genes is represented by the Expr gene. Each patient with HCC was assigned a risk score using this formula. The patients with HCC were divided into high-risk and low-risk groups based on the cut-off function in the survival package.
The Retrieval of Interacting Genes (STRING; http://stringdb.org/) database online tool was used to analyse protein-protein interactions (PPIs) of the DEGs, and experimentally validated interactions with a combined score > 0.4 were selected as significant. The screened networks were visualized with Cytoscape 3.7.0. Molecular Complex Detection (MCODE) was performed to establish PPI network modules, with degree cut-off = 2, node score cut-off = 0.2, k-core = 2, and max depth = 100 as selected.
Kaplan-Meier plots were created, and the log-rank test was performed using a survival package. Analyses of Cox regression using multivariate variables were performed to determine which genes affect patient outcomes. Prognostic factors selected by the lasso-Cox model were included in multivariate Cox regression analysis. The forest map was shown using the “survminer” R package.
All statistical tests were performed using R 4.2.1. Differences between groups were compared using the Wilcoxon rank-sum test or Student’s t test, as appropriate. Correlations were determined using Pearson or Spearman correlation tests, as appropriate. Kaplan-Meier plots were produced, and log-rank tests were performed to identify the significance of the difference between survival curves. Statistical significance was determined at a two-sided P value of 0.05.
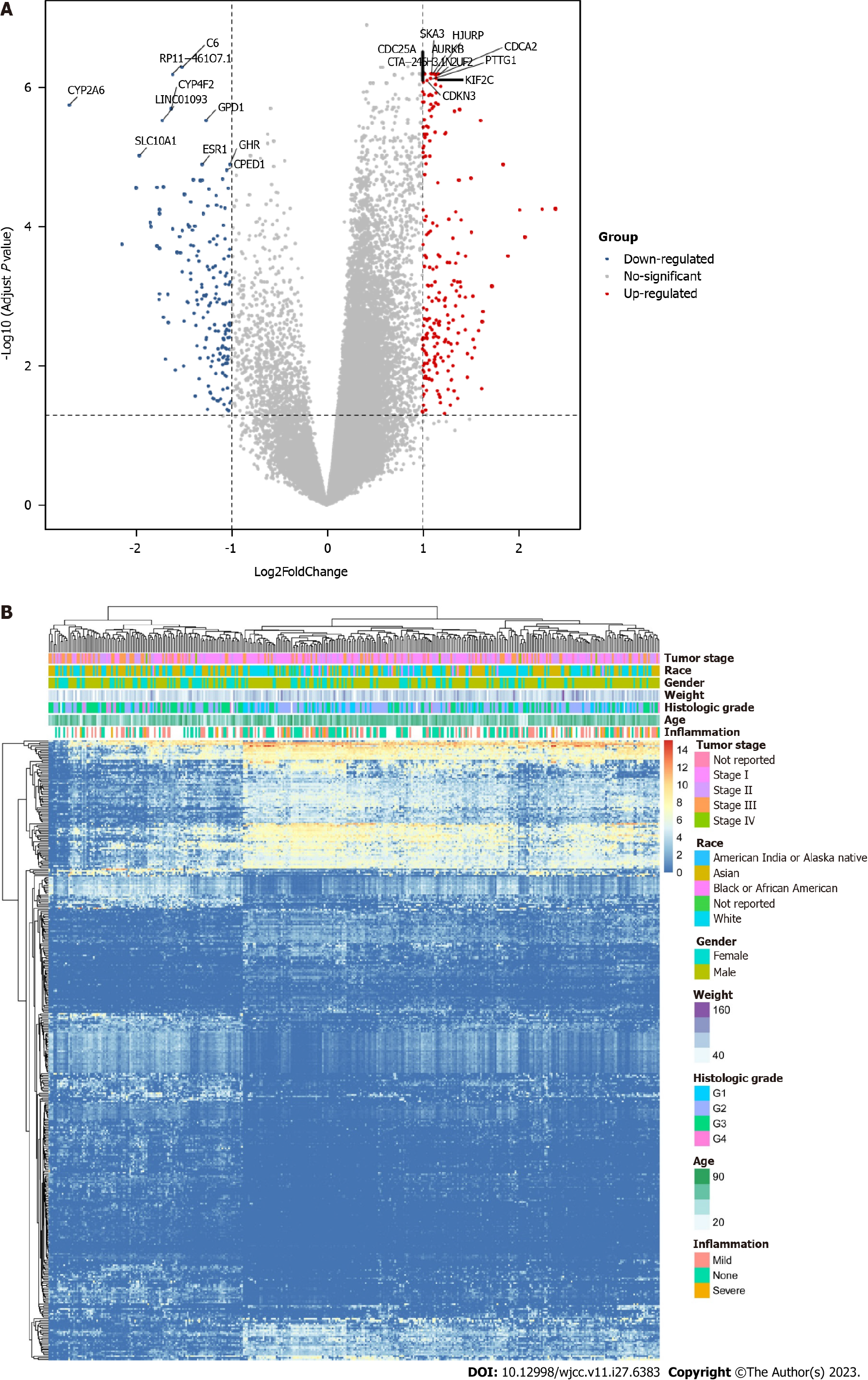
As shown in Figure 1, we assigned grade I and grade II patients with HCC as low-grade patients and grade III and grade IV patients with HCC as high-grade patients and conducted differential analysis on the two types of patients. A total of 382 DEGs were obtained. As shown in Figure 1A, the most significantly upregulated genes in high-grade patients included SKA3, HJURP, CDCA2, CDC25A, AURKB, CTA-246H3.12, NUF2, PTTG1, KIF2C, and CDKN3. Downregulated genes included CYP2A6, SLC10A1, LINC01093, CYP4F2, RP11-461O7.1, C6, GPD1, ESR1, GHR, and CPED1. According to the obtained list of 382 genes, we also drew an expression heatmap of these genes, as shown in Figure 1B. There were significant differences in expression of these 382 DEGs in patients with different disease stages.
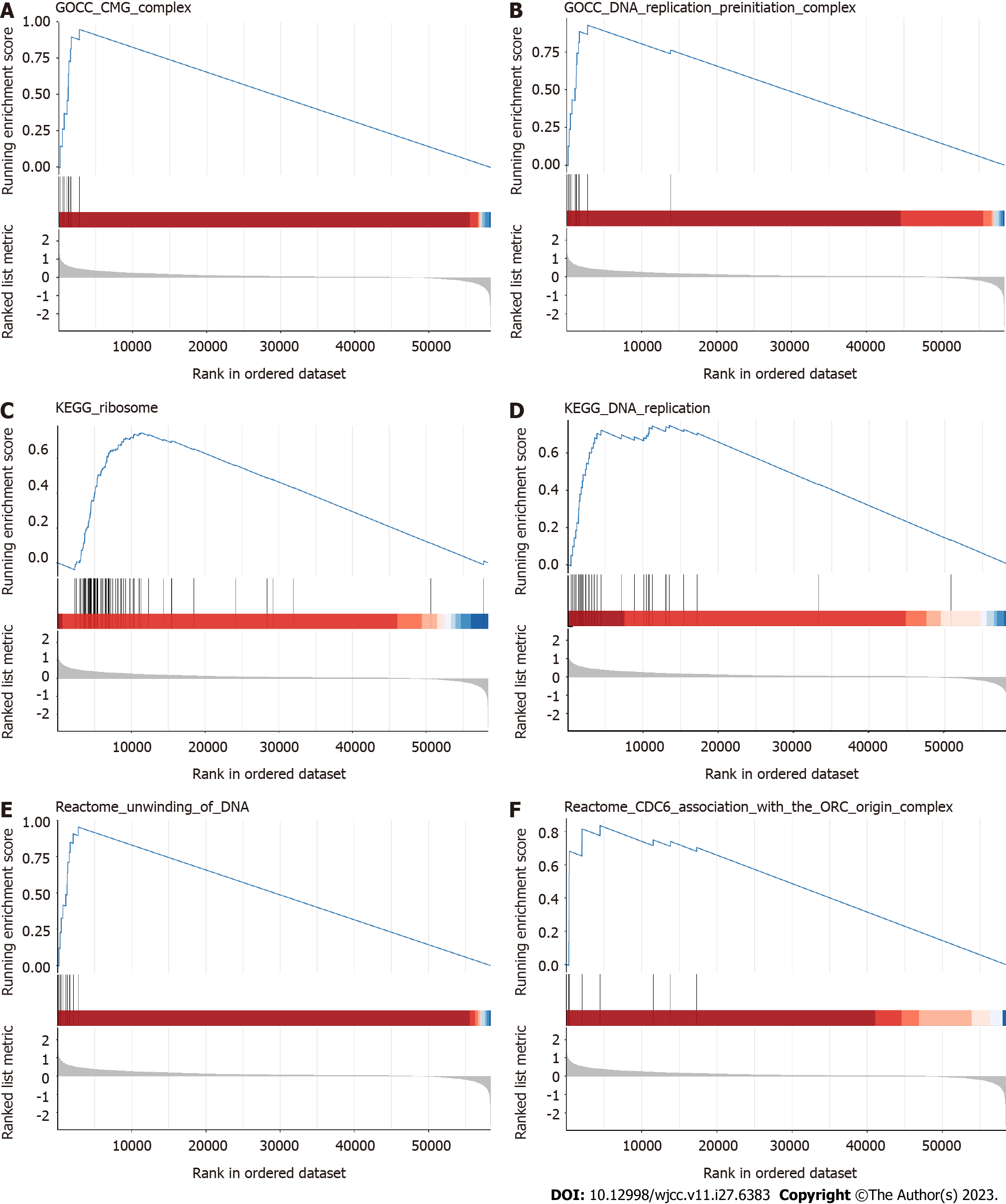
To better understand the biological functions of these 382 DEGs, we analysed the signalling pathways that these 382 DEGs may be involved in regulating by GSEA and assessed their roles in the occurrence and development of HCC (Figure 2). We selected relevant pathways in the GO, KEGG, and REACTOM databases for analysis. The results of GO analysis showed upregulated expression of the CMG COMPLEX pathway and the DNA REPLICATION PREINITIATION COMPLEX pathway. In the KEGG database, DEGs were involved in upregulation of the RIBOSOME pathway and DNA REPLICATION pathways. In the REACTOM database, the UNWINDING OF DNA pathway and CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX pathway were upregulated. These DEGs are generally involved in regulation of cell cycle-related signalling pathways, so these DEGs are likely to be involved in the regulation of tumour cell proliferation. Thus, we can observe the differences in the regulation of biological functions of these differential genes between patients with low grade liver cancer and those with high grade liver cancer. It is clear that differential genes in high-grade patients can promote cell proliferation and thus lead to a worse prognosis for patients.
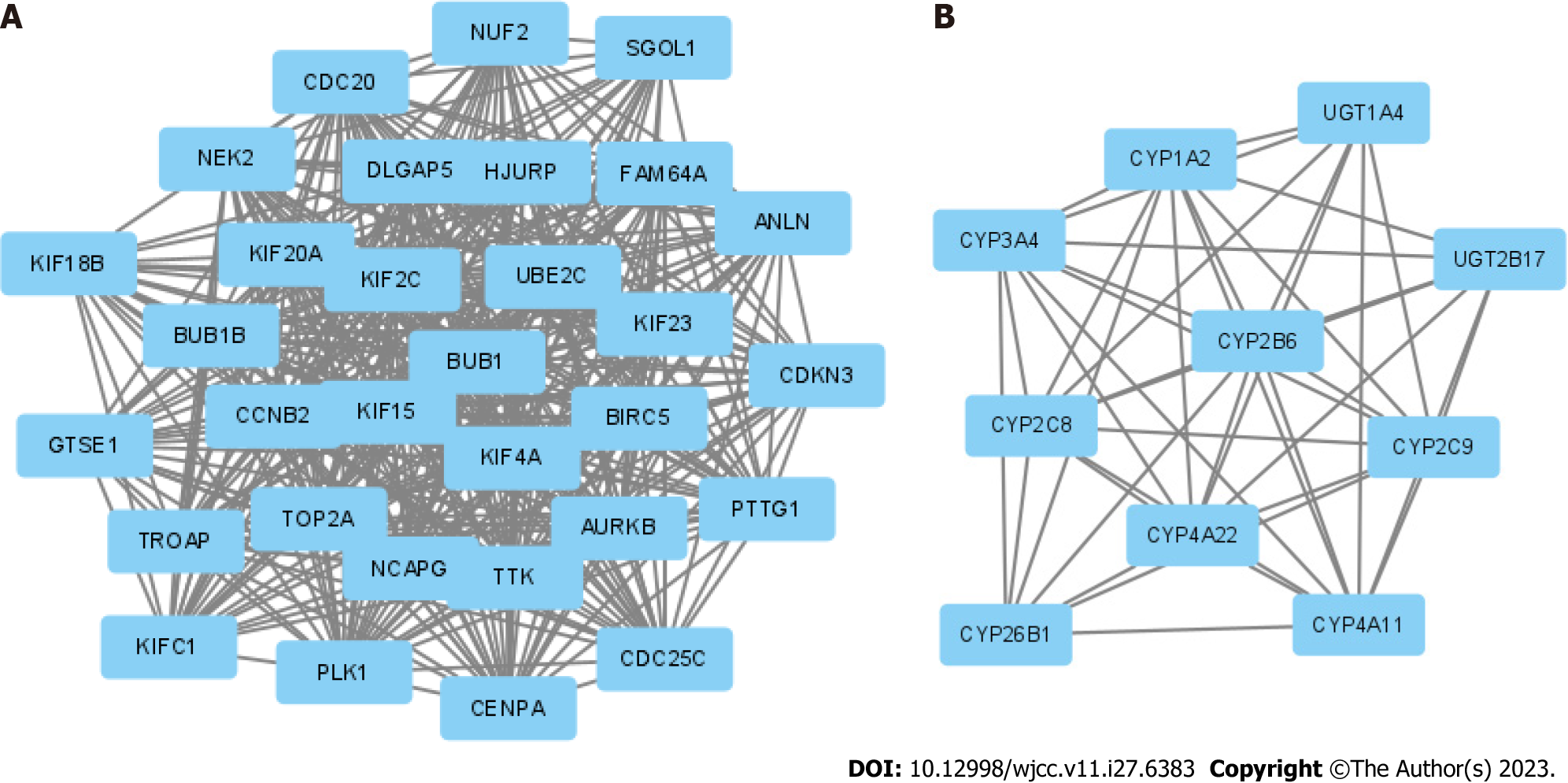
We obtained 382 DEGs through differential analysis. To investigate the interaction of these genes, we applied a PPI network analysis and network clustering (Figure 3). The top 2 PPI networks contained a total of 41 genes, and these 41 genes were selected for downstream analysis as hub genes. What we hoped to do is to eliminate the collinearity of these 41 genes and preserve the characteristic genes with prognostic value, and build a prognostic model based on these.
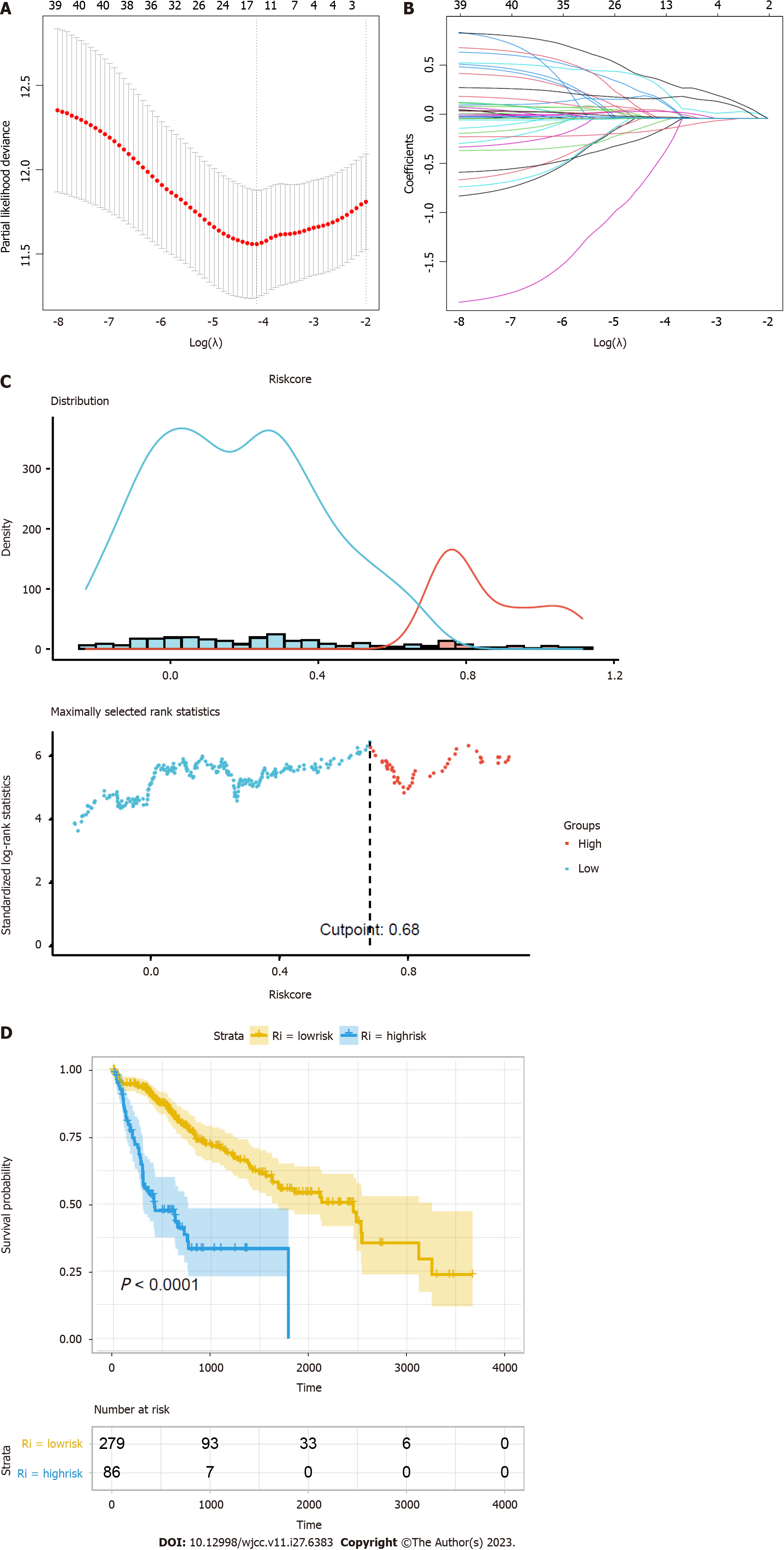
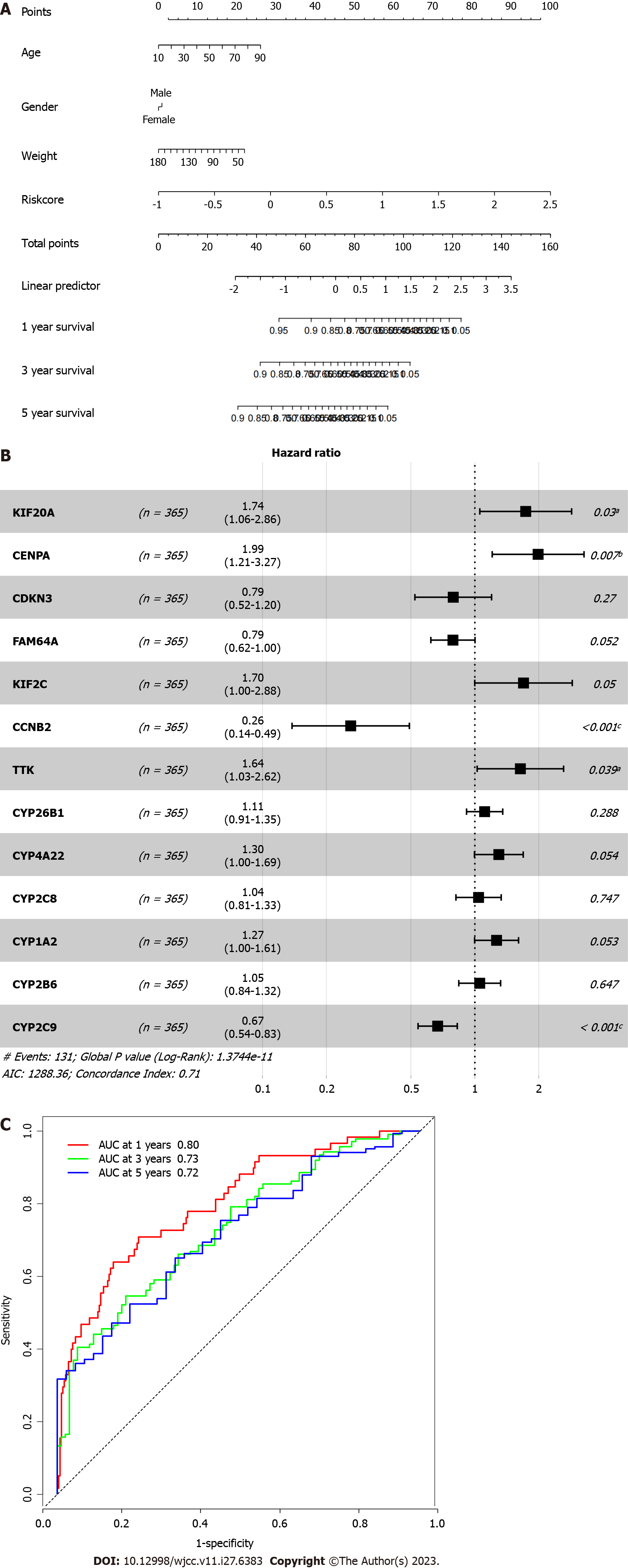
Cox-LASSO regression was used to fit the expression data of 41 hub genes with the prognostic survival data and delete the redundant genes with high collinearity (Figure 4). A total of 13 prognostic genes were obtained, namely, KIF20A, CENPA, CDKN3, FAM64A, KIF2C, CCNB2, TTK, CYP26B1, CYP4A22, CYP2C8, CYP1A2, CYP2B6, and CYP2C9 (Table 1). Risk factors were assessed by multiplying the expression levels of these prognostic genes with regression coefficients. Some prognostic genes related to cell proliferation were obtained. This is the same as the result in the functional analysis.
| Gene | Description | Coefficient |
| KIF20A | Enables protein kinase binding activity. Involved in microtubule bundle formation; midbody abscission; and regulation of cytokinesis | 0.207160352 |
| CENPA | This gene encodes a centromere protein which contains a histone H3 related histone fold domain that is required for targeting to the centromere. The protein is a replication-independent histone that is a member of the histone H3 family | 0.335103720 |
| CDKN3 | The protein encoded by this gene belongs to the dual specificity protein phosphatase family. It was identified as a cyclin-dependent kinase inhibitor | -0.037558302 |
| FAM64A | Predicted to be involved in cell division | -0.134768381 |
| KIF2C | This gene encodes a kinesin-like protein that functions as a microtubule-dependent molecular motor. The encoded protein can depolymerize microtubules at the plus end, thereby promoting mitotic chromosome segregation | 0.187028347 |
| CCNB2 | Cyclin B2 is a member of the cyclin family, specifically the B-type cyclins. The B-type cyclins, B1 and B2, are essential components of the cell cycle regulatory machinery | -0.438267747 |
| TTK | The protein encoded by this gene is essential for chromosome alignment at the centromere during mitosis and is required for centrosome duplication. Tumorigenesis may occur when this protein fails to degrade and produces excess centrosomes resulting in aberrant mitotic spindles | 0.400136922 |
| CYP26B1 | This gene encodes a member of the cytochrome P450 superfamily. The encoded protein functions as a critical regulator of all-trans retinoic acid levels by the specific inactivation of all-trans retinoic acid to hydroxylated forms | 0.205800982 |
| CYP4A22 | This gene encodes a member of the cytochrome P450 superfamily of enzymes. This gene is part of a cluster of cytochrome P450 genes on chromosome 1p33 | 0.084476409 |
| CYP2C8 | This gene encodes a member of the cytochrome P450 superfamily of enzymes. The enzyme is known to metabolize many xenobiotics, including the anticonvulsive drug mephenytoin, benzo(a)pyrene, 7-ethyoxycoumarin, and the anti-cancer drug taxol | 0.002430565 |
| CYP1A2 | This gene encodes a member of the cytochrome P450 superfamily of enzymes. It is able to metabolize some PAHs to carcinogenic intermediates | 0.072320556 |
| CYP2B6 | This gene encodes a member of the cytochrome P450 superfamily of enzymes. The enzyme is known to metabolize some xenobiotics, such as the anti-cancer drugs cyclophosphamide and ifosphamide | 0.013546756 |
| CYP2C9 | This gene encodes a member of the cytochrome P450 superfamily of enzymes. The enzyme is known to metabolize many xenobiotics, including phenytoin, tolbutamide, ibuprofen, and S-warfarin | -0.117163123 |
To understand the prognostic value of the risk factor model, we prepared a nomogram, conducted multiple Cox regression for the 13 genes included in the model, and evaluated the prediction accuracy of the model through a receiver operating characteristic curve (Figure 5). As shown in Figure 5, among the 11 characteristic genes, four had a statistically significant influence on prognosis: KIF20A, CENPA, CCNB2, and CYP2C9. The accuracy of risk factors in predicting the one-year prognosis of patients was as high as 80%, that of predicting the three-year prognosis was as high as 73%, and that of predicting the five-year prognosis was as high as 72%.
To validate the accuracy of the prognostic model, we applied it to the HCC dataset from the ICGC database (Figure 6). As the results showed, there was a trend of worse prognosis with a higher risk score. The trend did not show statistical significance, and the reason might be that the validation dataset does not contain the expression information of FAM64A, which might have caused inaccuracy of the score.
The grading information of liver cancer has very important guiding significance for predicting prognosis, and it is an important guiding index in the clinical diagnosis and treatment of liver cancer. Tumour grading describes how normal or abnormal cancer cells appear under the microscope. The higher the level is, the more abnormal the cells appear and the faster they grow and spread[20]. We identified some of characteristic genes by comparing the gene expression levels of liver cancer patients with different grades. These genes may be involved in the development of liver cancer. Through differential analysis, we obtained 382 DEGs, among which a large number of genes are involved in DNA replication, ribosomes, and other cell cycle-related signalling pathways. This suggests that these genes play a regulatory role in the proliferation of cancer cells. It is well known that cancer cells are characterized by uncontrolled mass proliferation, and higher cancer grade means more aggressive cancer cells[21,22]. We believe that these DEGs may play a role in this process. The biological functions of these DEGs are consistent with the proliferative characteristics of cancer cells of different grades.
Then, we extracted 13 prognostic genes by LASSO regression: KIF20A, CENPA, CDKN3, FAM64A, KIF2C, CCNB2, TTK, CYP26B1, CYP4A22, CYP2C8, CYP1A2, CYP2B6, and CYP2C9. We constructed a risk factor score by summing the product of the expression levels of these 13 genes and the regression coefficients. The nomogram, Cox regression, and external validation demonstrated the prognostic value and clinical significance of the model. The model maintained a high level of accuracy in predicting 1-year, 3-year, and even 5-year survival rates. According to the results of Cox regression, the 13 genes of the model have appreciated value for predicting the prognosis of patients with liver cancer. Therefore, we were interested in the related biological functions of these genes and sought to explore and explain the mechanism of this model. We searched for the biological function of these genes. A large proportion of these genes belong to the cytochrome P450 (CYP450) superfamily, such as CYP26B1, CYP4A22, CYP2C8, CYP1A2, CYP2B6, and CYP2C9. There are many members of the superfamily, and most are involved in regulating biological processes such as the cell cycle. For example, in colorectal cancer, CYP1B1, a member of the P450 family, has been found to promote resistance to ferroptosis in colorectal cancer cells[23]. Patients with high expression of CYP1B1 are predicted to have a worse prognosis, and the CYPB1 gene is the first CYP superfamily member found to be involved in mediating ferroptosis and resistance to programmed cell death protein 1 (PD-1) therapy[23]. We believe that since many members of the CYP450 superfamily are included in this model, the calculated risk factor score may reflect the cell cycle status to some extent. At present, the role of the CYP450 family in the occurrence and development of cancer is not completely clear. However, it is foreseeable that the role of the CYP450 superfamily in the occurrence and development of liver cancer is extremely complex.
Except for the possible correlation of the P450 family in liver cancer, among the 13 characteristic genes, KIF20A, CENPA, CCNB2, and TTK were statistically significant in relation to prognosis. CCNB2 is a protective factor for liver cancer; KIF20A, CENPA, and TTK are risk factors for liver cancer. Studies have revealed that reduced expression levels of KIF20A in liver cancer can inhibit cell proliferation and improve the prognosis of patients[24]. In addition, KIF20A has been reported to be associated with a poor prognosis in prostate cancer, renal cancer, and fibrosarcoma[25-27]. Abnormal expression of the CENPA gene is also found in multiple cancer species, such as lung cancer, colon cancer, breast cancer, and liver cancer[28-32]. In addition, studies have found that the CENPA gene is involved in the regulation of the cell cycle, gene stability, and other biological processes[28,33-35]. The CCNB2 gene encodes cyclin B2, which is involved in cell cyclin-related regulation. In glioma, a higher CCNB2 expression level predicts a worse prognosis[36]. Overexpression of CCNB2 is also thought to be associated with a poor prognosis in liver cancer, but CCNB2 was shown to be a protective factor in this study, which is contrary to the results of a previous study[37]. The TTK gene is associated with a poor prognosis in non-small cell lung cancer, and its main biological function is to induce cell proliferation and invasiveness, thereby promoting tumour growth[38,39]. There is sufficient research evidence on the role of these genes in cancer. We believe that the prognostic model constructed by integrating the expression data of these genes is of great significance for predicting the prognosis of patients with liver cancer. By integrating the related functions of all 13 genes, we can further explain the biological mechanism of the model, and the proliferation intensity of cancer cells can be reflected to some extent by calculating the sum of the product of the expression levels of these 13 genes and their weights. We hope that this mechanism will be verified experimentally in future studies.
Subsequently, we performed corresponding validation analysis using ICGC data. The gene expression data for the validation set were extracted to calculate the risk score grouping and compare differences in survival between groups. The trend of the survival curve and training data were the same for the two groups. Although not statistically significant, this finding suggests that patients in the high-risk group tended to have worse outcomes than those in the low-risk group. Clinically, this model may reflect the ability of cancer cells to proliferate to a certain extent because several genes included in the model are involved in the cell cycle regulation of cancer cells. Through the above verification, the results show that the model has certain clinical application value. According to in-depth analysis and discussion of the genes in the model, it was found that many of these genes are likely to participate in the biological process of the occurrence and development of cancer, which provides a guiding direction for future clinical practice and laboratory research. This model not only has reference value for predicting the prognosis of patients but also has great exploration potential for related research on the function of these genes. These genes have the potential to be therapeutic targets in cancer treatment.
However, our study also had several limitations. First, the sample size that we selected was not large enough for specific analysis of subtypes in HCC, which may lead to inaccurate hub gene screening and insufficient coverage. Second, the reliability of hub genes has not been verified experimentally, and the specific mechanism by which hub genes regulate HCC is not fully understood and needs further exploration in the future. Finally, the correlation between the high expression of these targets and the clinical manifestations and stage of HCC was not thoroughly clarified, which may be directly related to the insufficient size of our sample data. In future research, we will incorporate more diverse data to make our analysis more comprehensive.
In the future, we will include a more diverse and richer sample of liver cancer and analyse it for different grades of liver cancer and those caused by different aetiologies. These findings will provide further support for early liver cancer diagnosis and targeted treatment of liver cancer.
By comparing the gene profiles of patients with different stages of HCC, we hoped to select the characteristic genes that are significantly associated with the prognosis of patients and construct a prognosis model consisting of 13 genes, most of which have the function of regulating cell proliferation. This model has good application value and can be explained clinically. And these member genes have important prognostic value.
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer. With highly invasive biological characteristics and a lack of obvious clinical manifestations, HCC usually has a poor prognosis and ranks fourth in cancer mortality. The aetiology and exact molecular mechanism of primary HCC are still unclear.
To explore genomic characteristics between different grades of HCC.
To select the characteristic genes that are significantly associated with the prognosis of HCC patients and construct a prognosis model of this malignancy.
By comparing gene expression levels of patients with different cancer grades of HCC, we screened out differentially expressed genes associated with tumour grade. By PPI network analysis, we obtained the top 2 protein-protein interaction networks and hub genes from these differentially expressed genes. By using least absolute shrinkage and selection operator Cox regression, 13 prognostic genes were selected for feature extraction, and a prognostic risk model of HCC was established.
The model had significant prognostic ability in HCC. We also analysed the biological functions of these prognostic genes. These prognostic genes and the prognostic model based on these genes can be used in clinical practice.
By comparing the gene profiles of patients with different stages of HCC, we have constructed a prognostic model based on 13 genes. This model has good application value and can be explained clinically.
The component genes of the prognostic model are expected to be candidate targets for liver cancer treatment in future clinical therapy.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Alkhateeb A, Jordan; Emran TB, Bangladesh; Gupta MK, Germany S-Editor: Chen YL L-Editor: Wang TQ P-Editor: Yuan YY
| 1. | Asafo-Agyei KO, Samant H. Hepatocellular Carcinoma. 2023 Mar 27. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan-. [PubMed] |
| 2. | Legry V, Schaap FG, Delire B, Horsmans Y, Leclercq IA. Yin Yang 1 and farnesoid X receptor: a balancing act in non-alcoholic fatty liver disease? Gut. 2014;63:1-2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 3. | Yamamoto M, Oshita A, Nishisaka T, Nakahara H, Itamoto T. Synchronous double primary hepatic cancer consisting of hepatocellular carcinoma and cholangiolocellular carcinoma: a case report. J Med Case Rep. 2018;12:224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 4. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9215] [Cited by in RCA: 9856] [Article Influence: 821.3] [Reference Citation Analysis (4)] |
| 5. | Du ZG, Wei YG, Chen KF, Li B. Risk factors associated with early and late recurrence after curative resection of hepatocellular carcinoma: a single institution's experience with 398 consecutive patients. Hepatobiliary Pancreat Dis Int. 2014;13:153-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 6. | Poon RT. Prevention of recurrence after resection of hepatocellular carcinoma: a daunting challenge. Hepatology. 2011;54:757-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 7. | Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S, Sugawara Y, Minagawa M, Takayama T, Kawasaki S, Makuuchi M. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38:200-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1061] [Cited by in RCA: 1234] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 8. | Bruix J, Sherman M; Practice Guidelines Committee, American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4333] [Cited by in RCA: 4507] [Article Influence: 225.4] [Reference Citation Analysis (0)] |
| 9. | Zhang BH, Yang BH, Tang ZY. Randomized controlled trial of screening for hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130:417-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 960] [Cited by in RCA: 945] [Article Influence: 45.0] [Reference Citation Analysis (1)] |
| 10. | Choi D, Kim SH, Lim JH, Cho JM, Lee WJ, Lee SJ, Lim HK. Detection of hepatocellular carcinoma: combined T2-weighted and dynamic gadolinium-enhanced MRI versus combined CT during arterial portography and CT hepatic arteriography. J Comput Assist Tomogr. 2001;25:777-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Arguedas MR, Chen VK, Eloubeidi MA, Fallon MB. Screening for hepatocellular carcinoma in patients with hepatitis C cirrhosis: a cost-utility analysis. Am J Gastroenterol. 2003;98:679-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 169] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 12. | Masuzaki R, Karp SJ, Omata M. New serum markers of hepatocellular carcinoma. Semin Oncol. 2012;39:434-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 13. | Wang W, Wei C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020;7:308-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 292] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 14. | Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10573-10583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 294] [Cited by in RCA: 382] [Article Influence: 38.2] [Reference Citation Analysis (7)] |
| 15. | Zhou L, Rui JA, Wang SB, Chen SG, Qu Q, Chi TY, Wei X, Han K, Zhang N, Zhao HT. Factors predictive for long-term survival of male patients with hepatocellular carcinoma after curative resection. J Surg Oncol. 2007;95:298-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Jonas S, Bechstein WO, Steinmüller T, Herrmann M, Radke C, Berg T, Settmacher U, Neuhaus P. Vascular invasion and histopathologic grading determine outcome after liver transplantation for hepatocellular carcinoma in cirrhosis. Hepatology. 2001;33:1080-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 723] [Cited by in RCA: 706] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 17. | Zhou L, Rui JA, Ye DX, Wang SB, Chen SG, Qu Q. Edmondson-Steiner grading increases the predictive efficiency of TNM staging for long-term survival of patients with hepatocellular carcinoma after curative resection. World J Surg. 2008;32:1748-1756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 18. | Alkhateeb A, Rezaeian I, Singireddy S, Cavallo-Medved D, Porter LA, Rueda L. Transcriptomics Signature from Next-Generation Sequencing Data Reveals New Transcriptomic Biomarkers Related to Prostate Cancer. Cancer Inform. 2019;18:1176935119835522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 19. | Kwa M, Makris A, Esteva FJ. Clinical utility of gene-expression signatures in early stage breast cancer. Nat Rev Clin Oncol. 2017;14:595-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 185] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 20. | National Cancer Institute. Tumor Grade. 2013. [citd 7 May 2023]. Available from: https://www.cancer.gov/about-cancer/diagnosis-staging/diagnosis/tumor-grade. |
| 21. | Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 469] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 22. | Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719-724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2798] [Cited by in RCA: 2424] [Article Influence: 151.5] [Reference Citation Analysis (0)] |
| 23. | Chen C, Yang Y, Guo Y, He J, Chen Z, Qiu S, Zhang Y, Ding H, Pan J, Pan Y. CYP1B1 inhibits ferroptosis and induces anti-PD-1 resistance by degrading ACSL4 in colorectal cancer. Cell Death Dis. 2023;14:271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 24. | Wu C, Qi X, Qiu Z, Deng G, Zhong L. Low expression of KIF20A suppresses cell proliferation, promotes chemosensitivity and is associated with better prognosis in HCC. Aging (Albany NY). 2021;13:22148-22163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 25. | Copello VA, Burnstein KL. The kinesin KIF20A promotes progression to castration-resistant prostate cancer through autocrine activation of the androgen receptor. Oncogene. 2022;41:2824-2832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 26. | Liu B, Su J, Fan B, Ni X, Jin T. High expression of KIF20A in bladder cancer as a potential prognostic target for poor survival of renal cell carcinoma. Medicine (Baltimore). 2023;102:e32667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 27. | Jin Z, Tao S, Zhang C, Xu D, Zhu Z. KIF20A promotes the development of fibrosarcoma via PI3K-Akt signaling pathway. Exp Cell Res. 2022;420:113322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 28. | Sun X, Clermont PL, Jiao W, Helgason CD, Gout PW, Wang Y, Qu S. Elevated expression of the centromere protein-A(CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int J Cancer. 2016;139:899-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 29. | Wu Q, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012;77:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 30. | Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511-3516. [PubMed] |
| 31. | Li YM, Liu XH, Cao XZ, Wang L, Zhu MH. [Expression of centromere protein A in hepatocellular carcinoma]. Zhonghua Bing Li Xue Za Zhi. 2007;36:175-178. [PubMed] |
| 32. | Rajput AB, Hu N, Varma S, Chen CH, Ding K, Park PC, Chapman JA, Sengupta SK, Madarnas Y, Elliott BE, Feilotter HE. Immunohistochemical Assessment of Expression of Centromere Protein-A (CENPA) in Human Invasive Breast Cancer. Cancers (Basel). 2011;3:4212-4227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 33. | Li Y, Zhu Z, Zhang S, Yu D, Yu H, Liu L, Cao X, Wang L, Gao H, Zhu M. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS One. 2011;6:e17794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 34. | Li YM, Zhu Z, Chen Y, Luo ZG, Shi M, Zhu MH. [Effect of siRNA targeting centromere protein-A gene on biological behavior of HepG2 cells]. Zhonghua Bing Li Xue Za Zhi. 2008;37:124-128. [PubMed] |
| 35. | Behnan J, Grieg Z, Joel M, Ramsness I, Stangeland B. Gene knockdown of CENPA reduces sphere forming ability and stemness of glioblastoma initiating cells. Neuroepigenetics. 2016;7:6-18. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 36. | Wang D, Sun H, Li X, Wang G, Yan G, Ren H, Hou B. CCNB2 is a novel prognostic factor and a potential therapeutic target in low-grade glioma. Biosci Rep. 2022;42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | Li R, Jiang X, Zhang Y, Wang S, Chen X, Yu X, Ma J, Huang X. Cyclin B2 Overexpression in Human Hepatocellular Carcinoma is Associated with Poor Prognosis. Arch Med Res. 2019;50:10-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 38. | Chen J, Wu R, Xuan Y, Jiang M, Zeng Y. Bioinformatics analysis and experimental validation of TTK as a biomarker for prognosis in non-small cell lung cancer. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Liu XD, Yao DW, Xin F. TTK contributes to tumor growth and metastasis of clear cell renal cell carcinoma by inducing cell proliferation and invasion. Neoplasma. 2019;66:946-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |