Published online May 26, 2023. doi: 10.12998/wjcc.v11.i15.3444
Peer-review started: December 16, 2022
First decision: February 28, 2023
Revised: March 8, 2023
Accepted: April 12, 2023
Article in press: April 12, 2023
Published online: May 26, 2023
Processing time: 160 Days and 4.2 Hours
Regulatory T cells (Tregs) and natural killer (NK) cells play an essential role in the development of bladder urothelial carcinoma (BUC).
To construct a prognosis-related model to judge the prognosis of patients with bladder cancer, meanwhile, predict the sensitivity of patients to chemotherapy and immunotherapy.
Bladder cancer information data was obtained from The Cancer Genome Atlas and GSE32894. The CIBERSORT was used to calculate the immune score of each sample. Weighted gene co-expression network analysis was used to find genes that will have the same or similar expression patterns. Subsequently, multivariate cox regression and lasso regression was used to further screen prognosis-related genes. The prrophetic package was used to predict phenotype from gene expression data, drug sensitivity of external cell line and predict clinical data.
The stage and risk scores are independent prognostic factors in patients with BUC. Mutations in FGFR3 lead to an increase in Tregs percolation and affect the prognosis of the tumor, and additionally, EMP1, TCHH and CNTNAP3B in the model are mainly positively correlated with the expression of immune checkpoints, while CMTM8, SORT1 and IQSEC1 are negatively correlated with immune checkpoints and the high-risk group had higher sensitivity to chemotherapy drugs.
Prognosis-related models of bladder tumor patients, based on Treg and NK cell percolation in tumor tissue. In addition to judging the prognosis of patients with bladder cancer, it can also predict the sensitivity of patients to chemotherapy and immunotherapy. At the same time, patients were divided into high and low risk groups based on this model, and differences in genetic mutations were found between the high and low risk groups.
Core Tip: Tregs are thought to be connected to tumor cells evading the immune system, which is linked to cancer patients' poor prognoses. Natural killer (NK) cells control different immune responses and show antitumor cytotoxicity without being previously sensitized. We determined the immunological scores of several cell types in bladder urothelial carcinoma, discovered a gene set that was favorably connected with the Tregs score and negatively correlated with the NK cells score, and built a model that was related to prognosis. The model can predict the sensitivity of patients to chemotherapy and immunotherapy in addition to their prognosis for bladder cancers.
- Citation: Yang YJ, Xu XQ, Zhang YC, Hu PC, Yang WX. Establishment of a prognostic model related to tregs and natural killer cells infiltration in bladder cancer. World J Clin Cases 2023; 11(15): 3444-3456
- URL: https://www.wjgnet.com/2307-8960/full/v11/i15/3444.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i15.3444
Bladder urothelial carcinoma (BUC) is a typical malignant growth of the urinary tract that has a high likelihood of return and death[1]. Immunotherapy has lately attracted more research attention than traditional surgery methods and chemotherapy[2]. Understanding the changes in the tumor microenvironment and finding potential immunotherapy strategies have received a lot of attention from scientific specialists. Thus, it is crucial to research how different immune cells infiltrate the tumor's inflammatory environment.
The start and progression of BUC are influenced by three processes: Immune monitoring, immunological balance, and immune escape. T cells (Tregs) are currently believed to be implicated in tumor cell immune evasion; Tregs are frequently found in tumors and are typically identified by the expression of CD4+ and Foxp3+, which is linked to the bad outcome of cancer patients[3]. The grade and stage of BUC were considerably correlated with the proportion of Foxp3+ Tregs[4]. Inhibiting Tregs can improve the treatment of BUC patients. Natural killer (NK) cells are mainly related to killing infected microorganisms and malignant transformed allogeneic and autologous cells. NK cells exhibit antitumor cytotoxicity without prior sensitization and production of cytokines and chemokines that regulate various immune responses[5]. NK cells patrol the body and are recruited to tumor sites to destroy malignant cells[6].
Based on the infiltration of Treg cells and NK cells into the tumor tissue, we built a prognosis-related model of bladder tumor patients for this investigation. It can predict the susceptibility of patients to chemotherapy and immunotherapy in addition to assessing the prognosis of bladder cancer patients.
The validated database is derived from GSE32894. The platform of the data set is GPL6947. In the Gene Expression Omnibus (GEO) database, the end-point event is determined according to the disease-specific survival. GSE32894 included 308 patients with BUC. The data source of the training data set is the transcriptome data of The Cancer Genome Atlas (TCGA's) bladder cancer, including 19 patients' normal samples and 411 patients' cancer samples. The Data Category and Workflow Type of TCGA data are transcriptome profiling and Fragments per Kilobase Million (FPKM). Then, the TranscriptsPerKilobase of exonmodel per Million mapped reads (TPM) method was used to standardize the TCGA data for model verification. The mutation data were obtained from the simple nucleoside variation data of 409 patients with bladder urothelial carcinoma in TCGA database.
Each sample's immunological score was determined using "CIBERSORT". 22 immune cells in total were scored, and "perm = 1000, qn = true" was specified as the option in "CIBERSORT".
Finding genes that will have the same or comparable expression patterns may be done using the useful method known as weighted gene coexpression network analysis (WGCNA). These genes could have comparable physiological effects. WGCNA differs from other straightforward clustering methods in this way (such as based on Euclidean distance). It is a clustering technique with biologically relevant applications. In this work, we employed WGCNA to categorize the bladder cancer expression profile data into different groups.
Then, the prognosis-related diagnostic genes in the TCGA database were screened using the univariate cox regression model. Multivariate cox regression and lasso regression were utilized to further screen prognosis-related genes based on the findings of the univariate cox screening of the TCGA database and GEO data. We determined the riskcore for each sample using the multivariate Cox model (riskcore = gene expression * coef).
Prrophetic package is an ancient R package. Its main purpose is to predict phenotype from gene expression data (predict clinical results using CGP cell line data of cancer genome project), predict drug sensitivity of external cell line, and predict clinical data.
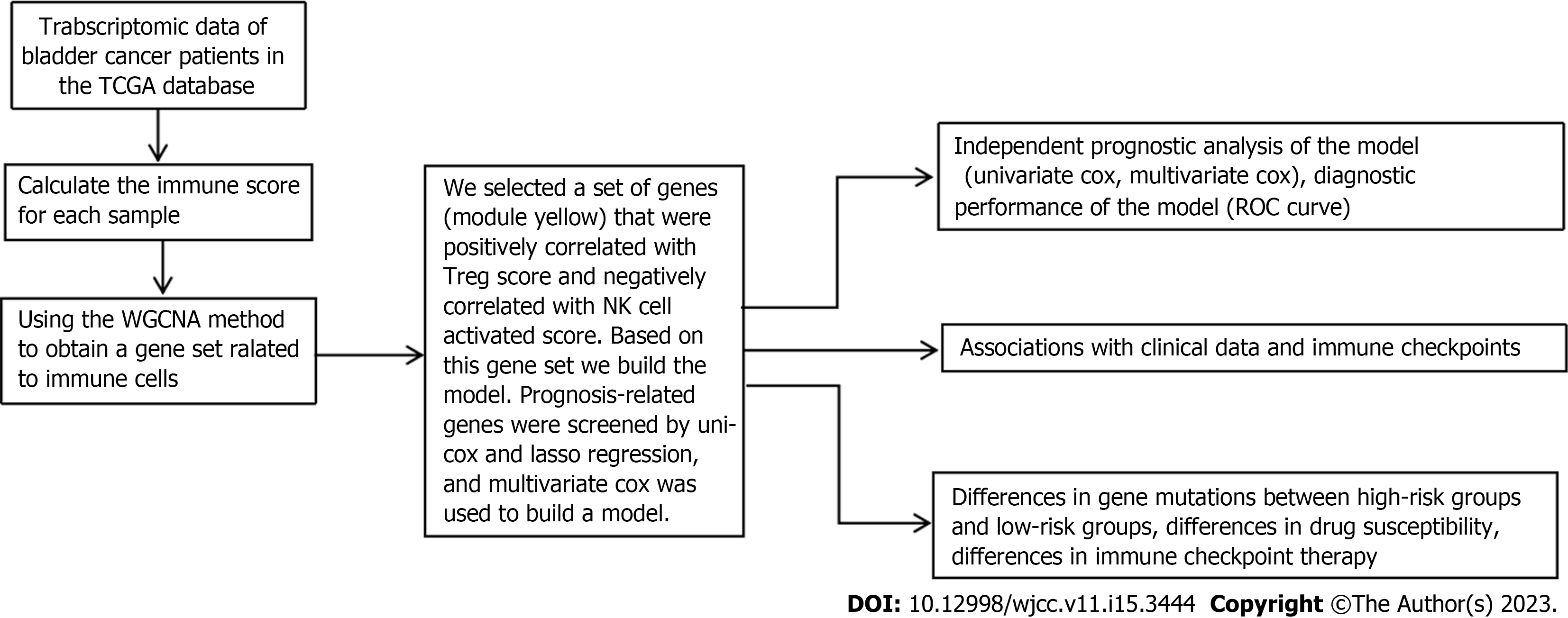
The clinical data were compared using the chi square test. The correlation between different values was determined using Pearson correlation analysis. The following is the study's flowchart (Figure 1).
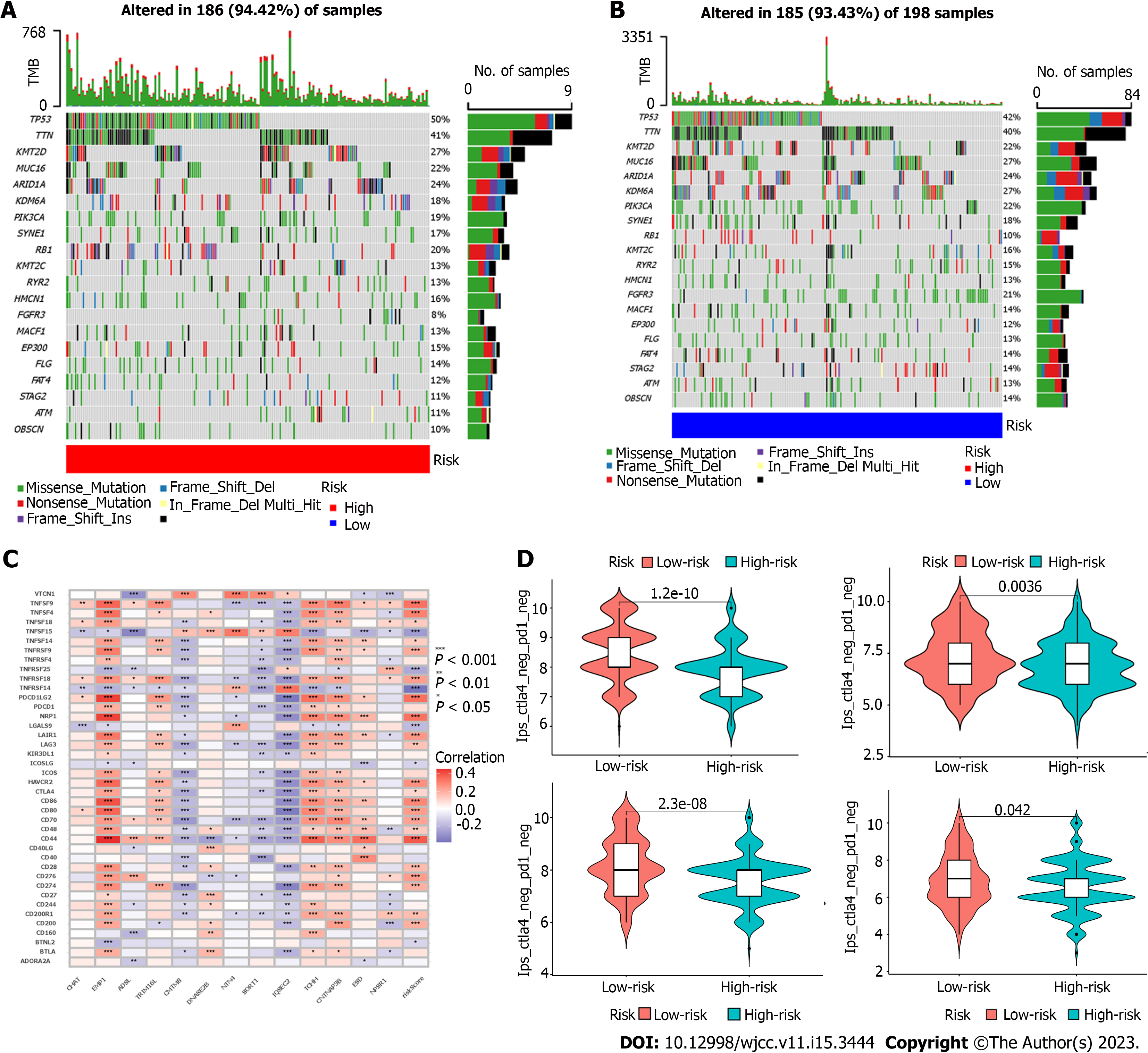
Maftools package is used to analyze the overall situation of gene mutation between high-risk and low-risk groups. The top 20 genes with the highest frequency of mutations in both high-risk and low-risk groups are displayed in a waterfall chart.
The Cancer Immunome Atlas (TCIA) is developed based on TCGA data. The difference is that TCIA only provides immune data analysis of 20 cancer species. The immunophenoscore (IPS) of each sample in the high and low risk categories, as well as the difference between them, were determined using TCIA. The reactivity of CTLA-4 and PD-1 can be accurately predicted by IPS.
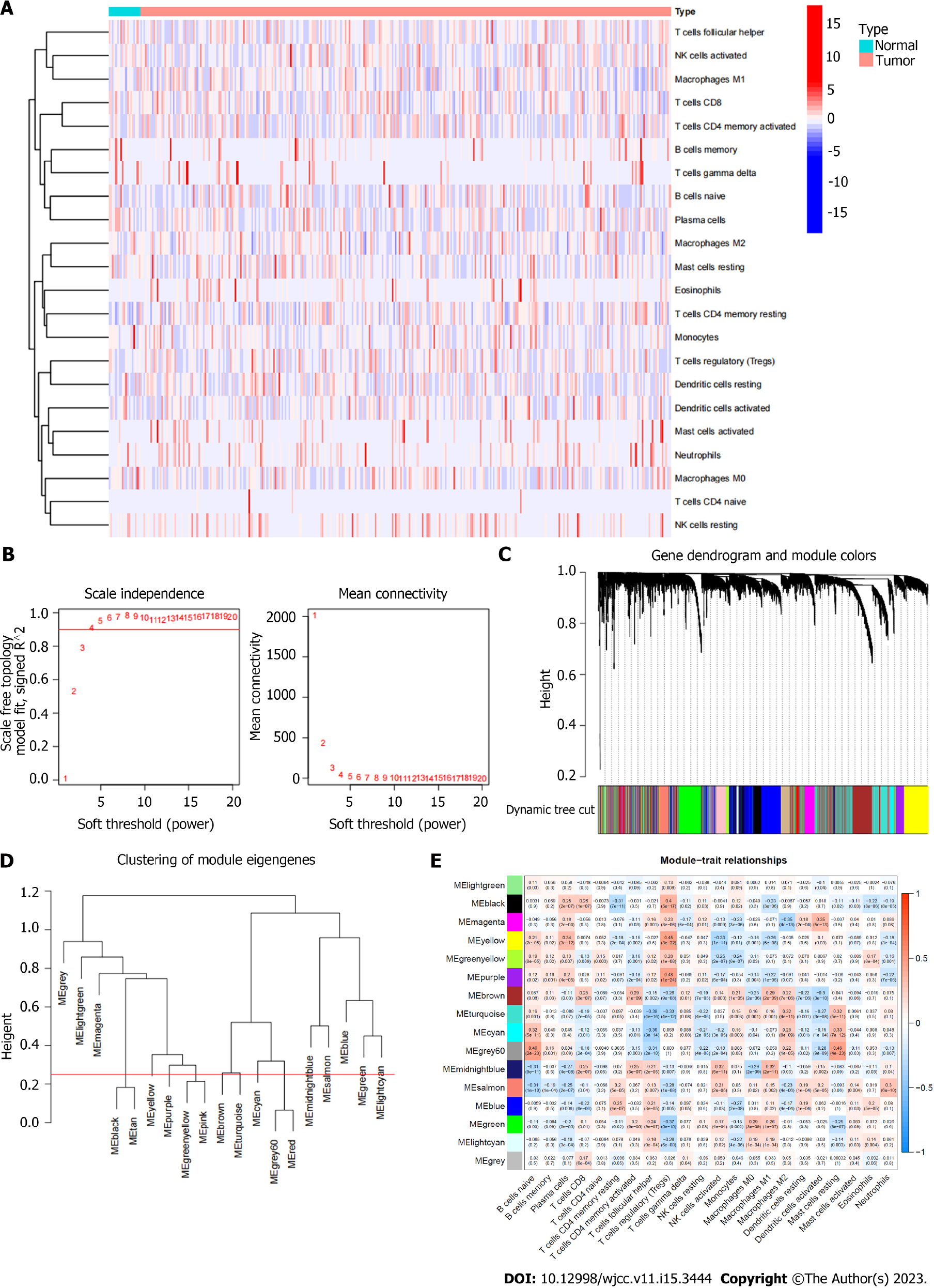
In the TCGA database, we first assigned a number to each sample's immune cells (Figure 2A). The total genes were divided into 9 gene sets based on the immune cell scores of each sample using the WGCNA technique, and the correlation between these 9 gene sets and the immune cell scores of each type was determined (Figure 2B-E). Then we chose a group of genes (module yellow) that had a positive correlation with the score for Regulatory T cells (Treg) (r = 0.45) and a negative correlation with the score for NK cell activation (r = -0.33). According to earlier studies, Tregs inhibit long-lasting immune reactions to viruses, tumors, and self-antigens; the infiltrate tumor tissue and are linked to a bad prognosis in cancer patients; the quantity of Treg was negatively correlated with recurrence-free survival in BUC and substantially increased in peripheral blood and tumor tissue[7]. The grade and stage of BUC were significantly associated with the proportion of Foxp3+ Tregs[8]. The most significant component of natural immunity is played by NK cells, which can directly destroy target cells like cancerous or virus-infected ones. They serve as the first line of protection against tumors and the first obstacle of human antibodies. From this perspective, the gene collection we chose is also consistent with the findings of earlier studies. More significantly, prior research has shown that tumor-infiltrating Treg cells can cause NK cells, B cells, Dendritic cells (DC) and cytotoxic T cells in the Tumor microenvironment (TME) to undergo death[9]. This strengthens the logic of our study concepts even more.
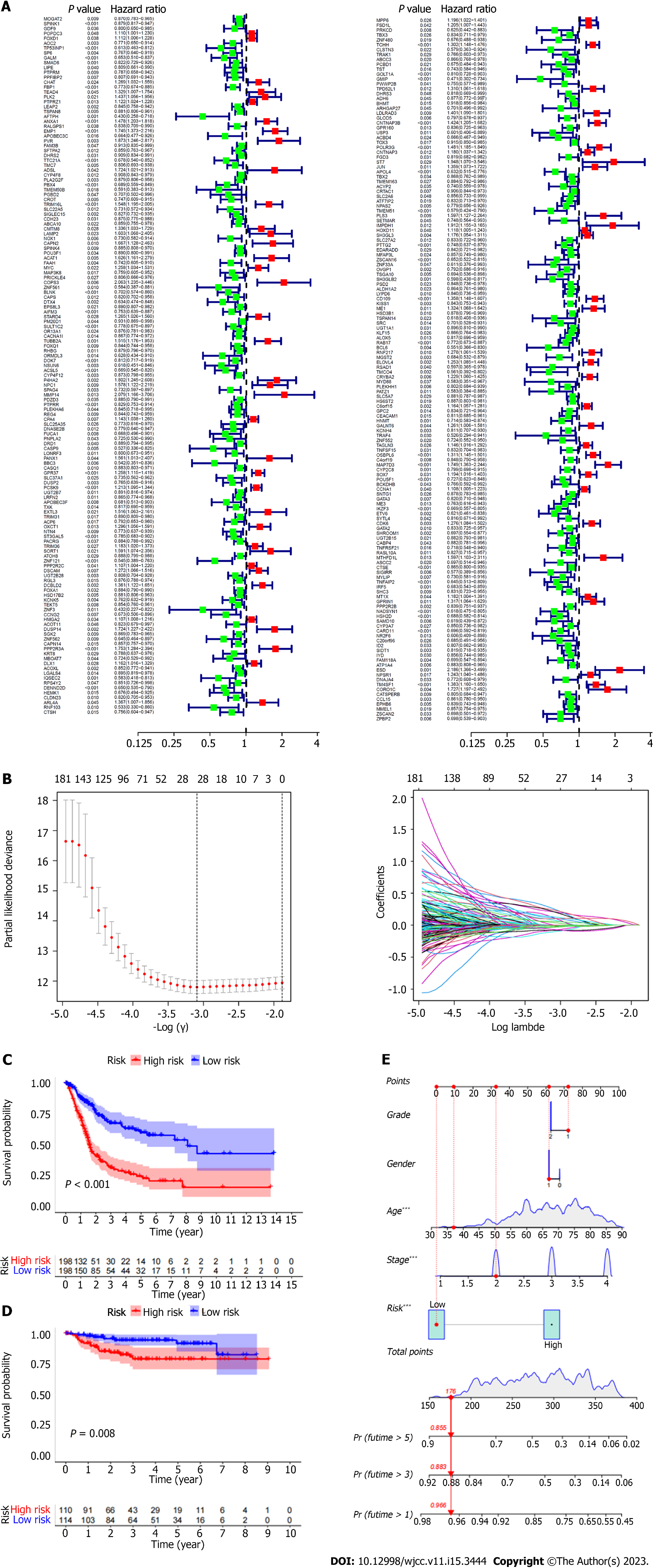
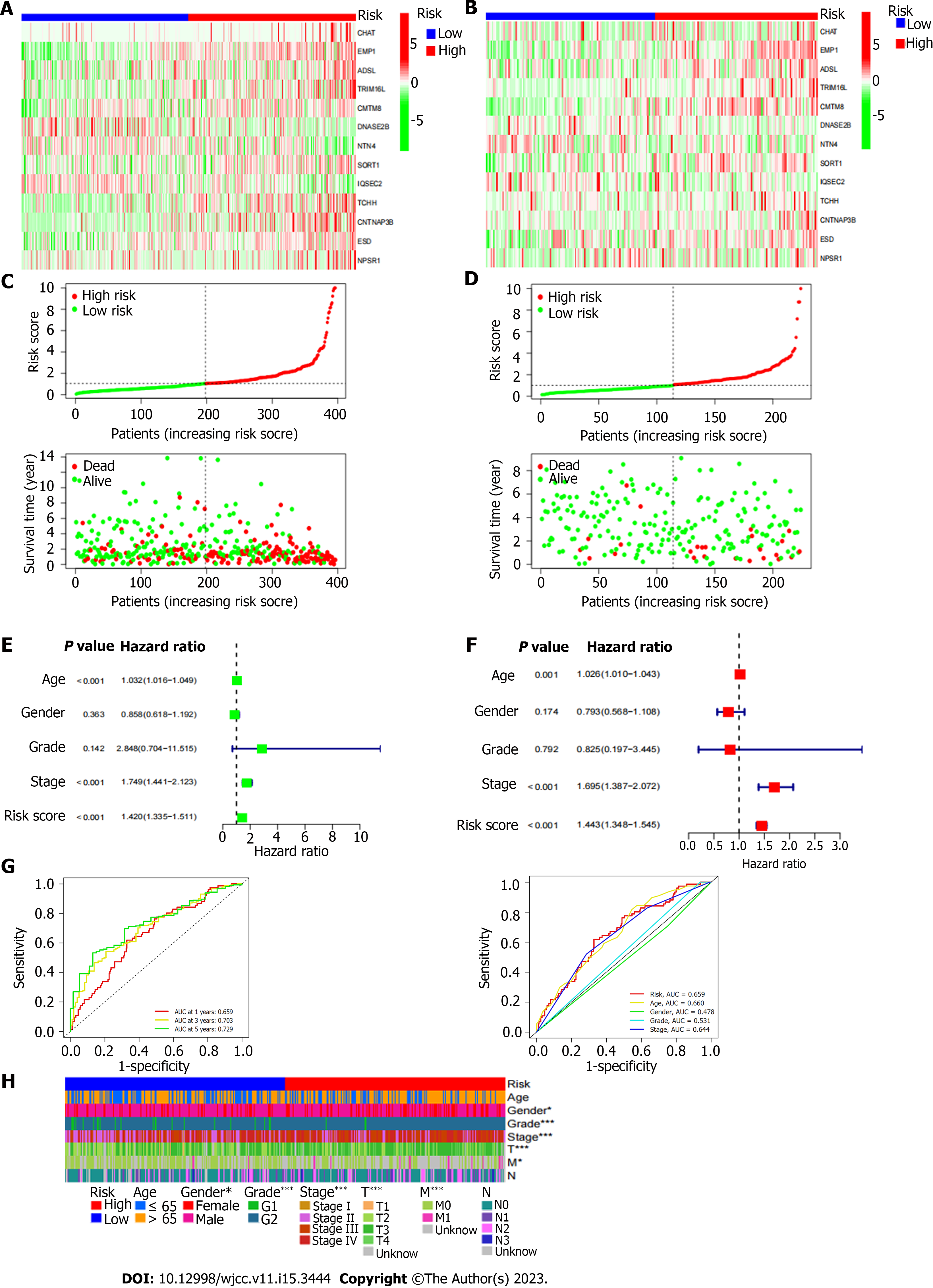
By using multivariate Cox regression analysis, the prognostic-related genes in the yellow gene group were chosen (Figure 3A). The alleles with high association were then eliminated using lasso regression (Figure 3B). Lastly, the prognostic association model was built using multivariate Cox regression. 13 genes in total (CHAT, EMP1, ADSl, TRIM16 L, CMTM8, DNASE2B, NTN4, SORT1, IQSEC2, TCHH, CNTNAP3B, ESD, NPSR1) were used to build this model (Figure 4A and B). The riskscore of each sample was then determined using the multivariate Cox model (riskscore = gene expression * coef). Samples that scored above the median for hazards were placed in the high-risk category, while those that scored below the median were placed in the low-risk group. Then it was looked at how long the high-risk group survived compared to the low-risk group. Clearly, the prognosis between the high-risk group and the low-risk group is extremely different (Figure 3C). In comparison to the high-risk group, the prognosis for the low-risk group was noticeably better. The GEO database has verified the model's efficacy in determining prognosis (Figure 4D). In the supporting materials, a survival analysis of individual genes will be presented. Furthermore, we determined the association (r > 0.4 and P value > 0.05 were deemed significant) between each gene's expression and immune cell infiltration in the model. The prognosis of patients was then predicted using Nomo map (Figure 2E; stage: "1","2","3","4" represent Stage I, Stage II, Stage III, and Stage IV, respectively; grade: "1" and "2"represent G1 and G2, respectively; gender: "0" and "1" represent females and males, respectively).
Cox regression analysis was conducted to determine whether our model could predict the prognosis of BUC patients independently of other clinical data. Stage and risk score were shown to be strongly correlated with the prognosis of BUC patients by univariate Cox regression analysis. Stage and risk score were identified as independent predictive variables in multivariate Cox regression analysis of BUC patients (Figure 4E and F). Furthermore, the scatter plot demonstrates that patients who scored highly for hazards have much poorer prognoses (Figure 4C and D). The receiver operating characteristic curve (AUC at 1 year: 0.699, AUC at 3 years: 0.745, AUC at 5 years: 0.753) further proves that the model can more accurately predict the prognosis of patients (Figure4G). We employed chi-square test to evaluate the difference in clinical data between high and low risk groups ("***" = P0.001, "**" = P0.01,
The variations in genetic mutations between high and low risk were then looked at. According to the findings, the prevalence of FGFR3 mutations varied considerably between the high-risk and low-risk groups (Figure 5A and B). One of the frequent mutations in bladder cancer is the FGFR3 mutation, which is also one of the key elements in carcinogenesis. Moreover, FGFR3-targeting medications have been utilized to treat BUC patients. It will be fascinating to explore if the FGFR3 mutation would boost Treg infiltration and have an impact on the prognosis of tumors.
The model and each immune checkpoint differ in that EMP1, TCHH, and CNTNAP3B are mostly favorably connected with the expression of immunological checkpoints, whereas CMTM8, SORT1, and IQSEC1 are adversely linked with immune checkpoints (Figure 5C). Moreover, the IPS between the high-risk and low-risk groups varied (Figure 5D). It is evident that the immune checkpoint inhibition may work better for the low-risk population.
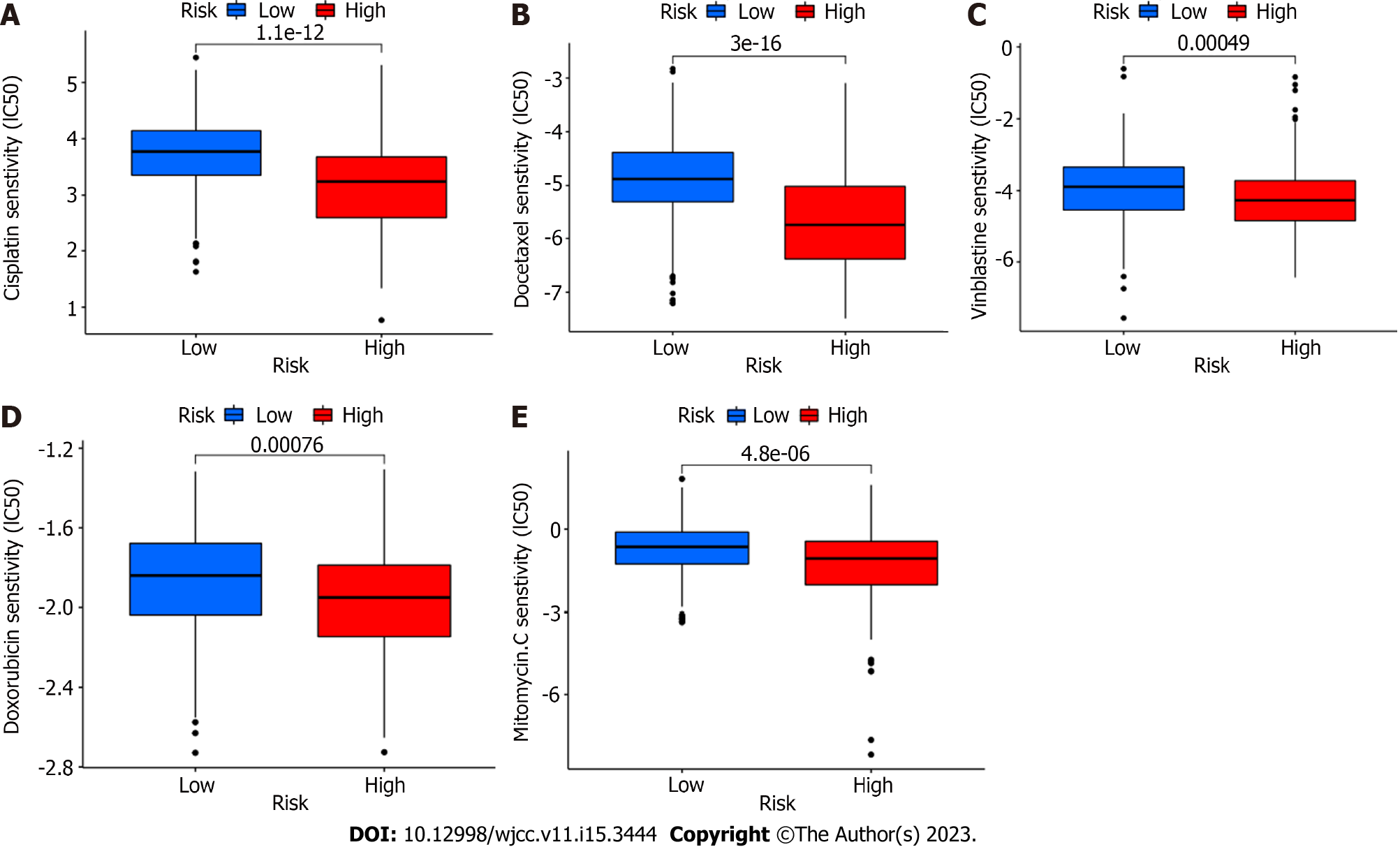
To distinguish between high and low risk groups in terms of medication sensitivity, we employed the prrophetic package (R package). Interestingly, chemotherapeutic medication sensitivity was greater in the high-risk group. These chemotherapy medications include vinblastine, cisplatin, docetaxel, mitomycin, and adriamycin (Figure 6). These are all chemotherapeutic medications that are frequently used to treat bladder cancer.
Tregs, an immunosuppressive subset of CD4+ T cells, play a crucial role in maintaining self-tolerance, avoiding autoimmune disorders and transplant rejection, it has been proposed as candidates of bio-markers because of their capability to control alloimmune responses[10]. In tumor immunity, Tregs compromise immune surveillance against cancer in healthy individuals and damage the antitumor immune response in tumor-bearing hosts[11]. Among different types of tumor infiltrating lymphocytes, the infiltration of cytotoxic CD8+T cells plays a positive role in the prognosis of tumor patients and the clinical response of immunotherapy[12,13], but the infiltration of tumor infiltrating lymphocytes called Treg commonly indicates poor prognosis and poor effect of immunotherapy. Tregs have been reported to increase infiltration in various tumor tissues, which is related to tumor immune escape.
Tregs can secrete anti-inflammatory mediators including Interleukin (IL)-10,Transforming factor β (TGF-β), and IL-35, which can suppress the immune system. TGF-β lowers the lethality of NK cells and induces them to transform into type 1 innate lymphocytes, which has further effects on tumor development and metastasis[14].
Due to their quick recognition and effective killing of tumor cells, NK cells, which are a crucial component of innate immunity, serve as the first line of defense against tumors[15]. Meanwhile, NK cells play a central role in cancer immune surveillance through express up to different activating inhibitory killer-cell immunoglobulin-like receptors[16]. The NK cells float throughout the body. NK cells are recruited to the cancer site when the cell becomes cancerous. It plays a positive role in the prognosis of tumor patients, as has been demonstrated in several cancer syndromes[17]. In addition to killing tumor cells directly, NK cells can also promote the recruitment of cdc1s to TME by producing CCL5, Xcl1/2 and FLT3LG. Cdc1s is a DC subgroup, which is responsible for cross presentation of tumor antigens to CD8+T cells, indicating that NK cells play a key role in enhancing anti-tumor CD8+T cell response[18,19].
The number of Treg in bladder cancer tissue is significantly higher than that in non-cancerous tissue; Fattahi et al[8] proved that conclusion through immunohistochemical.
EMP1 has been reported to be a prooncogene in many cancers, such as bladder cancer, childhood leukemia, non-small cell lung cancer and glioma. Miao et al[20] reported that EMP1 promotes tumor progression by targeting C-MYC. In BLCA, there was a moderate positive correlation between the number of Tregs infiltrating and EMP1 expression and this result was calculated[21,22]. Taha-Mehlitz et al[23] found that ADSL affects mitochondrial function by altering the Tricarboxylic acid cycle and mitochondrial respiratory injury, thus playing its role in promoting the progression of colon cancer. Gao et al[24] showed that the expression of CMTM8 was down regulated in bladder cancer and existed as a tumor suppressor gene. Overexpression of CMTM8 could improve the sensitivity of bladder cancer cell lines to epirubicin. Moreover, the expression of CMTM8 has been reported to be decreased in a variety of tumors, such as liver cancer, colon, lung adenocarcinoma and so on. At the same time, CMTM8 is also associated with EMT (epithelial mesenchymal transformation) and immune infiltration[25,26]. The conclusions drawn from this article contradict our results. DNASE2B may be associated with chemotherapy sensitivity of gastric cancer[27]. Nerve guidance factor 4 (Netrin-4, NTN4) is a ligand of neo1 (a transmembrane receptor). Its expression in gastric cancer, breast cancer and neuroblasts is related to the prognosis of tumor patients, and EMT is closely related[28-30]. Sort1 is involved in the occurrence, development, and drug resistance of a variety of tumors, such as colon cancer, breast cancer and gastric cancer[31,32]. The expression of TCHH was correlated with immune infiltration in Colon cancer and this result is based on TIMER database analysis[33]. Very little research has been done on the relationship between the genes included in the model and the infiltration of immune cells in tumor tissue. In addition, the risk score calculated by this model is positively correlated with PDCD2LG2 (PD-L1) and Cytotoxic T lymphocyte-4, suggesting that the model may be helpful to guide the treatment of immune examination sites.
Taha-Mehlitz et al[24] study showed that the mutation and overexpression of FGFR3 were related to the low immune score of bladder cancer. It has been reported that Treg cells are highly infiltrated into tumors, which may lead to immunosuppression of tumor microenvironment and increase tumor immune escape[9]. Based on our data and previous studies, we have obtained a hypothesis that the increase of FGFR3 mutation will lead to the increase of Treg infiltration (because the mutation frequency of FGFR3 is significantly increased in the high-risk group), which will lead to the immune escape of tumor cells and lead to the poor prognosis of patients. This hypothesis may be a good research idea. The prediction of drug susceptibility may be an unexpected finding of this study.
Based on our data analysis, this model related to Tregs and NK cells can predict the prognosis of BUC, correlate with clinical data, and predict the sensitivity of bladder patients to chemotherapeutic drugs. However, it is not clear whether the changes of these genes in tumor tissue are the cause or result of Tregs and NK cells participating in the formation of TME. Here we merely provide some reference data for future researchers.
We constructed a prognosis-related model for patients with bladder tumors. This model is based on the infiltration of Treg cells and NK cells in tumor tissue. In addition to judging the prognosis of bladder cancer patients, it can also predict how sensitive patients are to chemotherapy and immunotherapy. Furthermore, based on this model, patients were divided into high and low risk groups, and variations in genetic alterations were found between the high and low risk groups.
In summary, Treg cell and NK cell percolation in this model provides new targets for targeted therapy of BUC. This study does have certain restrictions, though. Because there are many types of bladder cancer, yet, other research aspects may be overlooked, due to the pursuit of "accuracy" in the study of differentially expressed genes. In follow-up studies, we would consider looking at other subtypes to determine if there are any changes and investigating other immune-related processes for prognosis-related factors.
The role and function of Tregs and NK cells need to be confirmed using more sophisticated methodologies and approaches as our analysis is based on the TCGA database, which is openly accessible but has been mined.
Prognosis-related models of bladder tumor patients, based on Treg and NK cell percolation in tumor tissue. In addition to judging the prognosis of patients with bladder cancer, it can also predict the sensitivity of patients to chemotherapy and immunotherapy. At the same time, patients were divided into high and low risk groups based on this model, and differences in genetic mutations were found between the high and low risk groups.
Regulatory T cells (Tregs) and natural killer (NK) cells play an essential role in the development of bladder urothelial carcinoma (BUC).
To identify genes associated with bladder cancer prognosis.
To construct a prognosis-related model to judge the prognosis of patients with bladder cancer, meanwhile, predict the sensitivity of patients to chemotherapy and immunotherapy.
In this study, we analyzed publicly available datasets from two databases (https://portal.gdc.cancer.gov/ and https://www.ncbi.nlm.nih.gov/geo/), both with authoritative data, to predict the diagnosis and prognosis of the disease by bioinformatic analysis.
The stage and risk scores are independent prognostic factors in patients with BUC. Mutations in FGFR3 lead to an increase in Tregs percolation and affect the prognosis of the tumor, and additionally, EMP1, TCHH and CNTNAP3B in the model are mainly positively correlated with the expression of immune checkpoints, while CMTM8, SORT1 and IQSEC1 are negatively correlated with immune checkpoints and the high-risk group had higher sensitivity to chemotherapy drugs.
The model can predict the sensitivity of patients to chemotherapy and immunotherapy in addition to their prognosis for bladder cancers.
Prognosis-related models of bladder tumor patients, based on Treg and NK cell percolation in tumor tissue. In addition to judging the prognosis of patients with bladder cancer, it can also predict the sensitivity of patients to chemotherapy and immunotherapy. At the same time, patients were divided into high and low risk groups based on this model, and differences in genetic mutations were found between the high and low risk groups.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cassell III AK, Liberia; Muro M, Spain S-Editor: Liu JH L-Editor: A P-Editor: Cai YX
| 1. | Martinez Rodriguez RH, Buisan Rueda O, Ibarz L. Bladder cancer: Present and future. Med Clin (Barc). 2017;149:449-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 2. | Rouanne M, Bajorin DF, Hannan R, Galsky MD, Williams SB, Necchi A, Sharma P, Powles T. Rationale and Outcomes for Neoadjuvant Immunotherapy in Urothelial Carcinoma of the Bladder. Eur Urol Oncol. 2020;3:728-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 3. | Toor SM, Murshed K, Al-Dhaheri M, Khawar M, Abu Nada M, Elkord E. Immune Checkpoints in Circulating and Tumor-Infiltrating CD4(+) T Cell Subsets in Colorectal Cancer Patients. Front Immunol. 2019;10:2936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 4. | Phé V, Rouprêt M, Cussenot O, Chartier-Kastler E, Gamé X, Compérat E. Forkhead box protein P3 (Foxp3) expression serves as an early chronic inflammation marker of squamous cell differentiation and aggressive pathology of urothelial carcinomas in neurological patients. BJU Int. 2015;115 Suppl 6:28-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Guillerey C. NK Cells in the Tumor Microenvironment. Adv Exp Med Biol. 2020;1273:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 76] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 6. | Shimasaki N, Jain A, Campana D. NK cells for cancer immunotherapy. Nat Rev Drug Discov. 2020;19:200-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 802] [Article Influence: 160.4] [Reference Citation Analysis (0)] |
| 7. | Hakimi AA, Voss MH, Kuo F, Sanchez A, Liu M, Nixon BG, Vuong L, Ostrovnaya I, Chen YB, Reuter V, Riaz N, Cheng Y, Patel P, Marker M, Reising A, Li MO, Chan TA, Motzer RJ. Transcriptomic Profiling of the Tumor Microenvironment Reveals Distinct Subgroups of Clear Cell Renal Cell Cancer: Data from a Randomized Phase III Trial. Cancer Discov. 2019;9:510-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 8. | Fattahi S, Karimi M, Ghatreh-Samani M, Taheri F, Shirzad H, Mohammad Alibeigi F, Anjomshoa M, Bagheri N. Correlation between aryl hydrocarbon receptor and IL-17(+) and Foxp3(+) T-cell infiltration in bladder cancer. Int J Exp Pathol. 2021;102:249-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Hatzioannou A, Boumpas A, Papadopoulou M, Papafragkos I, Varveri A, Alissafi T, Verginis P. Regulatory T Cells in Autoimmunity and Cancer: A Duplicitous Lifestyle. Front Immunol. 2021;12:731947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 10. | San Segundo D, Millán O, Muñoz-Cacho P, Boix F, Paz-Artal E, Talayero P, Morales JM, Muro M, De Cos MÁ, Guirado L, Llorente S, Pascual J, Arias M, Brunet M, López-Hoyos M. High proportion of pretransplantation activated regulatory T cells (CD4+CD25highCD62L+CD45RO+) predicts acute rejection in kidney transplantation: results of a multicenter study. Transplantation. 2014;98:1213-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 11. | Nishikawa H, Koyama S. Mechanisms of regulatory T cell infiltration in tumors: implications for innovative immune precision therapies. J Immunother Cancer. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 177] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 12. | Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, Nagase H, Nishimura J, Yamamoto H, Takiguchi S, Tanoue T, Suda W, Morita H, Hattori M, Honda K, Mori M, Doki Y, Sakaguchi S. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 653] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 13. | Saleh R, Elkord E. FoxP3(+) T regulatory cells in cancer: Prognostic biomarkers and therapeutic targets. Cancer Lett. 2020;490:174-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 221] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 14. | Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, Rautela J, Straube J, Waddell N, Blake SJ, Yan J, Bartholin L, Lee JS, Vivier E, Takeda K, Messaoudene M, Zitvogel L, Teng MWL, Belz GT, Engwerda CR, Huntington ND, Nakamura K, Hölzel M, Smyth MJ. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol. 2017;18:1004-1015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 518] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 15. | Marofi F, Abdul-Rasheed OF, Rahman HS, Budi HS, Jalil AT, Yumashev AV, Hassanzadeh A, Yazdanifar M, Motavalli R, Chartrand MS, Ahmadi M, Cid-Arreguid A, Jarahian M. CAR-NK cell in cancer immunotherapy; A promising frontier. Cancer Sci. 2021;112:3427-3436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 86] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 16. | Guillamón CF, Martínez-Sánchez MV, Gimeno L, Campillo JA, Server-Pastor G, Martínez-García J, Martínez-Escribano J, Torroba A, Ferri B, Abellán DJ, Legaz I, López-Álvarez MR, Moya-Quiles MR, Muro M, Minguela A. Activating KIRs on Educated NK Cells Support Downregulation of CD226 and Inefficient Tumor Immunosurveillance. Cancer Immunol Res. 2019;7:1307-1317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Demaria O, Gauthier L, Debroas G, Vivier E. Natural killer cell engagers in cancer immunotherapy: Next generation of immuno-oncology treatments. Eur J Immunol. 2021;51:1934-1942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 106] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 18. | Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, Nelson AE, Loo K, Kumar R, Rosenblum MD, Alvarado MD, Wolf DM, Bogunovic D, Bhardwaj N, Daud AI, Ha PK, Ryan WR, Pollack JL, Samad B, Asthana S, Chan V, Krummel MF. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med. 2018;24:1178-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 713] [Cited by in RCA: 759] [Article Influence: 108.4] [Reference Citation Analysis (0)] |
| 19. | Böttcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, Rogers NC, Sahai E, Zelenay S, Reis e Sousa C. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172:1022-1037.e14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1219] [Cited by in RCA: 1315] [Article Influence: 187.9] [Reference Citation Analysis (0)] |
| 20. | Miao L, Jiang Z, Wang J, Yang N, Qi Q, Zhou W, Feng Z, Li W, Zhang Q, Huang B, Chen A, Zhang D, Zhao P, Li X. Epithelial membrane protein 1 promotes glioblastoma progression through the PI3K/AKT/mTOR signaling pathway. Oncol Rep. 2019;42:605-614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 21. | Lai S, Wang G, Cao X, Li Z, Hu J, Wang J. EMP-1 promotes tumorigenesis of NSCLC through PI3K/AKT pathway. J Huazhong Univ Sci Technolog Med Sci. 2012;32:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Lin B, Zhang T, Ye X, Yang H. High expression of EMP1 predicts a poor prognosis and correlates with immune infiltrates in bladder urothelial carcinoma. Oncol Lett. 2020;20:2840-2854. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 23. | Taha-Mehlitz S, Bianco G, Coto-Llerena M, Kancherla V, Bantug GR, Gallon J, Ercan C, Panebianco F, Eppenberger-Castori S, von Strauss M, Staubli S, Bolli M, Peterli R, Matter MS, Terracciano LM, von Flüe M, Ng CKY, Soysal SD, Kollmar O, Piscuoglio S. Adenylosuccinate lyase is oncogenic in colorectal cancer by causing mitochondrial dysfunction and independent activation of NRF2 and mTOR-MYC-axis. Theranostics. 2021;11:4011-4029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 24. | Gao D, Hu H, Wang Y, Yu W, Zhou J, Wang X, Wang W, Zhou C, Xu K. CMTM8 inhibits the carcinogenesis and progression of bladder cancer. Oncol Rep. 2015;34:2853-2863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Kang N, Xie X, Zhou X, Wang Y, Chen S, Qi R, Liu T, Jiang H. Identification and validation of EMT-immune-related prognostic biomarkers CDKN2A, CMTM8 and ILK in colon cancer. BMC Gastroenterol. 2022;22:190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 26. | Zhang W, Qi H, Mo X, Sun Q, Li T, Song Q, Xu K, Hu H, Ma D, Wang Y. CMTM8 is Frequently Downregulated in Multiple Solid Tumors. Appl Immunohistochem Mol Morphol. 2017;25:122-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 27. | Ha YJ, Yoon SN, Jeon YJ, Cho DH, Roh SA, Kim BS, Kim HJ, Kim SY, Kim YS, Kim JC. Genome-wide identification of chemosensitive single nucleotide polymorphism markers in gastric cancer. Anticancer Res. 2011;31:4329-4338. [PubMed] |
| 28. | Lv B, Song C, Wu L, Zhang Q, Hou D, Chen P, Yu S, Wang Z, Chu Y, Zhang J, Yang D, Liu J. Netrin-4 as a biomarker promotes cell proliferation and invasion in gastric cancer. Oncotarget. 2015;6:9794-9806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Villanueva AA, Falcón P, Espinoza N, R LS, Milla LA, Hernandez-SanMiguel E, Torres VA, Sanchez-Gomez P, Palma V. The Netrin-4/ Neogenin-1 axis promotes neuroblastoma cell survival and migration. Oncotarget. 2017;8:9767-9782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Xu X, Yan Q, Wang Y, Dong X. NTN4 is associated with breast cancer metastasis via regulation of EMT-related biomarkers. Oncol Rep. 2017;37:449-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 31. | Demeule M, Charfi C, Currie JC, Larocque A, Zgheib A, Kozelko S, Béliveau R, Marsolais C, Annabi B. TH1902, a new docetaxel-peptide conjugate for the treatment of sortilin-positive triple-negative breast cancer. Cancer Sci. 2021;112:4317-4334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 32. | Liang M, Yao W, Shi B, Zhu X, Cai R, Yu Z, Guo W, Wang H, Dong Z, Lin M, Zhou X, Zheng Y. Circular RNA hsa_circ_0110389 promotes gastric cancer progression through upregulating SORT1 via sponging miR-127-5p and miR-136-5p. Cell Death Dis. 2021;12:639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 33. | Shi J, Jiang D, Yang S, Sun Y, Wang J, Zhang X, Liu Y, Lu Y, Yang K. Molecular profile reveals immune-associated markers of lymphatic invasion in human colon adenocarcinoma. Int Immunopharmacol. 2020;83:106402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |