Published online Jul 6, 2023. doi: 10.12998/wjcc.v11.i19.4655
Peer-review started: February 1, 2023
First decision: April 19, 2023
Revised: May 5, 2023
Accepted: May 31, 2023
Article in press: May 31, 2023
Published online: July 6, 2023
Processing time: 148 Days and 23.9 Hours
Cyclin-dependent kinase inhibitor 1C (CDKN1C) is a cell proliferation inhibitor that regulates the cell cycle and cell growth through G1 cell cycle arrest. CDKN1C mutations can lead to IMAGe syndrome (CDKN1C allele gain-of-function mutations lead to intrauterine growth restriction, metaphyseal dysplasia, adrenal hypoplasia congenital, and genitourinary malformations). We present a Silver-Russell syndrome (SRS) pedigree that was due to a missense mutation affecting the same amino acid position, 279, in the CDKN1C gene, resulting in the amino acid substitution p.Arg279His (c.836G>A). The affected family members had an SRS phenotype but did not have limb asymmetry or adrenal insufficiency. The amino acid changes in this specific region were located in a narrow functional region that contained mutations previously associated with IMAGe syndrome. In familial SRS patients, the PCNA region of CDKN1C should be analysed. Adrenal insufficiency should be excluded in all patients with functional CDKN1C variants.
We describe the case of an 8-year-old girl who initially presented with short stature. Her height was 91.6 cm, and her weight was 10.2 kg. Physical examination revealed that she had a relatively large head, an inverted triangular face, a protruding forehead, a low ear position, sunken eye sockets, and irregular cracked teeth but no limb asymmetry. Family history: The girl’s mother, great-grandmother, and grandmother’s brother also had a prominent forehead, triangular face, and severely proportional dwarfism but no limb asymmetry or adrenal insufficiency. Exome sequencing of the girl revealed a new heterozygous CDKN1C (NM_000076. 2) c.836G>A mutation, resulting in a variant with a predicted evolutionarily highly conserved arginine substituted by histidine (p.Arg279His). The same causative mutation was found in both the proband’s mother, great-grandmother, and grandmother’s brother, who had similar phenotypes. Thus far, we found an SRS pedigree, which was due to a missense mutation affecting the same amino acid position, 279, in the CDKN1C gene, resulting in the amino acid substitution p.Arg279His (c.836G>A). Although the SRS-related CDKN1C mutation is in the IMAGe-related mutation hotspot region [the proliferating cell nuclear antigen (PCNA) domain], no adrenal insufficiency was reported in this SRS pedigree. The reason may be that the location of the genomic mutation and the type of missense mutation determines the phenotype. The proband was treated with recombinant human growth hormone (rhGH). After 1 year of rhGH treatment, the height standard deviation score of the proband increased by 0.93 standard deviation score, and her growth rate was 8.1 cm/year. No adverse reactions, such as abnormal blood glucose, were found.
Functional mutations in CDKN1C can lead to familial SRS without limb asymmetry, and some patients may have glucose abnormalities. In familial SRS patients, the PCNA region of CDKN1C should be analysed. Adrenal insufficiency should be excluded in all patients with functional CDKN1C variants.
Core Tip: This is the fourth reported case of familial Silver–Russell syndrome (SRS) caused by a new missense mutation in the PCNA-binding domain of CDKN1C.The SRS pedigree, which was due to missense mutation affecting the amino acid position, 279, of the PCNA-binding domain of the CDKN1C gene, resulting in the amino acid substitution p.Arg279His (c.836G>A). Five affected family members also showed SRS phenotypes (small for gestational age, proportionately severe short stature, certain facial features (protruding forehead, triangular face, micrognathia), but without limb asymmetry or adrenal insufficiency. Initial efficacy and safety of growth hormone were observed in the proband treated with growth hormone.
- Citation: Li J, Chen LN, He HL. CDKN1C gene mutation causing familial Silver–Russell syndrome: A case report and review of literature. World J Clin Cases 2023; 11(19): 4655-4663
- URL: https://www.wjgnet.com/2307-8960/full/v11/i19/4655.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i19.4655
Cyclin-dependent kinase inhibitor 1C (CDKN1C), also known as p57/Kip2 (OMIM 600856), is active only when inherited maternally. The paternal copy is imprinted on the short arm of chromosome 11 (11p15.4) and is dose-sensitive. By binding to the cyclin/cyclin-dependent kinase complex, the CDKN1C protein prevents DNA replication and cell entry into S phase, arrests the cell cycle in G1 phase, and inhibits cell proliferation[1,2].
Up to 10%–15% of cases of Beckwith–Wiedemann Syndrome (BWS) are familial, and most cases are a result of CDKN1C loss-of-function mutations[3,4]. The clinical features of BWS include macrosomia, hyperinsulinemia, and adrenal tumours[5]. In contrast, gain-of-function variants of CDKN1C have been shown to cause conditions of growth restriction, including IMAGe (intrauterine growth restriction, metaphyseal dysplasia, adrenal hypoplasia congenital, and genital malformations) syndrome[6] and familial Silver–Russell syndrome (SRS)[7]. IMAGe syndrome is characterized by foetal/intrauterine growth restriction, adrenal dysplasia, metaphyseal dysplasia, genital abnormalities, and other characteristics, such as hypercalciuria and hearing loss[8,9]. Pathogenic single-nucleotide variations in a specific region of the PCNA-binding domain of CDKN1C have been found in children with IMAGe syndrome.
In 2013, Brioude et al[7] identified single-nucleotide variants in the PCNA-binding domain (p.Arg279Leu) of CDKN1C in patients with familial SRS for the first time. This type of SRS has a variety of clinical features, including foetal and postpartum growth restriction, particular facial features (triangular face, protruding forehead) and relative macrocephaly but no adrenal insufficiency or limb asymmetry. Two other familial SRSs resulting from mutations in this region have since been reported (p.Arg279Leu, p.Arg279ser)[10,11]. Inoue et al[12] recently examined the genes of 92 aetiology-unknown SRS patients and reported sporadic SRS cases caused by a new CDKN1C mutation, p.Arg316Gln. These cases met the four criteria of the Netchine–Harbison clinical scoring system, but there was no limb asymmetry and no adrenal insufficiency or metaphyseal dysplasia.
Here, we describe in detail a case of familial SRS caused by a new missense mutation in CDKN1C. This mutant gene resulted in an amino acid substitution (p.Arg279His) that was different from previous SRS mutations.
An 8-year-old girl complained of short stature for 8 years.
She was found to be severely short after birth, there was no vomiting, feeding difficulties, dizziness, headache, polydipsia, and polyuria. the growth rate was less than 5 cm per year.
There was no history of chronic disease.
The proband, born at 36+4 weeks’ gestational age, was delivered by caesarean section due to foetal hypoxia. Her birth weight was 1.44 kg, her body length was 39 cm (-6.22 standard deviation score, SDS), her head circumference was 31 cm, her sitting height was 26 cm, and her head was relatively large at birth. The anterior fontanelle was large (5 cm × 5 cm), and the anterior fontanelle was closed at 4 years of age. The proband could crawl at 10 mo, stand alone at 14 mo, walk at 24 mo, and consciously call her mum and dad at 15 mo.
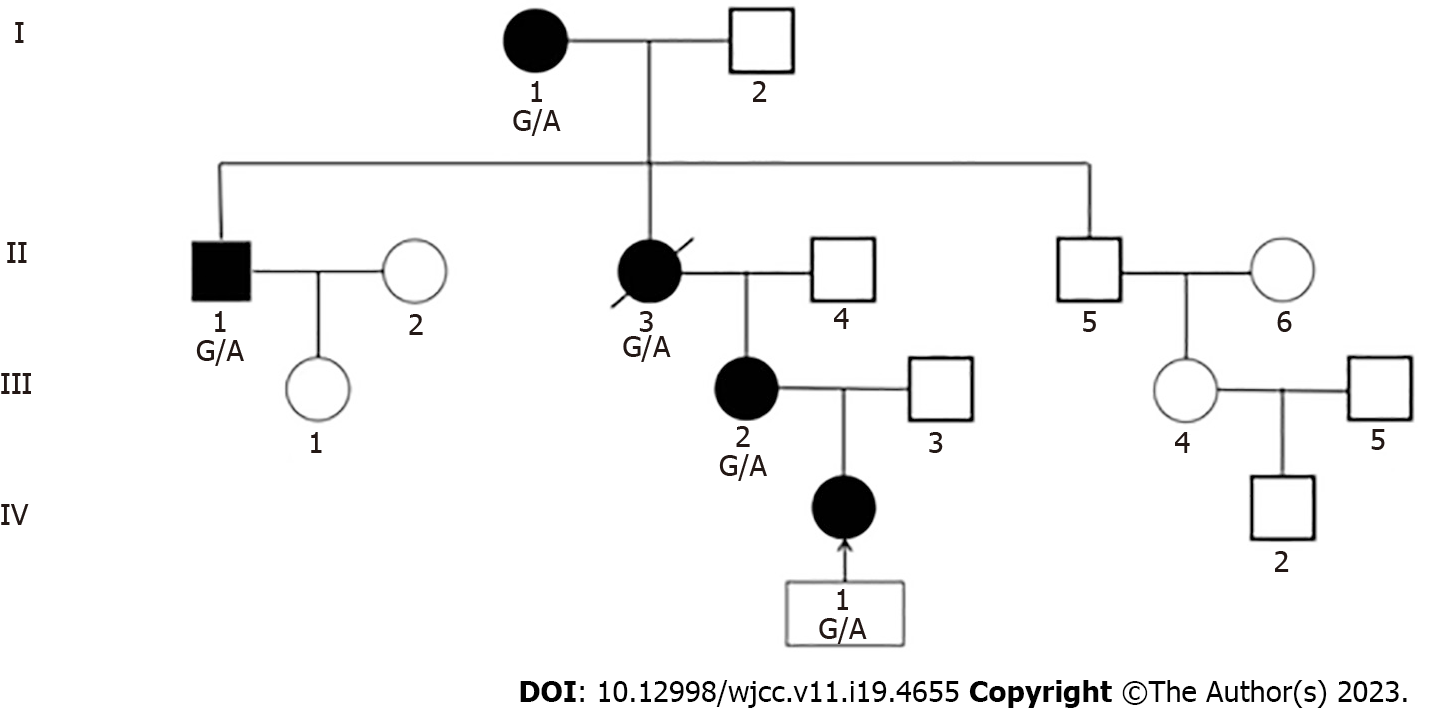
The mother of the proband was 33 years old (IV-2), with an unknown birth history, height 125 cm, weight before pregnancy 18 kg, body mass index (BMI) 11.5 kg/m2, head circumference 50 cm, sitting height 69.9 cm, and sitting height/height 0.56. Gestational diabetes was discovered during pregnancy, and she was later diagnosed with diabetes. The proband’s grandmother’s brother was 58 years old (III-1), with a height of 137 cm, a body weight of 28 kg, a BMI of 14.9 kg/m2, a head circumference of 52 cm, a sitting height of 76 cm, and a sitting height/height of 0.56. He was diagnosed with diabetes at the age of 45. The great-grandmother of the proband was 93 years old (I-1), with a height of 134 cm, body weight of 34 kg, BMI of 18.9 kg/m2, head circumference of 52 cm, sitting height of 73.7 cm, sitting height/height of 0.55, and no diabetes. All of them had an unknown birth history; however, they all mentioned being very thin and small at birth, and all three of them had a prominent forehead, triangular face, and severely proportional dwarfism but no limb asymmetry or adrenal insufficiency (Figure 1).
The proband’s late grandmother was 120 cm tall, and her appearance was similar to that of the proband. She passed away 10 years before due to an accident. She had no genetic testing, but we inferred that she had the same pathogenic mutation based on the genetic pedigree (Figure 2).
The girl had a relatively large head, an inverted triangular face, a protruding forehead, a low ear position, sunken eye sockets, and irregular cracked teeth but without limb asymmetry. She was 91.6 cm tall and weighed 10.2 kg, her head circumference was 48 cm, her sitting height was 54 cm, her sitting height/height was 0.58, and her BMI was 12.1 kg/m2. Her motor and language development was normal during treatment. She had no catch-up growth after birth.
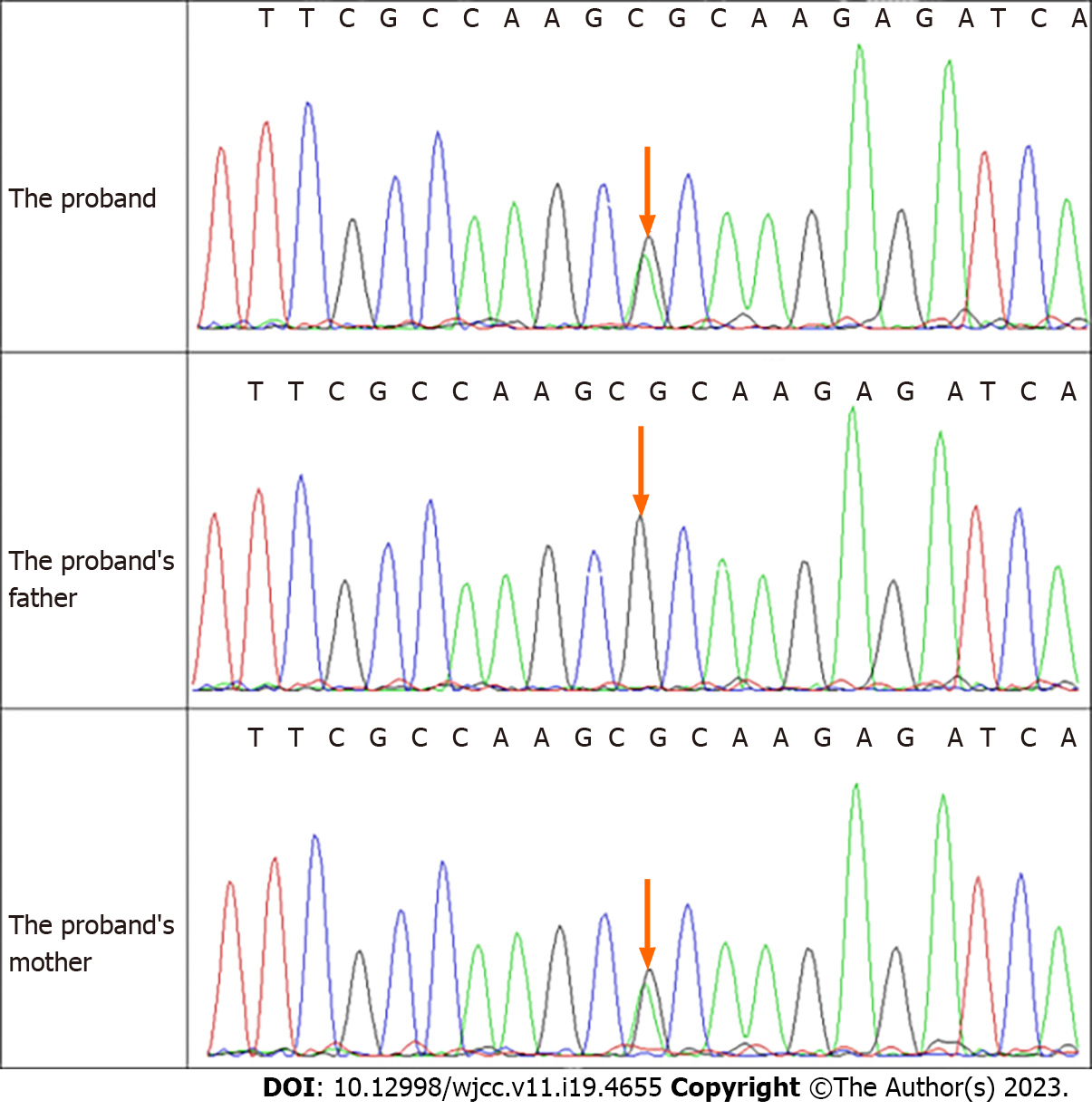
Serum insulin-like growth factor 1 (IGF1) was 244.08 ng/mL, adrenal cortex hormone ACTH was 19.3 pg/mL, cortisol rhythm (8 a.m.) was 6.29 µg/dL, blood glucose was 4.13 mmol/L, the growth hormone provocation test (arginine + levodopa) showed a peak value of growth hormone of 29.9 ng/mL. Exome sequencing revealed a new heterozygous CDKN1C (NM_000076. 2) c.836G>A mutation, resulting in a variant with a predicted evolutionarily highly conserved arginine substituted by histidine (p.Arg279His) (Figure 3).
Her bone age was 4.6 years, adrenal thin-slice computed tomography and pituitary magnetic resonance imaging were normal.
SRS.
She was treated with rhGH.
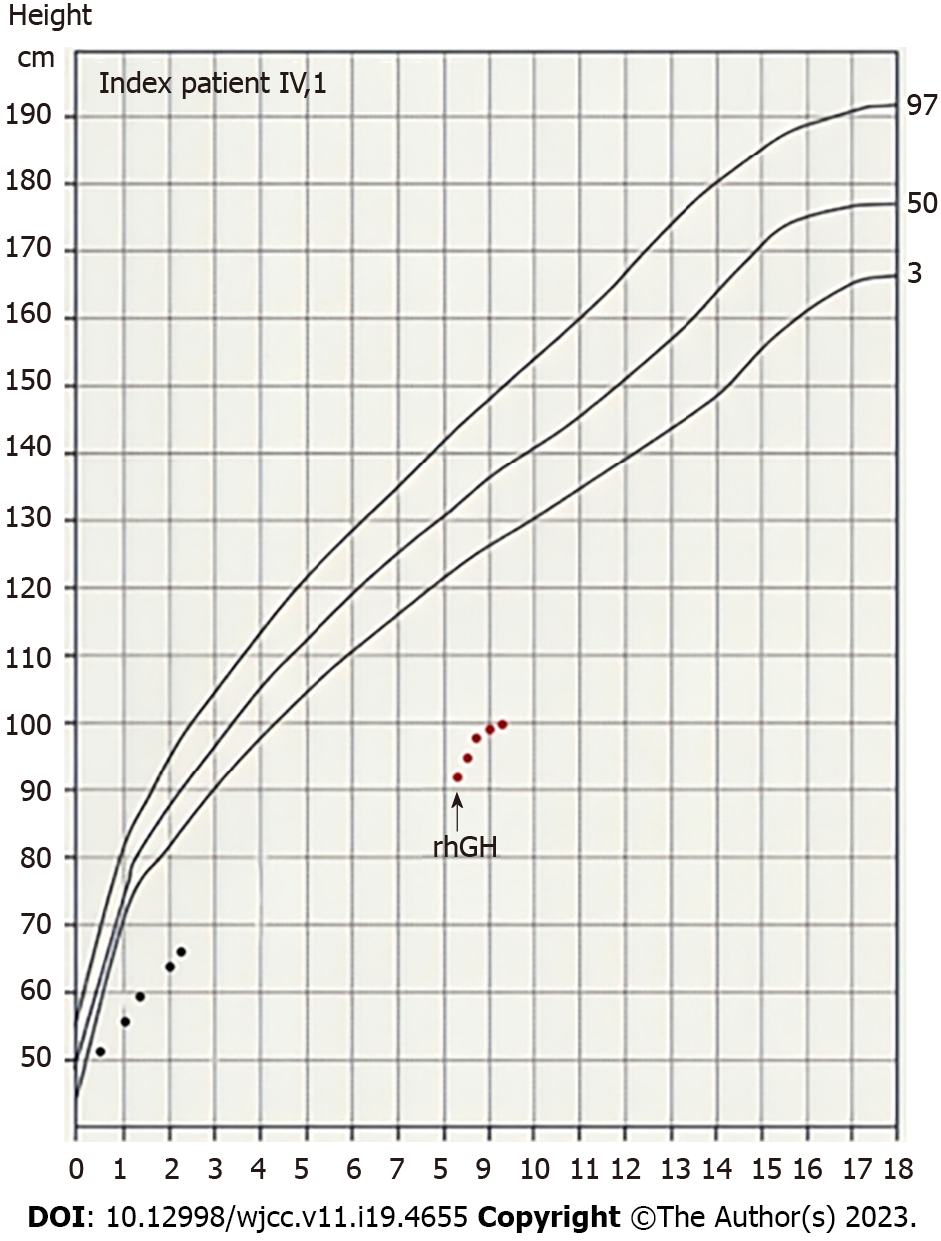
After 12 mo of treatment, the patient’s height was 99.7 cm (-5.9 SDS), her height standard deviation score increased by 0.93 SDS, and her growth rate was 8.1 cm/year (Figure 4). Blood glucose, insulin, thyroid function, and IGF-1 Levels were monitored every three months during treatment. No adverse reactions, such as abnormal blood glucose, were found.
This is the fourth reported case of familial SRS caused by a missense mutation in the PCNA-binding domain of CDKN1C, which was supported by the onset characteristics and genetic test results of the proband and the pedigree. CDKN1C, CDKN1A, and CDKN1B belong to the Cip/Kip family and are cyclin-dependent kinase (CDK) inhibitors[13]. The CDKN1C protein consists of three functional regions: (1) The N-terminal CDK inhibition domain (CdK); (2) The proline–alanine repeat (PAPA) domain[14,15].; and (3) The C-terminal PCNA-binding domain[1]. The C-terminal PCNA-binding domain binds to PCNA, a cofactor of DNA polymerases that encircles DNA and orchestrates the recruitment of factors to the replication fork[16,17].
CDKN1C mutations cause diseases with gain-of-function mutations such as IMAGe syndrome[6,8] and familial SRS[7,10,11]. These mutations are located in a small, conserved region of the gene (PCNA-binding domain containing 10 amino acid residues), and the common clinical manifestations of the two include foetal and postnatal growth restriction and forehead protrusion. However, none of the familial SRS patients who have been reported thus far have had adrenal insufficiency or limb asymmetry that is common in SRS. The PCNA-binding domain is a linear motif required for PCNA-dependent and crl4cdt2-mediated ubiquitination[18]. The proteins PCNA and CDKN1A associate closely to ensure the gradual ubiquitination and degradation of CDKN1A. The related motifs in CDKN1C are not perfect, resulting in low-affinity binding to PCNA. Low-affinity binding to PCNA is sufficient for monoubiquitination; however, it is not sufficient to carry out the polyubiquitination process required for protein degradation. CDKN1C monoubiquitination may have functions other than protein degradation[16,19]. Gain-of-function mutations affecting the 279th amino acid have been reported in both IMAGe syndrome and familial SRS (p.Arg279Pro, p.Arg279Ser, p.Arg279Leu)[7,10,11]. The Arg279 residue is highly conserved. However, in a flow cytometry study, the SRS-specific mutation p.Arg279Leu did not affect the cell cycle[7]. while the p.Arg279Pro mutation in IMAGe syndrome promoted cell cycle progression[7]. This finding was consistent with Hamajima's results[20]. Further research showed that p.Arg279Leu was associated with increased protein stability. These differences in amino acid changes (arginine to proline vs arginine to leucine) may be associated with a differential loss of binding to PCNA. In a recent study in Japan, the genes of 92 clinically diagnosed SRS patients with unknown aetiology were sequenced again. Sporadic SRS cases caused by the CDKN1C mutation Arg316Gln have been found. The clinical manifestations of the patients were consistent with SRS, but they had no limb asymmetry, adrenal insufficiency, or metaphyseal dysplasia. In vitro studies have shown that amino acid substitution leads to increased protein expression in vitro, and increased CDKN1C protein function leads to related phenotypes[12]. The SRS pedigree mutation (p.Arg279His) reported in this study has not been functionally verified. However, it can be speculated from the above studies that the p.Arg279His mutation increases CDKN1C protein stability.
In a study of IMAGe syndrome, mutations in the PCNA domain impaired the binding of PCNA and ubiquitin ligase to CDKN1C, thereby impairing PCDNA-dependent ubiquitination[6]. Monoubiquitination may have some functions in regulating protein localization, protein interaction, and protein chromosome degradation[21-23]; thus, impaired PCDNA-dependent ubiquitination might impair other functions of CDKN1C. Accordingly, it can also be speculated that mutations in the PCNA-binding domain may have different effects on ubiquitination, thereby affecting the regulatory characteristics of the domain.
After 1 year of rhGH treatment, the height standard deviation score of the proband increased by 0.93 SDS, and her growth rate was 8.1 cm/year, which was consistent with the first-year rhGH efficacy (growth velocity = 8.8 cm/year) of a proband’s mother (CDKN1C c.835C>T, p.Arg279Ser) reported by Binder et al[11] and was also consistent with the 1-year height standard deviation increase (0.75 ± 0.44 SDS) on rhGH treatment in children younger than gestational age[24]. The growth chart of the index patient is presented in Figure 4.
The proband’s grandmother’s brother (III.2) and mother (IV.1) both had diabetes, in line with the report of Kerns et al[25]. They found a variant in a pedigree with short stature syndrome in Ecuador (CDKN1C c.8433G>T, p.Arg281Leu). The affected family members all had intrauterine growth retardation, short stature, and normal adrenal function. Some patients in this pedigree had limb asymmetry, and eight of the 15 affected family members were diagnosed with diabetes before the age of 40.
CDKN1C plays a certain role in the proliferation of pancreatic β-cells. The loss of CDKN1C function leads to enhanced β-cell proliferation[26]. CDKN1C is highly expressed in pancreatic β cells, but its expression is absent in the pancreatic cell hyperplasia foci of infantile hyperinsulinemia patients with silencing of CDKN1C due to the loss of maternal 11p15 somatic cells[27]. Transplantation of short hairpin RNA-induced CDKN1C-silenced human islet cells into mice leads to the proliferation of transplanted β cells[28]. In addition, BWS patients often have hyperinsulinemia, and approximately 50% of BWS patients have hypoglycemia at birth[4,29,30]. A pathology study on the pancreas of four patients with BWS and hyperinsulinemia showed that the endocrine cells of the entire pancreas proliferated significantly, and the BWS-related CDKN1C loss-of-function mutation may be the main precipitating factor of β-cell proliferation[31]. CDKN1C (c.836G>A, p.Arg279His) is a gain-of-function mutation, which may be because this mutation leads to increased protein stability and produces the opposite phenotype from above: Decreased β-cell proliferation leads to decreased insulin secretion and the onset of diabetes.
CDKN1C (c.836G>A, p.Arg279Leu)- and (c.836G>A, p.Arg279Ser)-induced familial SRS members have not had diabetes[7,10,11]. In this study, the great-grandmother of the proband (I.1) was 91 years old and did not have diabetes. The blood glucose of the proband was normal, but long-term monitoring is needed. Seven of the people with mutations reported by Kerns et al[25] (p.Arg281Ile) were also temporarily free of diabetes. All of these mutations were located in the carboxy-terminal region of the "hot spot" region of the PCNA-binding domain. Kerns et al[25] demonstrated that the PCNA binding irregularities of p.Arg281Ile variants did not interfere with the ability of this CDKN1C mutant to associate with other proteins, such as the stress-activated protein kinase p38/SAPK, believed to interact with the N-terminus of CDKN1C.
Missense mutations in the highly conserved PCNA binding domain have been associated with clinical phenotypic heterogeneity (from growth restriction to skeletal abnormalities or no adrenal failure or diabetes in early adulthood)[13]. Further studies are needed to fully elucidate how CDKN1C variants defective only in PCNA binding regions lead to such a wide range of clinical manifestations.
In conclusion, gain-of-function mutations of CDKN1C are a rare cause of familial SRS. Its phenotype is similar to that of SRS, but there is no limb asymmetry, and some cases may be combined with abnormal blood glucose. In familial SRS cases, the PCNA region of CDKN1C should be analysed. Adrenal insufficiency should be excluded in all cases with functional CDKN1C variants.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kim BJ, South Korea; Nwabo Kamdje AH, Cameroon S-Editor: Li L L-Editor: A P-Editor: Li L
| 1. | Berland S, Haukanes BI, Juliusson PB, Houge G. Deep exploration of a CDKN1C mutation causing a mixture of Beckwith-Wiedemann and IMAGe syndromes revealed a novel transcript associated with developmental delay. J Med Genet. 2022;59:155-164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 2. | Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A, Harper JW, Elledge SJ. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 675] [Cited by in RCA: 699] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 3. | Brioude F, Kalish JM, Mussa A, Foster AC, Bliek J, Ferrero GB, Boonen SE, Cole T, Baker R, Bertoletti M, Cocchi G, Coze C, De Pellegrin M, Hussain K, Ibrahim A, Kilby MD, Krajewska-Walasek M, Kratz CP, Ladusans EJ, Lapunzina P, Le Bouc Y, Maas SM, Macdonald F, Õunap K, Peruzzi L, Rossignol S, Russo S, Shipster C, Skórka A, Tatton-Brown K, Tenorio J, Tortora C, Grønskov K, Netchine I, Hennekam RC, Prawitt D, Tümer Z, Eggermann T, Mackay DJG, Riccio A, Maher ER. Expert consensus document: Clinical and molecular diagnosis, screening and management of Beckwith-Wiedemann syndrome: an international consensus statement. Nat Rev Endocrinol. 2018;14:229-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 373] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 4. | DeBaun MR, King AA, White N. Hypoglycemia in Beckwith-Wiedemann syndrome. Semin Perinatol. 2000;24:164-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 51] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 5. | Elliott M, Bayly R, Cole T, Temple IK, Maher ER. Clinical features and natural history of Beckwith-Wiedemann syndrome: presentation of 74 new cases. Clin Genet. 1994;46:168-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 209] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 6. | Arboleda VA, Lee H, Parnaik R, Fleming A, Banerjee A, Ferraz-de-Souza B, Délot EC, Rodriguez-Fernandez IA, Braslavsky D, Bergadá I, Dell'Angelica EC, Nelson SF, Martinez-Agosto JA, Achermann JC, Vilain E. Mutations in the PCNA-binding domain of CDKN1C cause IMAGe syndrome. Nat Genet. 2012;44:788-792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 7. | Brioude F, Oliver-Petit I, Blaise A, Praz F, Rossignol S, Le Jule M, Thibaud N, Faussat AM, Tauber M, Le Bouc Y, Netchine I. CDKN1C mutation affecting the PCNA-binding domain as a cause of familial Russell Silver syndrome. J Med Genet. 2013;50:823-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 8. | Schrier Vergano SA, Deardorff MA. IMAGe Syndrome. 2014 Mar 13. In: GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–. [PubMed] |
| 9. | Kato F, Hamajima T, Hasegawa T, Amano N, Horikawa R, Nishimura G, Nakashima S, Fuke T, Sano S, Fukami M, Ogata T. IMAGe syndrome: clinical and genetic implications based on investigations in three Japanese patients. Clin Endocrinol (Oxf). 2014;80:706-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Sabir AH, Ryan G, Mohammed Z, Kirk J, Kiely N, Thyagarajan M, Cole T. Familial Russell-Silver Syndrome like Phenotype in the PCNA Domain of the CDKN1C Gene, a Further Case. Case Rep Genet. 2019;2019:1398250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Binder G, Ziegler J, Schweizer R, Habhab W, Haack TB, Heinrich T, Eggermann T. Novel mutation points to a hot spot in CDKN1C causing Silver-Russell syndrome. Clin Epigenetics. 2020;12:152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Inoue T, Nakamura A, Iwahashi-Odano M, Tanase-Nakao K, Matsubara K, Nishioka J, Maruo Y, Hasegawa Y, Suzumura H, Sato S, Kobayashi Y, Murakami N, Nakabayashi K, Yamazawa K, Fuke T, Narumi S, Oka A, Ogata T, Fukami M, Kagami M. Contribution of gene mutations to Silver-Russell syndrome phenotype: multigene sequencing analysis in 92 etiology-unknown patients. Clin Epigenetics. 2020;12:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 13. | Borges KS, Arboleda VA, Vilain E. Mutations in the PCNA-binding site of CDKN1C inhibit cell proliferation by impairing the entry into S phase. Cell Div. 2015;10:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Matsuoka S, Thompson JS, Edwards MC, Bartletta JM, Grundy P, Kalikin LM, Harper JW, Elledge SJ, Feinberg AP. Imprinting of the gene encoding a human cyclin-dependent kinase inhibitor, p57KIP2, on chromosome 11p15. Proc Natl Acad Sci U S A. 1996;93:3026-3030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 224] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 15. | Romanelli V, Belinchón A, Benito-Sanz S, Martínez-Glez V, Gracia-Bouthelier R, Heath KE, Campos-Barros A, García-Miñaur S, Fernandez L, Meneses H, López-Siguero JP, Guillén-Navarro E, Gómez-Puertas P, Wesselink JJ, Mercado G, Esteban-Marfil V, Palomo R, Mena R, Sánchez A, Del Campo M, Lapunzina P. CDKN1C (p57(Kip2)) analysis in Beckwith-Wiedemann syndrome (BWS) patients: Genotype-phenotype correlations, novel mutations, and polymorphisms. Am J Med Genet A. 2010;152A:1390-1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 16. | Eggermann T, Binder G, Brioude F, Maher ER, Lapunzina P, Cubellis MV, Bergadá I, Prawitt D, Begemann M. CDKN1C mutations: two sides of the same coin. Trends Mol Med. 2014;20:614-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 17. | Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1248] [Cited by in RCA: 1375] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 18. | Lam WW, Hatada I, Ohishi S, Mukai T, Joyce JA, Cole TR, Donnai D, Reik W, Schofield PN, Maher ER. Analysis of germline CDKN1C (p57KIP2) mutations in familial and sporadic Beckwith-Wiedemann syndrome (BWS) provides a novel genotype-phenotype correlation. J Med Genet. 1999;36:518-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 19. | Riccio A, Cubellis MV. Gain of function in CDKN1C. Nat Genet. 2012;44:737-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Hamajima N, Johmura Y, Suzuki S, Nakanishi M, Saitoh S. Increased protein stability of CDKN1C causes a gain-of-function phenotype in patients with IMAGe syndrome. PLoS One. 2013;8:e75137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 759] [Cited by in RCA: 769] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 22. | Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 896] [Cited by in RCA: 935] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 23. | Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397-2406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 199] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 24. | Ranke MB, Lindberg A, Cowell CT, Wikland KA, Reiter EO, Wilton P, Price DA; KIGS International Board. Prediction of response to growth hormone treatment in short children born small for gestational age: analysis of data from KIGS (Pharmacia International Growth Database). J Clin Endocrinol Metab. 2003;88:125-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 133] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 25. | Kerns SL, Guevara-Aguirre J, Andrew S, Geng J, Guevara C, Guevara-Aguirre M, Guo M, Oddoux C, Shen Y, Zurita A, Rosenfeld RG, Ostrer H, Hwa V, Dauber A. A novel variant in CDKN1C is associated with intrauterine growth restriction, short stature, and early-adulthood-onset diabetes. J Clin Endocrinol Metab. 2014;99:E2117-E2122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 26. | Ou K, Yu M, Moss NG, Wang YJ, Wang AW, Nguyen SC, Jiang C, Feleke E, Kameswaran V, Joyce EF, Naji A, Glaser B, Avrahami D, Kaestner KH. Targeted demethylation at the CDKN1C/p57 locus induces human β cell replication. J Clin Invest. 2019;129:209-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 27. | Kassem SA, Ariel I, Thornton PS, Hussain K, Smith V, Lindley KJ, Aynsley-Green A, Glaser B. p57(KIP2) expression in normal islet cells and in hyperinsulinism of infancy. Diabetes. 2001;50:2763-2769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 72] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Avrahami D, Li C, Yu M, Jiao Y, Zhang J, Naji A, Ziaie S, Glaser B, Kaestner KH. Targeting the cell cycle inhibitor p57Kip2 promotes adult human β cell replication. J Clin Invest. 2014;124:670-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Wang KH, Kupa J, Duffy KA, Kalish JM. Diagnosis and Management of Beckwith-Wiedemann Syndrome. Front Pediatr. 2019;7:562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 30. | Toda N, Ihara K, Kojima-Ishii K, Ochiai M, Ohkubo K, Kawamoto Y, Kohno Y, Kumasaka S, Kawase A, Ueno Y, Futatani T, Miyazawa T, Nagaoki Y, Nakata S, Misaki M, Arai H, Kawai M, Sato M, Yada Y, Takahashi N, Komatsu A, Maki K, Watabe S, Sumida Y, Kuwashima M, Mizumoto H, Sato K, Hara T. Hyperinsulinemic hypoglycemia in Beckwith-Wiedemann, Sotos, and Kabuki syndromes: A nationwide survey in Japan. Am J Med Genet A. 2017;173:360-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Laje P, Palladino AA, Bhatti TR, States LJ, Stanley CA, Adzick NS. Pancreatic surgery in infants with Beckwith-Wiedemann syndrome and hyperinsulinism. J Pediatr Surg. 2013;48:2511-2516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |