Published online Jul 16, 2023. doi: 10.12998/wjcc.v11.i20.4763
Peer-review started: January 4, 2023
First decision: April 3, 2023
Revised: April 11, 2023
Accepted: June 6, 2023
Article in press: June 6, 2023
Published online: July 16, 2023
Processing time: 174 Days and 6.9 Hours
Gastric cancer (GC) is one of the most common malignant tumors with poor prognosis in terms of advanced stage. However, the survival-associated biomarkers for GC remains unclear.
To investigate the potential biomarkers of the prognosis of patients with GC, so as to provide new methods and strategies for the treatment of GC.
RNA sequencing data from The Cancer Genome Atlas (TCGA) database of STAD tumors, and microarray data from Gene Expression Omnibus (GEO) database (GSE19826, GSE79973 and GSE29998) were obtained. The differentially expressed genes (DEGs) between GC patients and health people were picked out using R software (x64 4.1.3). The intersections were underwent between the above obtained co-expression of differential genes (co-DEGs) and the DEGs of GC from Gene Expression Profiling Interactive Analysis database, and Gene Ontology (GO) analysis, Kyoto Encyclopedia of Gene and Genome (KEGG) pathway analysis, Gene Set Enrichment Analysis (GSEA), Protein-protein Interaction (PPI) analysis and Kaplan-Meier Plotter survival analysis were performed on these DEGs. Using Immunohistochemistry (IHC) database of Human Protein Atlas (HPA), we verified the candidate Hub genes.
With DEGs analysis, there were 334 co-DEGs, including 133 up-regulated genes and 201 down-regulated genes. GO enrichment analysis showed that the co-DEGs were involved in biological process, cell composition and molecular function pathways. KEGG enrichment analysis suggested the co-DEGs pathways were mainly enriched in ECM-receptor interaction, protein digestion and absorption pathways, etc. GSEA pathway analysis showed that co-DEGs mainly concentrated in cell cycle progression, mitotic cell cycle and cell cycle pathways, etc. PPI analysis showed 84 nodes and 654 edges for the co-DEGs. The survival analysis illustrated 11 Hub genes with notable significance for prognosis of patients were screened. Furtherly, using IHC database of HPA, we confirmed the above candidate Hub genes, and 10 Hub genes that associated with prognosis of GC were identified, namely BGN, CEP55, COL1A2, COL4A1, FZD2, MAOA, PDGFRB, SPARC, TIMP1 and VCAN.
The 10 Hub genes may be the potential biomarkers for predicting the prognosis of GC, which can provide new strategies and methods for the diagnosis and treatment of GC.
Core Tip: Gastric cancer (GC) is one of the most common leading cause of death worldwide. The cases with advanced GC usually have poor prognosis. To date, the prognostic biomarkers of GC remain unclear. In this article, we investigated the co-expression of differential genes (co-DEGs) between GC tissues and normal tissues based on the data from Gene Expression Omnibus, Gene Expression Profiling Interactive Analysis and The Cancer Genome Atlas. By using bioinformatics analysis, the signal pathways of co-DEGs involvement in GC were identified, and 10 Hub biomarkers for the survival of GC were screened.
- Citation: Yin LK, Yuan HY, Liu JJ, Xu XL, Wang W, Bai XY, Wang P. Identification of survival-associated biomarkers based on three datasets by bioinformatics analysis in gastric cancer. World J Clin Cases 2023; 11(20): 4763-4787
- URL: https://www.wjgnet.com/2307-8960/full/v11/i20/4763.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i20.4763
Gastric cancer (GC) is one of the most common malignant tumors with high morbidity and mortality[1-3]. It is known that the occurrence of GC is the result of multiple factors. The genetic factors, dietary habits and Helicobacter pylori infection play a very important role in the occurrence and development of GC[4-6]. The most common screening methods for GC are gastroscopy and pathological examination, which can effectively improve the detection rate of early GC[7,8]. However, those patients, who with advanced GC, have poor treatment effect and poor prognosis[9].
The development of GC is a complex pathological process involving changes of various genes and pathways[10]. Previous studies have shown that some changes were happened between the GC tissues and normal tissues, especially the expression of the genes[11]. The biomarkers measured in different stages of GC are helpful as indicators of early diagnosis, routine screening, postoperative monitoring or pharmacological response to a therapeutic intervention[12]. Therefore, exploring the survival related biomarkers of GC may provide more approaches for the treatment of GC, improving the overall survival time of the patients. To date, the survival related biomarkers for GC remains unclear. Currently, the development of high-throughput sequencing technology has generated a large number of functional genomic data[13], making it possible to reveal the survival related biomarkers of GC by analyzing the differential gene expression data between the GC tissues and the normal tissues. In recent years, bioinformatics is widely used to analyze the genomic and proteomic data of tumors, and to reveal the function of gene products at the molecular level for cancer[14].
In this study, bioinformatics strategy was used to obtain data from Gene Expression Omnibus (GEO), Gene Expression Profiling Interactive Analysis (GEPIA) and The Cancer Genome Atlas (TCGA). In briefly, the software of Gene Expression Profiling Interactive Analysis (GEPIA), R software (x64.1.3), STRING, Kaplan-Meier plotter and Human Protein Atlas (HPA), were performed to analyze and integrate the mRNA expression data of GC tissues and adjacent tissues or normal gastric tissues to explore the molecular functions (MF) of differential genes and signal pathways of GC. Finally, we successfully screened the genes of 10 key biomarkers for the survival of GC. The present study analyzed the high-throughput data of multi database and multi-chip datasets, which could more accurately reveal the potential prognosis biomarkers of GC. The detailed analysis workflow is as shown in Figure 1.
GDC TCGA Stomach Cancer (STAD) related datasets were downloaded from UCSC Xena (https://xenabrowser.net/) database[15], including 373 GC tissue samples and 32 normal gastric tissue samples. R software (x64 4.1.3) was used to download and analyze datasets of gene expression profiles from GEO database (GSE19826, GSE79973 and GSE29998) with Tidyverse and query packages, including 72 GC tissue samples and 74 normal gastric tissue samples.
Based on TCGA data sets, the Tidyverse and DESeq2R packages of software (x64 4.1.3) were used for differential gene expression analysis. RNA sequencing data from normal and tumor tissue samples were extracted for analysis. Volcano map was drawn to show the folding changes and P values of differentially expressed genes (DEGs) (|LogFC| ≥ 1, adjusted P value < 0.05). R software dealt with GEO data sets (GSE19826, GSE79973 and GSE29998, respectively), then the ID was converted, and the adjust P value < 0.05 and |LogFC| ≥ 1 were set as the cut off criterion. Subsequently, a Venn diagram method was used to screen out co-expression of differential genes (co-DEGs). Of these co-DEGs, only protein-coding genes were further analyzed.
GEPIA[16] is a newly developed interactive web server for analyzing the RNA sequencing expression data from the TCGA and the GTEx projects. The GEPIA data sets was analyzed using the stat packet of R software to predict the potential functions of the co-DEGs. The functional analyses of Gene Ontology (GO) analysis, Kyoto Encyclopedia of Gene and Genome (KEGG) pathway analysis and Gene Set Enrichment Analysis (GSEA) pathway analysis were performed on the co-DEGs by using R software. P < 0.05 was considered statistically significant.
Protein-protein interaction (PPI) Analysis of the identified DEGs was constructed by STRING 11.5. The STRING Database[17] (https://cn.string-db.org/) is a database of known and predicted PPIs. Using STRING 11.5, the interaction network between the above co-DEGs and the related genes was presented by setting the maximum confidence at 0.9. Furthermore, and Cytoscape_v3.9.1 software was carried out for analyzing and mapping.
Using Boxplot functions of GEPIA database, we set |Log2FC| cutoff ≥ 1, P-value cutoff < 0.05 as the cut off criterion. The tumor-related data from STAD database were selected to match the normal gastric tissue data of TCGA and GTEx, the genes with significant differences were screened out, and box-plot was performed for the screened Hub genes.
Kaplan-Meier plotter[18] was able to assess the association between 30 K gene (mRNA, microRNA, protein) expression and survival in 25 K + samples from 21 tumor types, including breast cancer, ovarian cancer, lung cancer, GC, etc. We used Kaplan-Meier plotter online tool to perform visualization analysis of GC related database again, and verified the above results.
Due to the highly specific characteristics of antigen and antibody binding, immunohistochemistry (IHC) can reveal the relative distribution and abundance of proteins. Then, through HPA[19] (https://www.proteinatlas.org), currently the largest and most comprehensive Human tissue Protein spatial database, we can more intuitively observe the difference of Hub gene expression between normal stomach tissue and GC tissue. χ2 test was used to compare the difference between the two groups, and P < 0.05 is statistically significant.
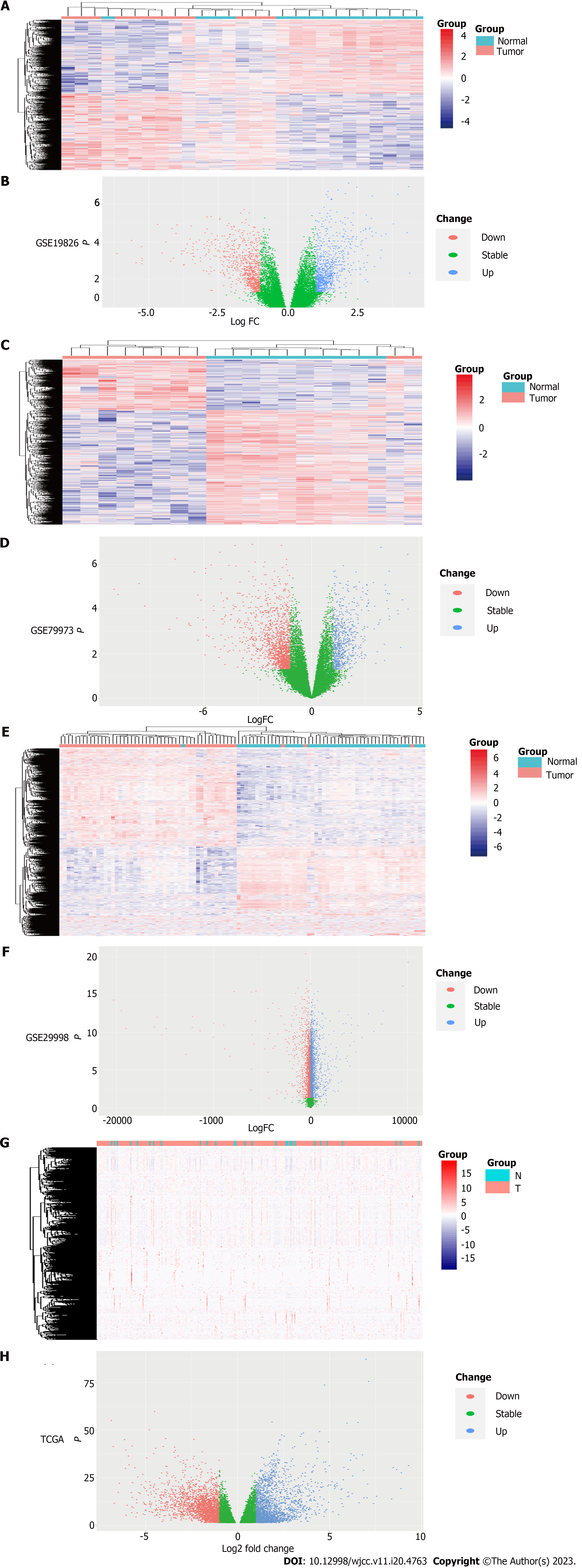
The TCGA STAD counts dataset was processed by using Tidyverse and DESeq2 packages of R software (x64 4.1.3), and DEGs was screened (|Log2FC| cutoff ≥ 1, P-value cutoff < 0.05 as the cut off criterion). Our results showed that there were 2133 up-regulated genes and 2349 down-regulated genes. R software (x64 4.1.3) was used to process data sets (GSE19826, GSE79973 and GSE29998), and then ID conversion was performed. Limma package was used to process the above three data sets respectively, and the screening criteria were |logFC| ≥ 1 and P-value < 0.05. There were 2202 up-regulated genes and 2700 down-regulated genes in GSE19826, 665 up-regulated genes and 1507 down-regulated genes in GSE79973, 4346 up-regulated genes and 3002 down-regulated genes in GSE29998, respectively. Heat maps were drawn by Pheatmap package of R software (x64 4.1.3) (Figures 2A, C and E), and volcano maps were drawn by ggplot2 package of R software (x64 4.1.3) (Figures 2B, D and F).
The TCGA STAD FPKM data set was processed by tidyverse and Pheatmap packages of R software (x64 4.1.3) combined with DEGs, and the heatmap was drawn (Figure 2G). Tidyverse, GGploT2 packages and DEGs were used to draw the volcano map (Figure 2H).
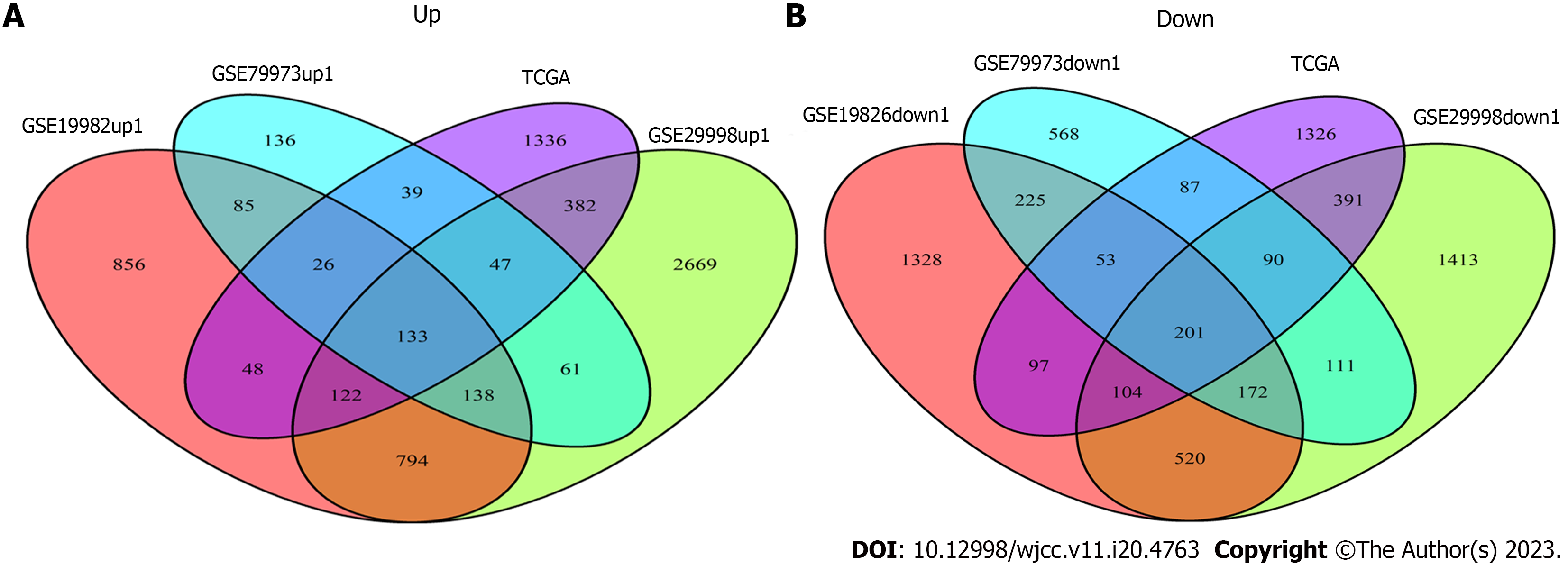
Venn Diagram package of R software (x64 4.1.3) was used to make Venn Diagram of DEGs in GEO and TCGA datasets. A total of 334 DEGs were obtained, including 133 up-regulated genes and 201 down-regulated genes (Figures 3A and B).
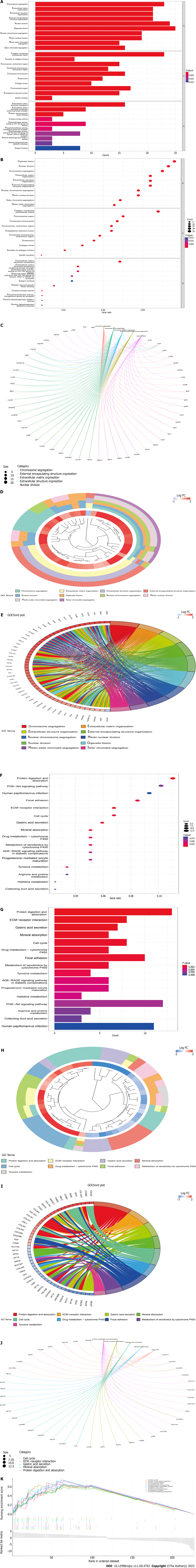
GO functional enrichment analysis showed that the GO annotation of co-DEGs was divided into three parts: Biological process (BP), cell composition (CC) and MF, and a diagram of which was shown in Figures 4A-E and Table 1 (list of top 5 GO pathways). Arraying the ascending order of P value (P < 0.05), our results revealed that the GO pathways of DEGs were enriched as the follows: Collagen fibrillary tissue, cell division, extracellular matrix organization, skeletal system development and copper ion detoxification, etc., were enriched in BP; extracellular space, extracellular matrix, extracellular region, collagen trimer and centromere, etc., were enriched in CC; extracellular matrix structural components, extracellular matrix structural components that give tensile strength, platelet-derived growth factor binding, zinc ion binding, creatine kinase activity and so on, were enriched in MF (details in Supplementary Tables 1 and 2).
| Category | Term | Count in gene set | P value | Genes |
| GOTERM_BP_DIRECT | GO:0030199: Collagen fibril organization | 10 | 2.08E-08 | COMP, COL3A1, ADAMTS2, FOXC1, COL1A2, COL5A1, COL12A1, COL5A2, SERPINH1, P3H4 |
| GO:0051301: Cell division | 20 | 7.76E-08 | CENPW, UBE2C, RCC2, CDCA7, KIF14, NCAPG, NDC80, CDC25B, CDC20, CCNB2, TPX2, CCNB1, PRC1, NUF2, CDK1, NEK2, KIF2C, BUB1, MAD2L1, SPC25 | |
| GO:0030198: Extracellular matrix organization | 13 | 2.67E-07 | OLFML2B, MMP7, MMP1, TNFRSF11B, COL3A1, ADAMTS2, COL1A2, COL5A1, COL4A1, COL5A2, COL4A5, COL8A1, COL10A1 | |
| GO:0001501: Skeletal system development | 12 | 3.61E-07 | TEAD4, COMP, COL3A1, VCAN, PKDCC, COL1A2, CDH11, COL5A2, COL10A1, TNFRSF11B, HOXA13, HOXC10 | |
| GO:0010273: Detoxification of copper ion | 6 | 7.19E-07 | MT2A, MT1M, MT1G, MT1H, MT1X, MT1E | |
| GOTERM_CC_DIRECT | GO:0005615: Extracellular space | 61 | 6.57E-14 | PIGR, SPARC, OLFML2B, CXCL8, COL12A1, CXCL17, COMP, VMO1, PLAU, CA2, FAM3B, COL10A1, SOSTDC1, CPXM1, TIMP1, CPA2, CHIA, MMP7, GPX3, GKN1, BGN, GKN2, PGC, PGF, SFRP4, ALDH3A1, VCAN, COL4A1, SCGB2A1, SFRP5, ANOS1, COL4A5, COL8A1, TFF2, TFF1, CELA3B, CPB1, TNFRSF11B, LRP8, SCUBE2, SELENBP1, CST2, CHAD, SPP1, SERPINH1, CKB, APOE, WNT2, CTHRC1, LINGO1, ANGPT2, CKM, SULF1, KLK11, ATP4A, COL3A1, COL1A2, COL5A1, FAP, COL5A2, ADA |
| GO:0031012: Extracellular matrix | 21 | 1.06E-11 | LINGO1, OLFML2B, MMP7, MMP1, BGN, TNFRSF11B, COMP, COL3A1, ADAMTS2, VCAN, COL1A2, COL5A1, COL4A1, COL5A2, CHAD, ANOS1, COL4A5, COL8A1, COL10A1, TIMP1, APOE | |
| GO:0005576: Extracellular region | 57 | 5.21E-10 | SPARC, OLFML2B, CXCL8, PSCA, COL12A1, AQP4, COMP, LIPF, ADAMTS2, PLAU, FAM3B, COL10A1, OLR1, TIMP1, CPA2, CHIA, MMP7, GPX3, MMP1, GKN1, MAMDC2, BGN, PGF, SFRP4, VCAN, FNDC1, COL4A1, NRG4, ANOS1, COL4A5, COL8A1, TFF1, LY6E, PKDCC, TNFRSF11B, THY1, THBS2, PLA2G7, SCUBE2, PTPRZ1, SPP1, APOE, METTL7A, WNT2, GPIHBP1, CTHRC1, ANGPT2, B3GAT1, GUCA2B, COL3A1, AKR1B10, COL1A2, COL5A1, QPCT, COL5A2, APOC1, CNTN3 | |
| GO:0005581: Collagen trimer | 12 | 7.15E-09 | COL3A1, COL1A2, COL5A1, COL4A1, MMP1, COL12A1, COL5A2, SERPINH1, COL10A1, COL4A5, TIMP1, CTHRC1 | |
| GO:0000776: Kinetochore | 11 | 4.23E-06 | CENPW, NUF2, HJURP, KIF2C, CENPN, NEK2, CENPA, BUB1, NDC80, MAD2L1, SPC25 | |
| GOTERM_MF_DIRECT | GO:0005201: Extracellular matrix structural constituent | 14 | 7.23E-09 | SPARC, BGN, THBS2, COMP, COL3A1, COL1A2, COL5A1, COL4A1, COL5A2, ANOS1, COL4A5, COL8A1, COL10A1, CTHRC1 |
| GO:0030020: Extracellular matrix structural constituent conferring tensile strength | 9 | 1.87E-08 | COL3A1, COL1A2, COL5A1, COL4A1, COL12A1, COL5A2, COL10A1, COL4A5, COL8A1 | |
| GO:0048407: Platelet-derived growth factor binding | 5 | 5.29E-06 | PDGFRB, COL3A1, COL1A2, COL5A1, COL4A1 | |
| GO:0008270: Zinc ion binding | 23 | 4.29E-04 | CPA2, TRIM50, CPB1, MMP7, ADH1C, MMP1, MT1M, ESRRB, MT1X, ESRRG, MYRIP, ADH7, ADAMTS2, MT2A, CA2, QPCT, MT1G, ZNF385B, CPXM1, MT1H, TIMP1, ADA, MT1E | |
| GO:0004111: Creatine kinase activity | 3 | 0.001918971 | CKMT2, CKM, CKB |
Using KEGG functional enrichment analysis, the results of pathways were arranged in ascending order of P value, and the P < 0.05 is the cutoff value (as shown in Figures 4F-J). As shown in Table 2 (list of top 5 KEGG pathways), our findings suggested that co-DEGs pathways were mainly enriched in ECM-receptor interaction, protein digestion and absorption, gastric acid secretion, mineral absorption and cell cycle pathways, etc. (detailed in Supplementary Tables 1 and 2).
| Category | Term | Count in gene set | P value | Genes |
| KEGG_pathway | hsa04974: Protein digestion and absorption | 13 | 2.23E-08 | CPA2, CELA3B, CPB1, COL12A1, COL3A1, COL1A2, SLC7A8, COL5A1, COL4A1, COL5A2, COL4A5, COL8A1, COL10A1 |
| hsa04512: ECM-receptor interaction | 8 | 2.57E-04 | COMP, COL1A2, COL4A1, ITGA2, CHAD, SPP1, COL4A5, THBS2 | |
| hsa04971: Gastric acid secretion | 7 | 7.46E-04 | ATP4B, ATP4A, KCNE2, CCKBR, CA2, KCNJ15, KCNJ16 | |
| hsa04978: Mineral absorption | 6 | 0.001635962 | MT2A, MT1M, MT1G, MT1H, MT1X, MT1E | |
| hsa04110: Cell cycle | 8 | 0.00218248 | CDC20, CCNB2, CCNB1, ORC1, CDK1, BUB1, CDC25B, MAD2L1 |
The results of pathways were arranged in ascending order of P value (as shown in Figure 4k, P < 0.05). As shown in Table 3 (list of top 5 GSEA pathways), GSEA pathway analysis illustrated that co-DEGs mainly concentrated in cell cycle progression, mitotic cell cycle, cell cycle, organelle fission and mitosis pathways, etc. (detailed in Supplementary Tables 1 and 2).
| ID | Description | Set size | P value |
| GO | GO_CELL_CYCLE_PROCESS | 41 | 1.00E-10 |
| GO_MITOTIC_CELL_CYCLE | 37 | 1.40E-10 | |
| GO_CELL_CYCLE | 44 | 1.49E-10 | |
| GO_ORGANELLE_FISSION | 24 | 9.65E-08 | |
| GO_MITOTIC_NUCLEAR_DIVISION | 19 | 1.20E-07 | |
| GO_CELL_CYCLE_PHASE_TRANSITION | 21 | 1.82E-07 |
STRING11.5 was conducted for PPI analysis on the above-mentioned differential expressed genes. The maximum confidence was set to 0.9, and the isolated genes without interaction were deleted (Figure 5A). Cytoscape_v3.9.1 software was used for further analyzing and mapping, showing 84 nodes and 654 edges (Figure 5B).
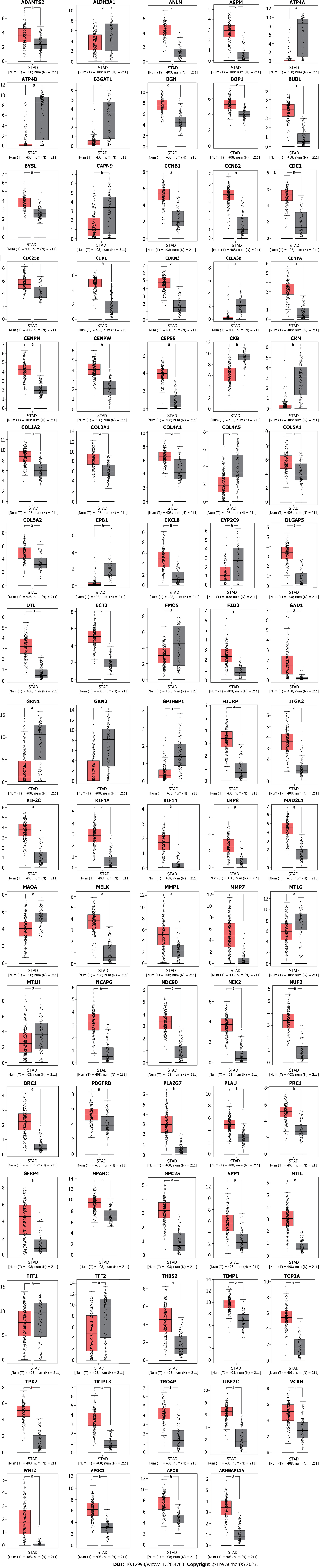
The Boxplot tool of GEPIA database was used for analyzing the above 84 selected gene nodes, and there was significant expression difference between the tumor tissues and normal tissues (Figure 6). Then, GEPIA’s survival tool was used for visual analysis of the selected genes [the cutoff value is Logrank P < 0.05 and hazard ratio (HR) < 0.05]. Our results suggested that 12 genes with notable significance for prognosis of patients were screened, including CEP55, COL1A2, COL3A1, gpihbp1, Vcan, TIMP1, SPARC, PDGFRb, MAOA, fzd2, COL4A1 and BGN. Visualized analysis of survival curve showed that CEP55 was the protective factor (Logrank P < 0.05, HR < 1). COL1A2, COL3A1, GPIHBP1, VCAN, TIMP1, SPARC, PDGFRB, MAOA, FZD2, COL4A1 and BGN were the risk factors (Logrank P < 0.05, HR > 1) (Figure 7).
For the above results, Kaplan-Meier plotter online network was carried out for visual analysis and verification. Excluding COL3A1 (Logrank P > 0.05), 11 key genes, including CEP55, COL1A2, GPIHBP1, VCAN, TIMP1, SPARC, PDGFRB, MAOA, FZD2, COL4A1 and BGN, were obtained (Logrank P < 0.05). The results confirmed that CEP55 was a protective factor (Logrank P < 0.05, HR < 1); COL1A2, GPIHBP1, VCAN, TIMP1, SPARC, PDGFRB, MAOA, FZD2, COL4A1 and BGN were risk factors (Logrank P < 0.05, HR > 1) (Figure 8).
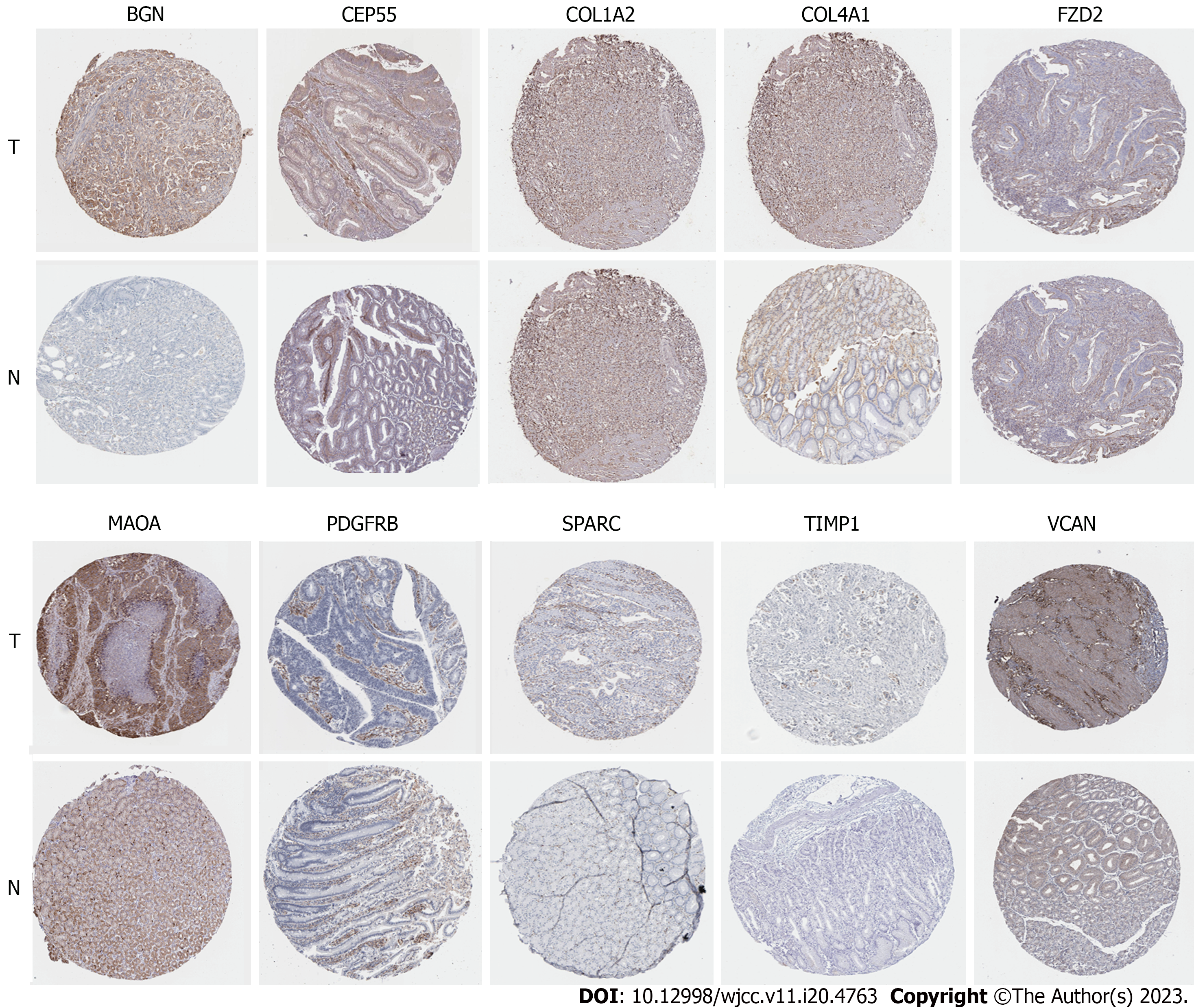
Compared with normal gastric tissues, the expressed proteins of 10 genes of BGN, CEP55, COL1A2, COL4A1, FZD2, MAOA, PDGFRB, SPARC, TIMP1 and VCAN were up-regulated in GC tissues by HPA IHC database (P < 0.05) (Figure 9). Therefore, in this study, 10 Hub genes related to the prognosis of GC were finally screened.
Based on GC gene expression data from GEO and TCGA databases, in this study, bioinformatics was conducted to screen differential expressed genes with relation to survival prognosis of GC. A total of 334 DEGs were analyzed by GO, KEGG and GSEA enrichment, respectively. By GO analysis (Figure 4), these DEGs were found to be enriched in BP, CC and MF. In KEGG analysis, ECM-receptor interaction, protein digestion and absorption, gastric acid secretion, mineral absorption, cell cycle and other signal pathways were enriched. GSEA pathway analysis showed that these DEGs were mainly concentrated in cell cycle progression, mitotic cell cycle, cell cycle, organelle fission, mitosis and other signaling pathways (Table 3). The GEPIA database was then used to verify the differences in expression of these key genes between tumors and normal tissues. The PPI network of these DEGs was analyzed and constructed by STRING11.5, and the credibility was set as the highest: 0.9. Eighty-four Hub genes were screened. GEPIA and Kaplan-Meier plotter identified 11 Hub genes (CEP55, COL1A2, GPIHBP1, VCAN, TIMP1, SPARC, PDGFRB, MAOA, FZD2, COL4A1, BGN) that were associated with GC prognosis. The combination of three databases (GEO, TCGA and GEPIA) makes our results more credible, which is a prominent feature of this study. Subsequently, 10 key genes involved in the prognosis of GC, including BGN, TIMP1, VCAN, COL1A2, COL4A1, FZD2, MAOA, PDGFRB, SPARC and CEP55, were screened by HPA immunohistochemical analysis.
The results of this study showed that BGN and VCAN genes, encoding multifunctional proteoglycans, which were highly expressed in GC tissues and associated with poor prognosis of patients. In this article, by GO, KEGG and GSEA analysis, we found that the two genes (BGN and VCAN) are mainly enriched in BP and MF such as extracellular matrix, extracellular matrix structural components, extracellular space and glycosaminoglycan binding (shown in Supplemen
As shown in Supplementary Tables 1 and 2, these findings also revealed that TIMP1 and MAOA genes, encoding enzyme proteins, predicted poor survival for patients; TIMP1 mainly participates in the degradation of extracellular matrix, promotes cell proliferation, and have anti-apoptosis function in tumors. Previous studies have found that TIMP1 is positively correlated with the pathological N stage of GC, which may inhibit the growth and metastasis of GC cells through mir-6745-TIMP1 axis[26,27], and Chemerin receptor antagonists down-regulate the expression of TIMP1 and TIMP2 through chemokine-like receptor-1 and G-protein coupled receptor 1 pathways, reducing the metastatic and invasive ability of GC cells[28]. Thus, TIMP1 could promote the progression of GC and shorten the survival period of patients. MAOA, encoding a mitochondrial enzyme, catalyzes the oxidative deamination of amines. The loss or decrease of MAOA expression can be used as a marker to monitor the immunotherapeutic effect of GC[29]; MAOA can also facilitate the proliferation and metastatic ability of gastric tumor cells by regulating mitochondrial function and aerobic glycolysis[30]. In the present study, we found that MAOA may affect the molecular functions of cells, such as protein binding, flavin adenine dinucleotide binding, oxidoreductase and so on. Therefore, it might affect the survival and prognosis of patients with GC by regulating the metabolic function of cells.
In addition, the results of this article suggested that two high-expressed genes of FZD2 and PDGFRB, encoding receptor proteins, were related to the prognosis of patients. It has been reported that a protein encoded by FZD2 is involved in binding to β-catenin typical signaling pathways and participates in regulation β-catenin dependent pathways. At present, only few literatures indicate that FZD 2 may play a key role in the occurrence and development of GC[31]. Our results demonstrated that FZD2 may act on: Atypical Wnt signaling pathway, classical Wnt signaling pathway, Wnt protein binding and other BPs. Previous studies have reported that PDGFRB is related to immune cell infiltration in GC, and may serve as a potential prognosis biomarker for GC[32]. PDGFRB is significantly correlated with the malignant phenotype of tumors, and the high expression of PDGFRB significantly reduced the overall survival of patients with GC[33,34]. In this study, we found that PDGFRB might promote tumor angiogenesis, cell proliferation and cell migration, and inhibit the aging and apoptosis of tumor cells, increasing the possibility of GC metastasis (details in Supplemen
The other four genes (COL1A2, COL4A1, SPARC and CEP55) are related to the prognosis of GC, which mainly encode collagen, acidic matrix related protein and centrosome protein of GC cells. The previous studies have found that collagen-encoding genes COL1A1 and P4HA3 may be related to the prognosis of GC[35]. At the present study, we found that COL4A1 and COL1A2 may have some effects on extracellular matrix and its interaction with receptors, protein binding, protein digestion and absorption, and regulate PI3K Akt signaling pathway. For the CEP55, some researchers have reported that it participates in promoting the malignant biological behavior of GC cells[36]. The expression of CEP55 in GC tissues is elevated, and CEP55 can also be a potential therapeutic target for GC[37]. The high expression of SPARC is associated with poor prognosis and shorter overall survival in patients with GC[38-41]. Our results suggested that CEP55 and SPARC could affect cell mitosis, cytokinesis and extracellular matrix structure, but its mechanisms are still remaining unclear (details in Supplementary Tables 1 and 2).
To sum up, this Bioinformatics present that BGN, CEP55, COL1A2, COL4A1, FZD2, MAOA, PDGFRB, SPARC, TIMP1 and VCAN, identified as the prognosis of GC, are involved in the occurrence and development of GC, and thus affect the survival and prognosis of patients by themselves or the encoded proteins. In this study, the 10 Hub genes obtained by comprehensive analysis of multiple databases and datasets may be used as survival biomarkers. Nevertheless, there are still many shortcomings in this study, for example, our data is currently limited to the online database, the selection of data may be biased or incomplete, and the relevant molecular mechanism needs further experimental research to verify.
In this study, we analyzed the gene expression profiles and sequencing data of GC tissues and adjacent or normal gastric tissues to explore the pathogenesis of GC using bioinformatics, investigated the signal pathways of co-DEGs involved in GC, and identified the 10 Hub genes correlated with the prognosis of patients with GC. The 10 key genes obtained through the analysis of multiple databases and datasets may be used as objective and reliable biomarkers for the survival analysis of patients. In addition, these genes or their encoded proteins can also be used as potential therapeutic targets for GC, improving the survival time of patients with GC. However, the mechanisms of 10 Hub genes in GC is still unclear, which needs further confirmation through molecular biology and clinical experiments.
Gastric cancer (GC) is one of the most common malignant tumors, and its pathogenesis and biomarkers are still unclear.
The present study for the first time investigated the 10 Hub genes as the potential biomarkers of the prognosis of patients using bioinformatics.
The aims of this study are to explore the potential biomarkers of the prognosis of patients with GC, so as to provide new strategies for the treatment of GC.
In this study, bioinformatics strategy was used to obtain Datasets from The Cancer Genome Atlas, Gene Expression Omnibus and Gene Expression Profiling Interactive Analysis. The software of R software, STRING, Kaplan-Meier plotter and Human Protein Atlas, were performed to analyze and integrate the mRNA datasets, respectively.
The signal pathways of the involvement of the co-expression of differential genes in GC were screened out, and the 10 Hub genes, including BGN, CEP55, COL1A2, COL4A1, FZD2, MAOA, PDGFRB, SPARC, TIMP1 and VCAN, were associated with prognosis of GC and identified as the potential prognostic biomarkers of GC.
The 10 key genes obtained through the analysis of multiple datasets may be used as objective and reliable biomarkers for the survival analysis of patients. In addition, these genes or their encoded proteins can also be as potential therapeutic targets for GC, improving the survival time of patients with GC.
The mechanisms of 10 Hub genes in GC is still unclear, which needs further confirmation through molecular biology and clinical experiments.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lucke-Wold B, United States; Zamani M, Iran S-Editor: Wang JJ L-Editor: A P-Editor: Cai YX
| 1. | Liu K, Yang K, Wu B, Chen H, Chen X, Jiang L, Ye F, He D, Lu Z, Xue L, Zhang W, Li Q, Zhou Z, Mo X, Hu J. Tumor-Infiltrating Immune Cells Are Associated With Prognosis of Gastric Cancer. Medicine (Baltimore). 2015;94:e1631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 2. | Zhao B, Zhang J, Chen X, Xu H, Huang B. Mir-26b inhibits growth and resistance to paclitaxel chemotherapy by silencing the CDC6 gene in gastric cancer. Arch Med Sci. 2019;15:498-503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 3. | Su W, Zhou B, Qin G, Chen Z, Geng X, Chen X, Pan W. Low PG I/II ratio as a marker of atrophic gastritis: Association with nutritional and metabolic status in healthy people. Medicine (Baltimore). 2018;97:e10820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Paoluzi OA, Del Vecchio Blanco G, Visconti E, Coppola M, Fontana C, Favaro M, Pallone F. Low efficacy of levofloxacin-doxycycline-based third-line triple therapy for Helicobacter pylori eradication in Italy. World J Gastroenterol. 2015;21:6698-6705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 5. | Deng X, Zheng H, Li D, Xue Y, Wang Q, Yan S, Zhu Y, Deng M. MicroRNA-34a regulates proliferation and apoptosis of gastric cancer cells by targeting silent information regulator 1. Exp Ther Med. 2018;15:3705-3714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 6. | Xie C, Yang Z, Hu Y, Cao X, Chen J, Zhu Y, Lu N. Expression of c-Met and hepatocyte growth factor in various gastric pathologies and its association with Helicobacter pylori infection. Oncol Lett. 2017;14:6151-6155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 7. | Yamamoto H, Watanabe Y, Sato Y, Maehata T, Itoh F. Non-Invasive Early Molecular Detection of Gastric Cancers. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Zhou CM, Wang Y, Ye HT, Yan S, Ji M, Liu P, Yang JJ. Machine learning predicts lymph node metastasis of poorly differentiated-type intramucosal gastric cancer. Sci Rep. 2021;11:1300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Zhai J, Wu J, Wang Y, Fan R, Xie G, Wu F, He Y, Qian S, Tan A, Yao X, He M, Shen L. Prediction of Sensitivity and Efficacy of Clinical Chemotherapy Using Larval Zebrafish Patient-Derived Xenografts of Gastric Cancer. Front Cell Dev Biol. 2021;9:680491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | Tao W, Li Y, Zhu M, Li C, Li P. LncRNA NORAD Promotes Proliferation And Inhibits Apoptosis Of Gastric Cancer By Regulating miR-214/Akt/mTOR Axis. Onco Targets Ther. 2019;12:8841-8851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 11. | Sung H, Hu N, Yang HH, Giffen CA, Zhu B, Song L, Su H, Wang C, Parisi DM, Goldstein AM, Taylor PR, Hyland PL. Association of high-evidence gastric cancer susceptibility loci and somatic gene expression levels with survival. Carcinogenesis. 2017;38:1119-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 12. | Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroenterol. 2018;24:2818-2832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 230] [Cited by in RCA: 317] [Article Influence: 45.3] [Reference Citation Analysis (7)] |
| 13. | Xie J, Wang M, Xu S, Huang Z, Grant PW. The Unsupervised Feature Selection Algorithms Based on Standard Deviation and Cosine Similarity for Genomic Data Analysis. Front Genet. 2021;12:684100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Tao Z, Shi A, Li R, Wang Y, Wang X, Zhao J. Microarray bioinformatics in cancer- a review. J BUON. 2017;22:838-843. [PubMed] |
| 15. | Chinni E, Tiscia G, Favuzzi G, Cappucci F, Malcangi G, Bagna R, Izzi C, Rizzi D, De Stefano V, Grandone E. Identification of novel mutations in patients with fibrinogen disorders and genotype/phenotype correlations. Blood Transfus. 2019;17:247-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Yao H, Yang L, Tian L, Guo Y, Li Y. LncRNA MSC-AS1 aggravates nasopharyngeal carcinoma progression by targeting miR-524-5p/nuclear receptor subfamily 4 group A member 2 (NR4A2). Cancer Cell Int. 2020;20:138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 17. | Kapitansky O, Gozes I. ADNP differentially interact with genes/proteins in correlation with aging: a novel marker for muscle aging. Geroscience. 2019;41:321-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 18. | Stielow B, Simon C, Liefke R. Making fundamental scientific discoveries by combining information from literature, databases, and computational tools - An example. Comput Struct Biotechnol J. 2021;19:3027-3033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 19. | Schaschl H, Wallner B. Population-specific, recent positive directional selection suggests adaptation of human male reproductive genes to different environmental conditions. BMC Evol Biol. 2020;20:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | Chen W, Yang Z. Identification of Differentially Expressed Genes Reveals BGN Predicting Overall Survival and Tumor Immune Infiltration of Gastric Cancer. Comput Math Methods Med. 2021;2021:5494840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Zhang S, Yang H, Xiang X, Liu L, Huang H, Tang G. BGN May be a Potential Prognostic Biomarker and Associated With Immune Cell Enrichment of Gastric Cancer. Front Genet. 2022;13:765569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 22. | Huang G, Xiang Z, Wu H, He Q, Dou R, Yang C, Song J, Huang S, Wang S, Xiong B. The lncRNA SEMA3B-AS1/HMGB1/FBXW7 Axis Mediates the Peritoneal Metastasis of Gastric Cancer by Regulating BGN Protein Ubiquitination. Oxid Med Cell Longev. 2022;2022:5055684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Liu H, Xiang Y, Zong QB, Zhang XY, Wang ZW, Fang SQ, Zhang TC, Liao XH. miR-6745-TIMP1 axis inhibits cell growth and metastasis in gastric cancer. Aging (Albany NY). 2021;13:24402-24416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 24. | Peduk S, Tatar C, Dincer M, Ozer B, Kocakusak A, Citlak G, Akinci M, Tuzun IS. The Role of Serum CK18, TIMP1, and MMP-9 Levels in Predicting R0 Resection in Patients with Gastric Cancer. Dis Markers. 2018;2018:5604702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Kumar JD, Aolymat I, Tiszlavicz L, Reisz Z, Garalla HM, Beynon R, Simpson D, Dockray GJ, Varro A. Chemerin acts via CMKLR1 and GPR1 to stimulate migration and invasion of gastric cancer cells: putative role of decreased TIMP-1 and TIMP-2. Oncotarget. 2019;10:98-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Li W, Han F, Fu M, Wang Z. High expression of VCAN is an independent predictor of poor prognosis in gastric cancer. J Int Med Res. 2020;48:300060519891271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 27. | Cheng Y, Sun H, Wu L, Wu F, Tang W, Wang X, Lv C. VUp-Regulation of VCAN Promotes the Proliferation, Invasion and Migration and Serves as a Biomarker in Gastric Cancer. Onco Targets Ther. 2020;13:8665-8675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 28. | Jiang K, Liu H, Xie D, Xiao Q. Differentially expressed genes ASPN, COL1A1, FN1, VCAN and MUC5AC are potential prognostic biomarkers for gastric cancer. Oncol Lett. 2019;17:3191-3202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 29. | Pan H, Ding Y, Jiang Y, Wang X, Rao J, Zhang X, Yu H, Hou Q, Li T. LncRNA LIFR-AS1 promotes proliferation and invasion of gastric cancer cell via miR-29a-3p/COL1A2 axis. Cancer Cell Int. 2021;21:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 30. | Hu Y, Li J, Luo H, Song W, Yang J. Differential Expression of COL1A1, COL1A2, COL6A3, and SULF1 as Prognostic Biomarkers in Gastric Cancer. Int J Gen Med. 2021;14:5835-5843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Ding YL, Sun SF, Zhao GL. COL5A2 as a potential clinical biomarker for gastric cancer and renal metastasis. Medicine (Baltimore). 2021;100:e24561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 32. | Rong L, Huang W, Tian S, Chi X, Zhao P, Liu F. COL1A2 is a Novel Biomarker to Improve Clinical Prediction in Human Gastric Cancer: Integrating Bioinformatics and Meta-Analysis. Pathol Oncol Res. 2018;24:129-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Li Z, Liu Z, Shao Z, Li C, Li Y, Liu Q, Zhang Y, Tan B, Liu Y. Identifying multiple collagen gene family members as potential gastric cancer biomarkers using integrated bioinformatics analysis. PeerJ. 2020;8:e9123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | Cao L, Chen Y, Zhang M, Xu DQ, Liu Y, Liu T, Liu SX, Wang P. Identification of hub genes and potential molecular mechanisms in gastric cancer by integrated bioinformatics analysis. PeerJ. 2018;6:e5180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 35. | Niu X, Ren L, Hu A, Zhang S, Qi H. Identification of Potential Diagnostic and Prognostic Biomarkers for Gastric Cancer Based on Bioinformatic Analysis. Front Genet. 2022;13:862105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 36. | Tao F, Qi L, Liu G. Long intergenic non-protein coding RNA 662 accelerates the progression of gastric cancer through up-regulating centrosomal protein 55 by sponging microRNA-195-5p. Bioengineered. 2022;13:3007-3018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 37. | Tao J, Zhi X, Tian Y, Li Z, Zhu Y, Wang W, Xie K, Tang J, Zhang X, Wang L, Xu Z. CEP55 contributes to human gastric carcinoma by regulating cell proliferation. Tumour Biol. 2014;35:4389-4399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 38. | Li L, Zhu Z, Zhao Y, Zhang Q, Wu X, Miao B, Cao J, Fei S. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci Rep. 2019;9:7827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 39. | Liao P, Li W, Liu R, Teer JK, Xu B, Zhang W, Li X, Mcleod HL, He Y. Genome-scale analysis identifies SERPINE1 and SPARC as diagnostic and prognostic biomarkers in gastric cancer. Onco Targets Ther. 2018;11:6969-6980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 40. | Shan Z, Wang W, Tong Y, Zhang J. Genome-Scale Analysis Identified NID2, SPARC, and MFAP2 as Prognosis Markers of Overall Survival in Gastric Cancer. Med Sci Monit. 2021;27:e929558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 41. | Ma Y, Zhu J, Chen S, Ma J, Zhang X, Huang S, Hu J, Yue T, Zhang J, Wang P, Wang X, Rong L, Guo H, Chen G, Liu Y. Low expression of SPARC in gastric cancer-associated fibroblasts leads to stemness transformation and 5-fluorouracil resistance in gastric cancer. Cancer Cell Int. 2019;19:137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |