Published online Apr 26, 2023. doi: 10.12998/wjcc.v11.i12.2604
Peer-review started: January 28, 2023
First decision: March 10, 2023
Revised: March 18, 2023
Accepted: March 27, 2023
Article in press: March 27, 2023
Published online: April 26, 2023
Processing time: 87 Days and 11.3 Hours
Osteogenesis imperfecta (OI) is a genetically heterogeneous monogenic disease characterized by decreased bone mass, bone fragility, and recurrent fractures. The phenotypic spectrum varies considerably ranging from prenatal fractures with lethal outcomes to mild forms with few fractures and normal stature. The basic mechanism is a collagen-related defect, not only in synthesis but also in folding, processing, bone mineralization, or osteoblast function. In recent years, great progress has been made in identifying new genes and molecular mechanisms underlying OI. In this context, the classification of OI has been revised several times and different types are used. The Sillence classification, based on clinical and radiological characteristics, is currently used as a grading of clinical severity. Based on the metabolic pathway, the functional classification allows identifying regulatory elements and targeting specific therapeutic approaches. Genetic classification has the advantage of identifying the inheritance pattern, an essential element for genetic counseling and prophylaxis. Although genotype-phenotype correlations may sometimes be challenging, genetic diagnosis allows a personalized management strategy, accurate family planning, and pregnancy manage
Core Tip: Osteogenesis imperfecta (OI) is a genetically heterogeneous systemic collagenous disorder with high phenotypic variability. Recent discoveries of new genes and molecular mechanisms underlying the disease have led to revisions of classical classification. Identifying the causative gene and molecular mechanisms allows a personalized management strategy, accurate family planning, and pregnancy management decisions including options for mode of delivery, or early antenatal OI treatment.
- Citation: Panzaru MC, Florea A, Caba L, Gorduza EV. Classification of osteogenesis imperfecta: Importance for prophylaxis and genetic counseling. World J Clin Cases 2023; 11(12): 2604-2620
- URL: https://www.wjgnet.com/2307-8960/full/v11/i12/2604.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i12.2604
Osteogenesis imperfecta (OI) is a rare monogenic disorder with an incidence estimated at about 1 per 10000 individuals[1]. It is a genetically and clinically heterogeneous disease characterized by decreased bone mass and bone fragility. This generates susceptibility to fractures with minimal or no trauma, vertebral compressions, variable skeletal deformities, and growth deficiency. Bone tissue is characterized by alterations of trabecular architecture, thin cortex, high bone turnover, and hypermineralized matrix. Patients with OI have a broad phenotypic spectrum ranging from prenatal fractures and pre and perinatal lethal outcome to mild forms with few fractures and normal stature. This phenotypic variability is only partially explained by the type of mutation or causative gene. Patients with the same pathogenic variant may present variable degrees of phenotype expression. The basic mechanism is a collagen-related defect, not only in structure or production, but also in folding, posttranslational processing, bone mineralization or osteoblast differentiation[2]. The disorder is a systemic collagen disorder and it also has extra-skeletal manifestations like blue-gray sclera, dentinogenesis imperfecta, conductive or sensory hearing loss, ligamentous laxity, muscle weakness, respiratory impairment, increased fragility of vessels, and cardiac valve abnormalities[1,3-5].
Collagen is a major component of the extracellular matrix, with essential roles in mechanical resistance and regulation of several signaling pathways. Type I collagen is a crucial skin, bone, tendons, lungs, heart, and blood vessels constituent. This collagen is a heterotrimer synthesized in a precursor form, procollagen, containing two proα1(I) and one proα2(I) chains. The procollagen has a rod-like central triple-helical domain with globular extensions at the N- and C- ends. The helical core contains repeating Gly-Xaa-Yaa tripeptides, where X is often proline and Y hydroxyproline. Type I procollagen synthesis is a complex process, involving numerous phases and many proteins necessary for post-translational modifications, folding, transport, and secretion. The proα1(I) and proα2(I) polypeptide chains, encoded by the COL1A1 and COL1A2 genes, are translated in the rough endoplasmic reticulum (ER). The post-translational modifications include 4hydroxylation of most prolines in the Yaa position – essential for triple helical stability. The complex formed by P3H1 – CRTAP – PPIB and the FKBP10 protein have an important role in triple helix formation. Serpin H1 is involved in the stabilization of the triple helix and transport to the Golgi apparatus. The terminal procollagen extensions are cleaved by specific proteases: Disintegrin, metalloproteinase with thrombospondin motifs 2 (ADAMTS2) and bone morphogenetic protein 1 (BMP1). Three specific regions, relevant to the interaction of collagen with other collagen molecules or extracellular matrix proteins, were identified along the α1chain - major ligand-binding regions (MLBRs), which are very important for matrix quality[6-9]. Pathogenic variants in gene-encoding key players in these processes lead to collagen defects and OI phenotype.
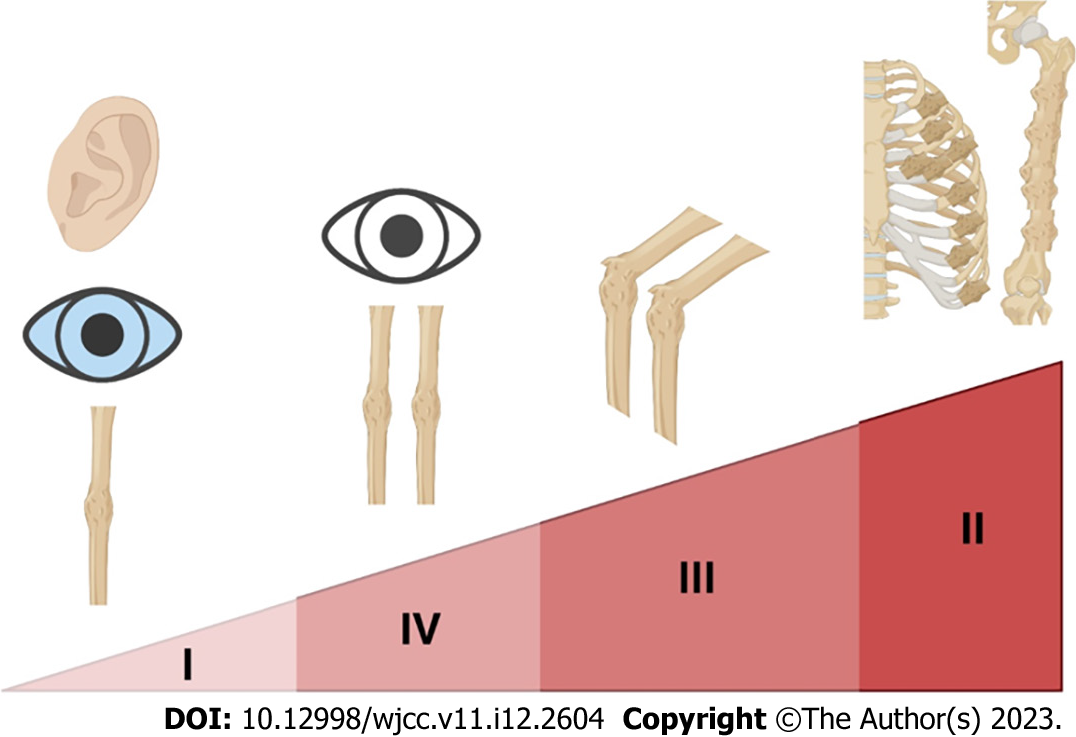
Lately, OI classification has been the subject of extensive debates and has been revised several times. In 1979, Sillence proposed a classification based on clinical/radiological characteristics and mode of inheritance: OI type I - autosomal dominant (AD) with blue sclerae, OI type II - perinatal lethal form with radiographically broad, crumpled femora and beaded ribs, OI type III - progressively deforming, and OI type IV – dominant with normal sclerae[10] (Figure 1). This classification included only patients with defects in the primary structure of collagen. The discovery of pathogenic variants in new genes with clinical overlap with previous types has caused many debates. In 2014, Van Dijk and Sillence suggested the addition of OI type V – a form with calcification in the intraosseous membranes[11]. The revised nosology and classification of genetic skeletal disorders recognizes these five clinical types (but Arabic numerals are used), with subclasses based on inheritance patterns and genes involved[12]. Some types (e.g. 3 or 4) have many genes and different inheritance (dominant/ recessive) resulting in challenges for genetic counseling. An alternative functional classification based on the metabolic mechanism was also proposed: Group A - defects in collagen synthesis, structure, or processing, group B - defects in collagen modification, group C - collagen folding and cross-linking defects, group D - compromised bone mineralization, and group E - defects in osteoblast development with collagen insufficiency[8,13]. The Online Mendelian Inheritance in Man database uses a mixed-genetic classification, with types I–IV according to the Sillence classification and pathogenic variants in COL1A1 or COL1A2, and the new gene-classified type[1] (Table 1). The advantages of genetic classification are the identification of the inheritance pattern for counseling, prophylaxis, and the possibility of grouping for etiology-based therapies research.
| OI type | OMIM | Gene symbol | Approved gene name | Location | Protein name | Functional group |
| I | 166200 | COL1A1 | Collagen type I alpha 1 chain | 17q21.33 | Collagen alpha-1(I) chain | A |
| II | 166210 | COL1A1 | Collagen type I alpha 1 chain | 17q21.33 | Collagen alpha-1(I) chain | A |
| COL1A2 | Collagen type I alpha 2 chain | 7q21.3 | Collagen alpha-2(I) chain | |||
| III | 259420 | COL1A1 | Collagen type I alpha 1 chain | 17q21.33 | Collagen alpha-1(I) chain | A |
| COL1A2 | Collagen type I alpha 2 chain | 7q21.3 | Collagen alpha-2(I) chain | |||
| IV | 166220 | COL1A1 | Collagen type I alpha 1 chain | 17q21.33 | Collagen alpha-1(I) chain | A |
| COL1A2 | Collagen type I alpha 2 chain | 7q21.3 | Collagen alpha-2(I) chain | |||
| V | 610967 | IFITM5 | Interferon induced transmembrane protein 5 | 11p15.5 | Interferon-induced transmembrane protein 5 | D |
| VI | 613982 | SERPINF1 | Serpin family F member 1 | 17p13.3 | Pigment epithelium-derived factor | D |
| VII | 610682 | CRTAP | Cartilage associated protein | 3p22.3 | Cartilage-associated protein | B |
| VIII | 610915 | P3H1 | Prolyl 3-hydroxylase 1 | 1p34.2 | Prolyl 3-hydroxylase 1 | B |
| IX | 259440 | PPIB | Peptidylprolyl isomerase B | 15q22.31 | Peptidyl-prolyl cis-trans isomerase B | B |
| X | 613848 | SERPINH1 | Serpin family H member 1 | 11q13.5 | Serpin H1 | C |
| XI | 610968 | FKBP10 | FKBP prolyl isomerase 10 | 17q21.2 | Peptidyl-prolyl cis-trans isomerase FKBP10 | C |
| XII | 613849 | SP7 | Sp7 transcription factor | 12q13.13 | Transcription factor Sp7 | E |
| XIII | 614856 | BMP1 | Bone morphogenetic protein 1 | 8p21.3 | Bone morphogenetic protein 1 | A |
| XIV | 615066 | TMEM38B | Transmembrane protein 38B | 9q31.2 | Trimeric intracellular cation channel type B | B |
| XV | 615220 | WNT1 | Wnt family member 1 | 12q13.12 | Proto-oncogene Wnt-1 | E |
| XVI | 616229 | CREB3L1 | cAMP responsive element binding protein 3 like 1 | 11p11.2 | Cyclic AMP-responsive element-binding protein 3-like protein 1 | E |
| XVII | 616507 | SPARC | Secreted protein acidic and cysteine rich | 5q33.1 | SPARC | E |
| XVIII | 617952 | TENT5A | Terminal nucleotidyltransferase 5A | 6q14.1 | Terminal nucleotidyltransferase 5A | Unclassified |
| XIX | 301014 | MBTPS2 | Membrane bound transcription factor peptidase, site 2 | Xp22.12 | Membrane-bound transcription factor site-2 protease | E |
| XX | 618644 | MESD | Mesoderm development LRP chaperone | 15q25.1 | LRP chaperone MESD | Unclassified |
| XXI | 619131 | KDELR2 | KDEL ER protein retention receptor 2 | 7p22.1 | ER lumen protein-retaining receptor 2 | C |
| XXII | 619795 | CCDC134 | Coiled-coil domain containing 134 | 22q13.2 | Coiled-coil domain-containing protein 134 | Unclassified |
Over 85% of OI cases are associated with pathogenic variants in COL1A1 and COL1A2 genes, which lead to quantitative or qualitative alterations of collagen. These mutations generate OI types I - IV. Pathogenic variants (mostly nonsense mutations) lead to haploinsufficiency and reduce the amount of normal collagen, thus generating a milder phenotype. In contrast, pathogenic variants leading to structural collagen defects cause a more severe phenotype. The most common mutations are single-nucleotide variants resulting in the substitution of a glycine residue. Substitutions in gene regions that encode branched-chain or charged amino acids interfere with triple helix folding and are associated with severe clinical consequences. Substitutions on the α1(I) chain have a more severe/Lethal outcome than those in α2(I). The nature of the substituting amino acid, the chain in which it is located, and its position along the chain influence the phenotype. Pathogenic variants in the 3' or 5' splice sites that produce exon skipping do not affect the Gly-Xaa-Yaa triplet pattern but may cause local looping out of chains[1,2]. Pathogenic variants in the C-terminal propeptide, which is cleaved from the mature collagen, impair chain association or delay the incorporation into the collagen trimer[14]. Deletions or duplications of the codons for one or two Gly-Xaa-Yaa triplets shift the chain alignment without interrupting the sequence and produce severe or lethal phenotypes[15]. Due to the direction of the zipper-like folding of the chains, pathogenic variants in the N-terminal region have minimal consequences, whereas those in the C-terminal region cause moderate to lethal outcomes. Substitutions in MLBR3 regions impair extracellular matrix organization and usually have lethal consequences. In the α2(I) chain, severe pathogenic variants are gathered in clusters that are correlated with the proteoglycan binding site on collagen fibrils[16-19] .
Interferon-induced transmembrane protein 5 (IFITM5), also known as bone-restricted IFITM-like, is a short transmembrane protein expressed specifically in osteoblasts and attached to the cell membrane by palmitoylation of cysteines 50 and 5, with a regulatory role in mineralization. IFITM5 plays a crucial role in the regulation of SERPINF1 expression and the resultant production of the protein pigment epithelium-derived factor (PEDF). Different pathogenic variants in IFITM5 gene lead to a distinct OI phenotype, named OI V. A heterozygous gain of (new) function variant in the 5' untranslated region (c.-14C>T) is associated with a moderate type of OI with distinctive radiographic findings. This pathogenic variant creates a new start codon, resulting in the elongation of the cytoplasmic N- terminus of IFITM5 protein by five amino acids and inducing increased bone formation[2,20,21]. The characteristic radiographic findings include hyperplastic callus formation, calcification of the interosseous membrane of the forearm, and hyperdense metaphyseal band. Some patients may present radial head dislocation. Histologic examination of bone under polarized light reveals a “mesh-like pattern” of irregularly arranged lamellar deposition[22]. Heterozygous missense variants lead to substitution of the serine at position 40 (c.119C>T and c.119C>G), impairment of the palmitoylation process, and are associated with a more severe phenotype. Patients present prenatal fractures or shortening/ bowing of long bones, a severe deforming course, and a fish-scale lamellar pattern at the bone examination under polarized light. Patients do not show radial head dislocation or signs from the radiographic triad. Lim et al[23] reported a case of gonadal mosaicism in the unaffected mother[24]. Bisphosphonates (BPs) are more effective in patients with c.119C>T variant than in cases with other variants[25].
PEDF, encoded by SERPINF1 gene, is a ubiquitously expressed protein, with anti-angiogenic, anti-tumorigenic, and anti-metastatic properties. The binding of PEDF to type I collagen is essential for anti-angiogenic properties. PEDF induces the expression of osteoprotegerin which interacts with the receptor activator of nuclear factor-κβ ligand (RANKL) pathway and regulates the activity of osteoclasts. Kang et al[26] suggest that antagonism between PEDF and TGF-β pathways controls osteogenesis and bone vascularization[8,27]. Patients with biallelic pathogenic variants in SERPINF1 have postnatal fractures, progressive skeletal deformity, vertebral compressions, and a fish-scale pattern at bone examination under polarized light (similar to patients with loss of function mutation in IFITM5). These mutations produce OI type VI. The RANKL-antibody is a potential therapeutic agent for this form of OI[1,4,28].
Cartilage-associated protein (CRTAP) forms a complex with prolyl-3-hydroxylase 1 (P3H1) and peptidyl-prolyl-cis-transisomerase B (PPIB). This complex is involved in 3-hydroxylation of specific proline residues. Chang et al[29] showed that CRTAP and P3H1 are mutually stabilized in the collagen prolyl 3-hydroxylation complex. Biallelic pathogenic variants in CRTAP or P3H1 genes lead to a marked decrease in proline hydroxylation and subsequently to a delay in collagen folding and are associated with a severe/lethal form of OI. Patients have neonatal fractures, rhizomelia, and undertubulation of the long bones[4,13,30-31]. Biallelic pathogenic variants in PPIB gene are associated with a low percentage of 3-hydroxylated proline at position 986, but higher than in CRTAP or P3H1 genes mutations. The phenotype overlaps with that caused by CRTAP or P3H1 genes mutation, with the exception of rhizomelia[32,33]. CRTAP genes mutations determine OI type VII, P3H1 genes mutations determine OI type VIII, while PPIB genes mutations determine OI type IX.
Serpin H1 is a collagen-specific chaperone, that localizes in the ER and is encoded by SERPINH1 gene. Serpin H1 binds to arginine-rich sequences of triple helical collagen and stabilizes it by preventing unfolding and aggregate formation. Also, serpin H1 participates in the shuttling of correctly folded collagen into the Golgi apparatus. Homozygous or compound heterozygous pathogenic variants in SERPINH1 gene lead to misfolding, intracellular aggregation, and delayed collagen secretion and are associated with a moderate to severe form of OI – OI type X[34-36].
Serpin H1 interacts with peptidyl-prolyl cis-trans isomerase FKBP10, another collagen chaperone, which provides mutual stability and allows for a synergistic effect during collagen folding. FKBP10 is also required for the activity of lysyl hydroxylase 2 (LH2, encoded by PLOD2 gene). Hydroxylation of the collagen telopeptide lysyl residues is essential in cross-linking. Recessive pathogenic variants in FKBP10 gene are associated with a wide clinical spectrum which includes a progressive deforming form of OI (OI type XI), Bruck syndrome, and Kuskokwim syndrome. Bruck syndrome is characterized by congenital contractures with pterygia, early onset of fractures, short stature, limb deformity, and progressive scoliosis. Bruck syndrome 2 presents a similar phenotype, but it is generated by a biallelic pathogenic variant in PLOD2 gene. Kuskokwim syndrome is a congenital contracture disorder with mild skeletal anomalies, occurring in Yup’ik Eskimos in Alaska. Bisphosphonate therapy reduces the fracture rate and pain but has no effect on joint abnormalities[4,37-39].
SP7 gene encodes transcription factor SP7, a key regulator of osteoblast differentiation and subsequently of osteocyte formation. Recessive pathogenic variants in SP7 are associated with increased bone porosity, recurrent fractures, skeletal deformities, delayed teeth eruption and hearing loss. This particular phenotype is characteristic to OI type XII. Recently, heterozygous (dominant) missense variants affecting a highly conserved zinc finger domain have been reported in cases with bone fragility, high bone turnover, and patchy sclerosis[40-43].
The major function of bone morphogenetic protein 1 (BMP1) involves procollagen I C-terminal propeptide processing, crucial for fibril formation. Other roles are activation of lysyl oxidases (involved in cross-linking), processing of small leucine-rich proteoglycans (e.g. decorin - important for collagen fibrillogenesis) and dentin matrix protein 1 involved in bone mineralization, and activation of TGFβ1, a key signaling molecule for bone remodeling. Patients with homozygous or compound heterozygous pathogenic variants in BMP1 gene present a variable phenotype ranging from mild to severe progressive deforming OI (OI type XIII). Individuals with BMP1 pathogenic variants with residual activity of the protein have a milder phenotype than cases with null pathogenic variants. In the majority of cases, increased bone mineral density (BMD) has been noticed. In this context, the indication, dose, and duration of antiresorptive therapy are questionable due to concerns about increasing bone stiffness leading to fracture[2,44-47].
TMEM38B gene encodes TRimeric Intracellular Cation channel type B (TRIC-B), specific for potassium, also known as transmembrane protein 38 (TMEM 38B). TMEM 38B is involved in the opening of cation channels, after calcium release, and thus the ER membrane potential is maintained. Disruption of intracellular calcium kinetics affects the activity of many proteins required for type I collagen synthesis and folding[48,49]. Webb et al[50] revealed characteristics of bone modifications in patients with biallelic TMEM38B pathogenic variants: Decreased hydroxylation of collagen helical lysine residues and intracellular retention and degradation of misfolded collagen. The phenotypic spectrum varies considerably ranging from mild scoliosis to severe cases with prenatal bowed femur, early-onset multiple fractures, and growth retardation (OI type XIV). The variability in phenotype severity indicates that other factors may be involved, including genetic modifiers, different genetic backgrounds, and other genes involved in intracellular calcium dynamics. Patients responded well to BPs, but, in some cases, cardiovascular abnormalities and hypotonia were reported (possibly related to abnormal ER calcium kinetics)[51-54].
Proto-oncogene Wnt-1 (the wingless-type mouse mammary tumor virus integration site family, member 1) is a glycoprotein with a key role in bone development. WNT1 interacts with the cell surface lipoprotein receptor-related protein 5 (LRP5) and Frizzled receptor leading to the translocation and accumulation of beta-catenin into the cell nucleus and subsequent transcriptional regulation of target genes. WNT/β-catenin signaling is involved in osteoblast progenitor proliferation, osteoblast differentiation, and regulation of osteoclastogenesis in mature osteoblasts and osteocytes through the secretion of osteoprotegerin[55-57]. The WNT/β-catenin pathway is an activator of BMP2 gene transcription, a member of the TGF-β gene superfamily essential for osteoblast differentiation and osteogenesis[57]. Biallelic pathogenic variants in WNT1 gene lead to moderate-severe OI, whereas heterozygous WNT1 gene mutations are reported in patients with ‘early-onset osteoporosis’ indicating a gene dose effect. Neurologic features such as hypotonia, ptosis, developmental delays, and brain anomalies have been reported in some patients with OI type XV[57-60].
Cyclic AMP-responsive element-binding protein 3-like protein 1 (CREB3L1), also known as old astrocyte specifically induced substance (OASIS), a leucine zipper transcription factor is encoded by CREB3L1 gene. In osteoblasts, CREB3L1 is activated by regulated intramembrane proteolysis (RIP) and induces the transcription of COL1A1 gene by binding to the UPR element-like sequence in its promoter. Also, CREB3L1 plays an important role in the expression of coat protein complex II component SEC24D, involved in procollagen export from ER[45,61,62]. Phenotype severity varies considerably ranging from forms with prenatal fractures, severe demineralization, and early lethal outcome to cases with severe bone deformities and survival to adulthood (OI type XVI). Severe phenotypes are caused by homozygous whole gene deletions or in frame deletions in a highly conserved DNA binding domain (loss of function). Siblings with heterozygous pathogenic variants are mildly affected: fractures with minimal trauma, blue sclerae, and osteopenia[61,63]. Truncating homozygous pathogenic variants (outside the highly conserved DNA binding domain) led to a non-lethal phenotype, while heterozygotes are unaffected[64,65].
Secreted protein acidic and rich in cysteine (SPARC), also known as osteonectin or basement membrane protein 40, has a collagen-binding domain and a hydroxyapatite (necessary for mineralization of the collagenous matrix) binding site. During bone development, SPARC is secreted by osteoblasts and has important roles in procollagen processing and assembly in the bone matrix, cross linking, and mineralization[66,67]. Pathogenic variants (substitution, nonsense or splice site) that affect the collagen-binding domain have been reported in patients with OI. Common features of this type of OI (OI type XVII) are multiple postnatal fractures, scoliosis, delayed motor development, neuromuscular weakness (especially of the lower extremities), and brain MRI abnormalities[68-70].
Terminal nucleotidyltransferase 5A (TENT5A) belongs to the nucleotidyltransferase fold superfamily proteins and acts as a noncanonical poly(A) polymerase. TENT5A forms a complex with SMAD proteins and induces transcription of BMP target genes. TENT5A is involved in embryonic development, adult bone formation, and hemin-induced hemoglobinization[2,71,72]. Doyard et al[73] reported biallelic pathogenic variants (nonsense or missense) in TENT5A genes in patients with a severe form of OI (OI type XVIII) with multiple fractures, severe bowing of lower limbs, joint hyperlaxity, vertebral collapses, and blue sclerae. Lin et al[71] considered TENT5A a molecular modulator and a future therapeutic target.
An X-linked form of OI (OI type XIX) is caused by a mutation in MBTPS2, a gene that encodes the membrane-bound transcription factor protease, site-2 (MBTPS2), also known as site-2 protease, a component of the RIP pathway. ER stress - due to retention of unfolded proteins - leads to translocation of CREB3L1, sterol regulatory element-binding protein, and activating transcription factor 6, from the ER membrane to the Golgi membrane, where they are cleaved by endoproteases MBTPS1 and MBTPS2. The resulting fragments regulate the production of collagen and matrix components in the nucleus[1,2,74]. Substitutions that affect a highly conserved MBTPS2 motif, essential for zinc ion coordinating site, are associated with reduced hydroxylation of lysine 87 in both α collagen chains and altered collagen cross-linking. Patients have a form of moderate-to-severe OI with prenatal fractures, generalized osteopenia, long bone bowing, short stature, pectus deformity, and scoliosis, but no dermatological features[75]. Missense pathogenic variants elsewhere in MBTPS2 have been associated with dermatological conditions - IFAP (ichthyosis follicularis, atrichia, and photophobia) syndrome with or without BRESHECK (Brain anomalies, severe mental Retardation, Ectodermal dysplasia, Skeletal deformities, Ear/eye and Kidney dysplasia/hypoplasia) syndrome, Olmsted syndrome (mutilating palmoplantar keratoderma with periorificial keratotic plaques), and keratosis follicularis spinulosa decalvans[76-78].
MESD gene (mesoderm development) encodes an ER chaperone for the WNT signaling receptors LRP5 and LRP6. Moosa et al[79] reported patients with biallelic pathogenic variants (frameshift predicted to result in a premature termination codon or substitution that removes a highly conserved domain) in exon 3 of MESD gene, with partial loss of function, and a progressively deforming type of OI with oligodontia and developmental delay. BPs were not effective in these patients, but antisclerostin antibodies that affect Wnt signaling could be a valid therapeutic option. Two infant deaths due to respiratory insufficiency were reported. Stürznickel et al[80] reported three stillbirths with multiple intrauterine fractures and compound heterozygous frameshift pathogenic variants in exon 2 and exon 3 of MESD gene. They blamed the lethal phenotype on a complete loss of function mutation, located within the chaperone domain of MESD (exon 2)[80].
KDELR2 gene encodes ER lumen protein-retaining receptor 2, involved in protein with a KDEL-like peptide traffic from the Golgi to the ER. The protein binds heat shock protein 47 (HSP47), with an essential role in the intracellular processing of procollagen. Biallelic pathogenic variants in KDELR2 gene are associated with abnormal collagen fibril formation due to the failure of HSP47 to dissociate from collagen type 1. Patients present a progressively deforming type of OI. Efthymiou et al[81] reported neurodevelopmental disorders (motor and speech delay) in three cases[82].
CCDC134 gene encodes a secreted coiled-coil domain-containing protein, involved in the regulation of MAPK pathway, especially phosphorylation of the extracellular signal-related kinase (Erk) or c-JUN N-terminal kinase (JUNK). Erk1/2 and JUNK have an important role in bone morphogenesis, by regulating osteoblast extracellular matrix protein deposition in response to stress. Loss of function pathogenic variants in CCDC134 gene were associated with reduced COL1A1 and SPP1 (ostepontin) mRNA expression and mineralization in osteoblasts. Dubail et al[83] reported patients with homozygous pathogenic variants in CCDC134 gene and a severe form of OI with intrauterine growth retardation, multiple pre and postnatal fractures, short stature, low mineral density, and no response to BPs[84,85].
The identification of the inheritance pattern is essential for genetic counseling and management. Up to 90% of OI patients have an AD pathogenic variant in COL1A1, COL1A2, or IFITM5 genes, with a 50% risk to transmit this variant to their offspring[86]. Nearly half of the OI cases are caused by de novo pathogenic variants[87]. Although advanced paternal age is associated with a high risk of de novo pathogenic variants in monogenic disorders, Mei et al[88] reported a significantly younger paternal and maternal age at conception in OI patients with a de novo mutation. Pyott et al[89] reported a 16% rate of parental mosaicism in couples with a child affected by lethal AD OI. In couples with two or more children with lethal AD OI, the recurrence rate of this mosaicism was 27%[89]. The rate of parental mosaicism is estimated at approximately 5–8% in all OI cases[86]. Persons with mosaicism for AD pathogenic variants are often asymptomatic or have subtle clinical findings depending on the percentage of cells that carry the pathogenic variant[88].
About 10% of OI patients have pathogenic variants with autosomal recessive (AR) inheritance. The distribution of these AR variants is different across populations due to consanguinity or founder effect. In a genetically isolated Dutch group, the carrier frequency of a CRTAP frameshift variant was 4.1% while in the general Dutch population is < 0.2%[90]. Founder pathogenic variants in other genes associated with recessive OI have been reported: P3H1 in West African, United States African American populations and ethnic Kinhs, TMEM38B in Palestinians, and Israeli Arab Bedouins, FKBP10 in indigenous southern Africans, Palestinians, Bavarians, and Samoan islanders, SEC24D in southwestern Germans, WNT1 in the Finnish Hmong group, and PPIB in Chinese people[33,91-95]. A few cases with X-linked pathogenic variants in MBTPS2 or PLS3 have also been reported[75,96].
Genetic testing is essential for the identification of pathogenic variants, inheritance pattern, and differential diagnosis. Based on the clinical and radiographic features, and family history either the sequencing of COL1A1 and COL1A2 or a comprehensive next-generation sequencing panel (all OI genes and genes associated with skeletal dysplasia) is initially recommended. The interpretation of genetic testing results can sometimes be challenging: Identifying unknown significance variants, or sequence variants in a new gene (not previously reported in OI cases). Genotype-phenotype correlations are sometimes difficult to establish, due to the wide OI phenotypical variability, in association with genetic or epigenetic modifiers[97]. Some OI lethality/ severity prediction algorithms were established with variable accuracy[98].
Preconception carrier screening is recommended in healthy couples in different circumstances: positive OI family history, consanguineous marriage, or members of founder populations. Carrier screening cannot detect parental germline mosaicism or predict the possibility of de novo pathogenic variants. A couple with a significant recurrence risk - affected parent(s), carriers of AR pathogenic variants - has many reproductive options: In vitro fertilization (IVF) with gamete or embryo donation, IVF with own cells, and preimplantation genetic diagnosis (PGD), adoption, or natural pregnancy with prenatal diagnosis. Gamete donation is usually recommended in couples with infertility or affected women, because repeated superovulation procedures are associated with an increased risk of osteoporosis and cardiovascular problems[99]. PGD has the advantage of also detecting aneuploidies, but the accuracy rate is 95 to 99.5%, so there is a small risk of false negative results. Moreover, the success rate for artificial reproductive techniques is below 30%. Prenatal testing should be recom
Prenatal genetic testing includes non-invasive prenatal testing (NIPT), and invasive techniques. NIPT has the advantages of early testing (first trimester), less invasive procedure (circulating cell-free fetal DNA extracted from maternal blood), and no associated miscarriage risk. The disadvantages of NIPT are the risk of false-positive, false-negative, or inconclusive results due to confined placental mosaicism (the trophoblastic origin of cell-free fetal DNA is associated with a much higher mutation rate than other fetus cells), or vanishing twin syndrome. NIPT is technically challenging for X-linked and for AR forms when both parents are carriers of the same pathogenic variant due to the presence of the relevant variant from maternal cells in the circulating cell-free DNA. Moreover, NIPT does not cover all the genes involved in OI pathogeny. NIPT results should be confirmed by invasive prenatal testing[102-106].
Invasive prenatal testing uses fetal cells extracted by chorionic villus sampling (CVS), amniocentesis, or cordocentesis, and is associated with an increased risk of pregnancy complications, including fetal loss. CVS has an associated risk of false results due to confined placental mosaicism but allows biochemical type I collagen analysis in extracted cells. Amniocentesis avoids misdiagnosis due to placental mosaicism or twin pregnancy but is performed in the second trimester, after 15 wk of pregnancy, and thus means a long distressful waiting period for the couple. Prenatal diagnosis allows pregnancy management decisions, including the alternative to terminate pregnancy or options for mode of delivery, early OI treatment, before (mesenchymal stem cell transplantation), or after birth[86,107,108].
Severe and lethal forms of OI could be detected by ultrasound screening in the second trimester. Abnormal ultrasound findings suggestive of severe OI include long bone shortening (especially femur length), bowing, and multiple fractures. Moreover, lethal forms have severe demineralization with a thin, easily compressible calvarium, and no posterior acoustic shadowing from long bones[107,109]. Femur length-to-abdominal circumference ratio < 0.16, fetal lung volume below the fifth percentile for gestational age (measured by ultrasound or MRI), and polyhydramnios were associated with lethal outcome. Ultrasound findings do not allow an accurate differential diagnosis with other skeletal dysplasias[107,110,111]. Three-dimensional helical computed tomography provides more accurate data about skeletal anomalies but there are concerns regarding the safety of radiation exposure (even to low doses)[112].
In the past, cesarean delivery was considered safer and more useful for the prevention of fractures at birth than vaginal delivery. Recent studies on babies with types I, III and IV of OI showed that the delivery mode does not influence the rate of fractures at birth. Also, the breech presentation seems to be more frequent in OI type III. Bellur et al suggested that cesarean delivery should be performed only for usual maternal or fetal indications, not for fracture prevention in OI. Pregnant women affected by OI require close monitoring to detect possible complications such as cardiorespiratory problems, bone loss, cephalopelvic disproportion, uterine and placenta rupture, and excessive bleeding at delivery[107,113-115].
The OI treatment includes physical therapy, medication, and surgical procedures. The major goals are the prevention of fractures, and deformities, maximizing the patient's functional ability, and reducing pain. Fracture healing might be delayed in cases with pathogenic WNT1 gene variants, or might be altered by hyperplastic callus formation in patients with pathogenic IFITM5 gene variants. Surgical procedures are used for complex fractures or when correction of deformities is necessary. Intramedullary telescopic rods are used during growth because these have the ability to lengthen. OI patients have anesthetic risks due to abnormal shape or airway, impaired lung function, or the possibility of cervical spine fracture during intubation[4,116]. The rate of fractures decreases in adulthood but the risk of joint osteoarthritis increases[13]. Physiotherapy is essential to improve mobility, due to hypotonia and ligament laxity. Obstructive pulmonary disease (type I collagen is present in lung parenchyma) and scoliosis lead to respiratory complications, a major cause of mortality in OI[117]. Cardiovascular complications include aortic root dilatation left valvular regurgitation, and aortic root dilatation with dissection risk[118]. A multi-disciplinary approach is recommended to address problems related to bone fragility, and also extra-skeletal manifestations.
BPs are currently the most commonly used pharmacological agents in the treatment of pediatric OI. BPs bind to the hydroxyapatite crystals, promote osteoclasts apoptosis, and decrease bone resorption and remodeling. BPs also interact with osteocytes and interfere with osteoblast recruitment on eroded surfaces[119]. Intravenous infusion is superior to oral administration in improving BMD and decreasing fracture rate. Studies reported that maximum benefits are obtained after 3 years of treatment, but there is no difference in adult fracture rates[120-123]. Long-term treatment is associated with microcrack accumulation and increased potential of progression into fractures, loss of microstructural integrity, and reduced mechanical strength[124]. Another disadvantage of BPs is their long half-life; BPs persist in the bone for years after drug discontinuation. Green et al[125] reported decreased birth weight and transient neonatal electrolyte abnormalities (hypocalcemia, hypercalcemia, hyperphosphatemia) associated with maternal use of BPs before or during pregnancy. Whether BPs should be used for a long time at similar or lower doses is debatable. Also, BPs do not have the same efficiency in all types of OI[13,126].
Denosumab is a monoclonal antibody that targets RANKL and inhibits osteoclast activity without binding to the bone. The mechanism of action is similar to BPs, antiresorptive. Denosumab has a shorter half-life (months) and showed promising results in increasing BMD in a few studies. Further studies are necessary to assess the efficiency of fracture prevention[126,127].
Osteo-anabolic therapies stimulate osteoblast activity and bone formation, instead of inhibiting osteoclast function as antiresorptive. Growth hormone (GH) has been used to stimulate long bone growth in GH deficiency, but GH therapy showed only limited benefits in increasing bone mass density compared to BPs (mostly in OI type IV). GH has been less efficient in the more severe forms of OI (type III)[128,129]. Teriparatide, a recombinant parathyroid hormone, leads to a significant increase in BMD in adults with type I OI but seems less effective in patients with types III and IV. Teriparatide has not been used for more than 24 mo and its use in children is contraindicated due to the concern of increased risk of osteosarcoma reported by animal studies[116,129-131]. Lately, the US FDA removed the warning because the risk was only confined to animal studies.
Sclerostin-inhibitory antibodies, romosozumab, and setrusumab, neutralize sclerostin, a negative regulator of Wnt signaling in osteoblasts. Studies revealed good responses of BMD and bone turnover markers to sclerostin-inhibitory antibody treatment in adults with OI[132,133]. Lv et al[134] revealed that romosozumab might increase the risk of cardiovascular adverse events in the elderly.
Animal studies have shown that TGF-β signaling is an essential element of pathogenesis, and blocking TGF-β improves bone mass and biomechanical properties, so anti-TGF-antibodies could represent a valuable therapeutic option. Song et al[135] reported an increase of BMD in children with type IV OI treated with fresolimumab (an anti-TGF-antibody), but no effect in type III and VIII OI. Losartan, an angiotensin II receptor blocker may also have anti-TGF properties[135]. Losartan increased bone mass and accelerated chondrocyte hypertrophy in the growth plate in an animal study[136].
Stem cell therapy is a promising pre and postnatal option based on the cells’ ability to differentiate into osteoblasts that produce normal collagen. Transplantation of bone marrow from HLA-matched siblings and prenatal and postnatal transplantation of mesenchymal stem cells have been associated with improved growth and reduction of fractures rate. In the first group (bone marrow from HLA-matched siblings), the effect was transient, the growth rate slowed over time and a second transplantation with bone marrow/mesenchymal stem cells has been used. There is limited experience in this area, so further trials are necessary[137,138]. The application of cellular reprogramming to create induced pluripotent stem cells (iPSCs) opens a new therapeutic approach.
Advances in gene editing technology bring the possibility of correcting the pathogenic variant. A recent approach involves the silencing of a dominant (gain of function) pathogenic variant, leading to allele suppression and converting the severe forms into a milder phenotype. Different strategies have been used: Antisense oligonucleotides, short interfering RNA, and hammerhead ribozymes. Another approach, gene addition therapy, involves the correction of the expression of deficient or absent alleles in affected cells. In cases where an abnormal collagen chain is produced and affects triple helix assembly, this method will not influence the phenotype. The efficiency of gene editing is still debatable, there are no data about the duration of the positive effects, and concerns regarding off-target effects, risks of an immune response, and genotoxicity are raised. Clinical trials are needed[126,139].
The combination of the CRISPR–Cas9 gene editing tool with induced pluripotent stem cells may improve therapeutic options. Jung et al[140] demonstrated the restoration of type I collagen expression in iPSCs in an OI patient corrected by the CRISPR–Cas9 system.
A new promising therapy is the chemical chaperone 4-phenylbutyrate (4-PBA), involved in protein folding and aggregation in ER. 4-PBA also has histone deacetylase inhibitor activity. Experimental studies reported the reduction of fracture rate and improvement of growth deficiency in animal OI models after 4-BPA treatment[141,142].
In recent decades, great progress has been made in identifying genes and molecular mechanisms underlying OI. These advances demonstrate that OI is an extremely heterogeneous collagen-related disease. The classical clinical Sillence classification is now partially revolute, and the involvement of different causative genes and the presence of different inheritance patterns generate challenges for genetic counseling. However, genetic classification allows an accurate identification of the inheritance for family planning, and offers the possibility of the development of genotype-based therapeutic approaches.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: Romania
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Muthu S, India; Stogov MV, Russia S-Editor: Liu XF L-Editor: A P-Editor: Yu HG
| 1. | Marini JC, Forlino A, Bächinger HP, Bishop NJ, Byers PH, Paepe A, Fassier F, Fratzl-Zelman N, Kozloff KM, Krakow D, Montpetit K, Semler O. Osteogenesis imperfecta. Nat Rev Dis Primers. 2017;3:17052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 370] [Cited by in RCA: 497] [Article Influence: 62.1] [Reference Citation Analysis (0)] |
| 2. | Jovanovic M, Guterman-Ram G, Marini JC. Osteogenesis Imperfecta: Mechanisms and Signaling Pathways Connecting Classical and Rare OI Types. Endocr Rev. 2022;43:61-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 95] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 3. | Chretien A, Couchot M, Mabilleau G, Behets C. Biomechanical, Microstructural and Material Properties of Tendon and Bone in the Young Oim Mice Model of Osteogenesis Imperfecta. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 4. | Etich J, Leßmeier L, Rehberg M, Sill H, Zaucke F, Netzer C, Semler O. Osteogenesis imperfecta-pathophysiology and therapeutic options. Mol Cell Pediatr. 2020;7:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (1)] |
| 5. | Marom R, Rabenhorst BM, Morello R. Osteogenesis imperfecta: an update on clinical features and therapies. Eur J Endocrinol. 2020;183:R95-R106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 153] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 6. | Amirrah IN, Lokanathan Y, Zulkiflee I, Wee MFMR, Motta A, Fauzi MB. A Comprehensive Review on Collagen Type I Development of Biomaterials for Tissue Engineering: From Biosynthesis to Bioscaffold. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 132] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 7. | Ishikawa Y, Taga Y, Zientek K, Mizuno N, Salo AM, Semenova O, Tufa SF, Keene DR, Holden P, Mizuno K, Gould DB, Myllyharju J, Bächinger HP. Type I and type V procollagen triple helix uses different subsets of the molecular ensemble for lysine posttranslational modifications in the rER. J Biol Chem. 2021;296:100453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Marini JC, Reich A, Smith SM. Osteogenesis imperfecta due to mutations in non-collagenous genes: lessons in the biology of bone formation. Curr Opin Pediatr. 2014;26:500-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 9. | Sharma U, Carrique L, Vadon-Le Goff S, Mariano N, Georges RN, Delolme F, Koivunen P, Myllyharju J, Moali C, Aghajari N, Hulmes DJ. Structural basis of homo- and heterotrimerization of collagen I. Nat Commun. 2017;8:14671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 10. | Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16:101-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1452] [Cited by in RCA: 1433] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 11. | Van Dijk FS, Sillence DO. Osteogenesis imperfecta: clinical diagnosis, nomenclature and severity assessment. Am J Med Genet A. 2014;164A:1470-1481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 472] [Article Influence: 42.9] [Reference Citation Analysis (1)] |
| 12. | Mortier GR, Cohn DH, Cormier-Daire V, Hall C, Krakow D, Mundlos S, Nishimura G, Robertson S, Sangiorgi L, Savarirayan R, Sillence D, Superti-Furga A, Unger S, Warman ML. Nosology and classification of genetic skeletal disorders: 2019 revision. Am J Med Genet A. 2019;179:2393-2419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 407] [Article Influence: 67.8] [Reference Citation Analysis (0)] |
| 13. | Forlino A, Marini JC. Osteogenesis imperfecta. Lancet. 2016;387:1657-1671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 643] [Article Influence: 71.4] [Reference Citation Analysis (0)] |
| 14. | Symoens S, Hulmes DJ, Bourhis JM, Coucke PJ, De Paepe A, Malfait F. Type I procollagen C-propeptide defects: study of genotype-phenotype correlation and predictive role of crystal structure. Hum Mutat. 2014;35:1330-1341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Cabral WA, Mertts MV, Makareeva E, Colige A, Tekin M, Pandya A, Leikin S, Marini JC. Type I collagen triplet duplication mutation in lethal osteogenesis imperfecta shifts register of alpha chains throughout the helix and disrupts incorporation of mutant helices into fibrils and extracellular matrix. J Biol Chem. 2003;278:10006-10012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Marini JC, Forlino A, Cabral WA, Barnes AM, San Antonio JD, Milgrom S, Hyland JC, Körkkö J, Prockop DJ, De Paepe A, Coucke P, Symoens S, Glorieux FH, Roughley PJ, Lund AM, Kuurila-Svahn K, Hartikka H, Cohn DH, Krakow D, Mottes M, Schwarze U, Chen D, Yang K, Kuslich C, Troendle J, Dalgleish R, Byers PH. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28:209-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 592] [Cited by in RCA: 526] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 17. | Online Mendelian Inheritance in Man, OMIM. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University Baltimore: Baltimore, MD, USA. 2022 [cited 2022 October 14]. Database: omim [Internet]. Available from: https://omim.org/. |
| 18. | HGNC. HUGO Gene Nomenclature Committee Home Page; 2022 [cited 2022 November 5]. Database: genenames [Internet]. Available from: http://www.genenames.org/. |
| 19. | UniProt: the Universal Protein Knowledgebase; 2023 [cited 2022 November 12]. Database: uniprot [Internet]. Available from: https://www.uniprot.org/. |
| 20. | Cho TJ, Lee KE, Lee SK, Song SJ, Kim KJ, Jeon D, Lee G, Kim HN, Lee HR, Eom HH, Lee ZH, Kim OH, Park WY, Park SS, Ikegawa S, Yoo WJ, Choi IH, Kim JW. A single recurrent mutation in the 5'-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Hum Genet. 2012;91:343-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 21. | Semler O, Garbes L, Keupp K, Swan D, Zimmermann K, Becker J, Iden S, Wirth B, Eysel P, Koerber F, Schoenau E, Bohlander SK, Wollnik B, Netzer C. A mutation in the 5'-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am J Hum Genet. 2012;91:349-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 171] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 22. | Shapiro F, Maguire K, Swami S, Zhu H, Flynn E, Wang J, Wu JY. Histopathology of osteogenesis imperfecta bone. Supramolecular assessment of cells and matrices in the context of woven and lamellar bone formation using light, polarization and ultrastructural microscopy. Bone Rep. 2021;14:100734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Lim JY, Bhatia NS, Vasanwala RF, Chay PL, Lim KBL, Khoo PC, Schwarze U, Jamuar SS. A novel Ser40Trp variant in IFITM5 in a family with osteogenesis imperfecta and review of the literature. Clin Dysmorphol. 2019;28:120-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Dagdeviren D, Tamimi F, Lee B, Sutton R, Rauch F, Retrouvey JM. Dental and craniofacial characteristics caused by the p.Ser40Leu mutation in IFITM5. Am J Med Genet A. 2019;179:65-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Zeitlin L, Rauch F, Travers R, Munns C, Glorieux FH. The effect of cyclical intravenous pamidronate in children and adolescents with osteogenesis imperfecta type V. Bone. 2006;38:13-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Kang H, Aryal Ac S, Barnes AM, Martin A, David V, Crawford SE, Marini JC. Antagonism Between PEDF and TGF-β Contributes to Type VI Osteogenesis Imperfecta Bone and Vascular Pathogenesis. J Bone Miner Res. 2022;37:925-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Amorim DMR, Koga GKC, Dos Santos RN, Secundo PFC, de Ávila Fernandes E, Cardili L, Maeda SS, da Rocha Corrêa Fernandes A, Lazaretti-Castro M. Rare Association Between Osteogenesis Imperfecta and Chondrosarcoma: Could a Pathogenic Variant in the Gene SERPINF1 Explain It? Calcif Tissue Int. 2023;112:118-122. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (1)] |
| 28. | Hoyer-Kuhn H, Netzer C, Koerber F, Schoenau E, Semler O. Two years' experience with denosumab for children with osteogenesis imperfecta type VI. Orphanet J Rare Dis. 2014;9:145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 84] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 29. | Chang W, Barnes AM, Cabral WA, Bodurtha JN, Marini JC. Prolyl 3-hydroxylase 1 and CRTAP are mutually stabilizing in the endoplasmic reticulum collagen prolyl 3-hydroxylation complex. Hum Mol Genet. 2010;19:223-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Barnes AM, Chang W, Morello R, Cabral WA, Weis M, Eyre DR, Leikin S, Makareeva E, Kuznetsova N, Uveges TE, Ashok A, Flor AW, Mulvihill JJ, Wilson PL, Sundaram UT, Lee B, Marini JC. Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. N Engl J Med. 2006;355:2757-2764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 277] [Cited by in RCA: 217] [Article Influence: 11.4] [Reference Citation Analysis (2)] |
| 31. | Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, Monticone M, Castagnola P, Rauch F, Glorieux FH, Vranka J, Bächinger HP, Pace JM, Schwarze U, Byers PH, Weis M, Fernandes RJ, Eyre DR, Yao Z, Boyce BF, Lee B. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127:291-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 365] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 32. | van Dijk FS, Nesbitt IM, Zwikstra EH, Nikkels PG, Piersma SR, Fratantoni SA, Jimenez CR, Huizer M, Morsman AC, Cobben JM, van Roij MH, Elting MW, Verbeke JI, Wijnaendts LC, Shaw NJ, Högler W, McKeown C, Sistermans EA, Dalton A, Meijers-Heijboer H, Pals G. PPIB mutations cause severe osteogenesis imperfecta. Am J Hum Genet. 2009;85:521-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 200] [Article Influence: 12.5] [Reference Citation Analysis (1)] |
| 33. | Zhytnik L, Duy BH, Eekhoff M, Wisse L, Pals G, Reimann E, Kõks S, Märtson A, Maugeri A, Maasalu K, Micha D. Phenotypic Variation in Vietnamese Osteogenesis Imperfecta Patients Sharing a Recessive P3H1 Pathogenic Variant. Genes (Basel). 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 34. | Ito S, Nagata K. Biology of Hsp47 (Serpin H1), a collagen-specific molecular chaperone. Semin Cell Dev Biol. 2017;62:142-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 174] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 35. | Schwarze U, Cundy T, Liu YJ, Hofman PL, Byers PH. Compound heterozygosity for a frameshift mutation and an upstream deletion that reduces expression of SERPINH1 in siblings with a moderate form of osteogenesis imperfecta. Am J Med Genet A. 2019;179:1466-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 36. | Syx D, Ishikawa Y, Gebauer J, Boudko SP, Guillemyn B, Van Damme T, D'hondt S, Symoens S, Nampoothiri S, Gould DB, Baumann U, Bächinger HP, Malfait F. Aberrant binding of mutant HSP47 affects posttranslational modification of type I collagen and leads to osteogenesis imperfecta. PLoS Genet. 2021;17:e1009339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 37. | Barnes AM, Duncan G, Weis M, Paton W, Cabral WA, Mertz EL, Makareeva E, Gambello MJ, Lacbawan FL, Leikin S, Fertala A, Eyre DR, Bale SJ, Marini JC. Kuskokwim syndrome, a recessive congenital contracture disorder, extends the phenotype of FKBP10 mutations. Hum Mutat. 2013;34:1279-1288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 38. | Lim J, Lietman C, Grol MW, Castellon A, Dawson B, Adeyeye M, Rai J, Weis M, Keene DR, Schweitzer R, Park D, Eyre DR, Krakow D, Lee BH. Localized chondro-ossification underlies joint dysfunction and motor deficits in the Fkbp10 mouse model of osteogenesis imperfecta. Proc Natl Acad Sci U S A. 2021;118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 39. | Otaify GA, Abdel-Hamid MS, Hassib NF, Elhossini RM, Abdel-Ghafar SF, Aglan MS. Bruck syndrome in 13 new patients: Identification of five novel FKBP10 and PLOD2 variants and further expansion of the phenotypic spectrum. Am J Med Genet A. 2022;188:1815-1825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 40. | Hojo H, Ohba S. Sp7 Action in the Skeleton: Its Mode of Action, Functions, and Relevance to Skeletal Diseases. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 41. | Lapunzina P, Aglan M, Temtamy S, Caparrós-Martín JA, Valencia M, Letón R, Martínez-Glez V, Elhossini R, Amr K, Vilaboa N, Ruiz-Perez VL. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am J Hum Genet. 2010;87:110-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 197] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 42. | Ludwig K, Ward LM, Khan N, Robinson ME, Miranda V, Bardai G, Moffatt P, Rauch F. Dominant osteogenesis imperfecta with low bone turnover caused by a heterozygous SP7 variant. Bone. 2022;160:116400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Lui JC, Raimann A, Hojo H, Dong L, Roschger P, Kikani B, Wintergerst U, Fratzl-Zelman N, Jee YH, Haeusler G, Baron J. A neomorphic variant in SP7 alters sequence specificity and causes a high-turnover bone disorder. Nat Commun. 2022;13:700. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 44. | Campanini EH, Baker D, Arundel P, Bishop NJ, Offiah AC, Keigwin S, Cadden S, Dall'Ara E, Nicolaou N, Giles S, Fernandes JA, Balasubramanian M. High bone mass phenotype in a cohort of patients with Osteogenesis Imperfecta caused due to BMP1 and C-propeptide cleavage variants in COL1A1. Bone Rep. 2021;15:101102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 45. | Claeys L, Storoni S, Eekhoff M, Elting M, Wisse L, Pals G, Bravenboer N, Maugeri A, Micha D. Collagen transport and related pathways in Osteogenesis Imperfecta. Hum Genet. 2021;140:1121-1141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 46. | Pollitt RC, Saraff V, Dalton A, Webb EA, Shaw NJ, Sobey GJ, Mughal MZ, Hobson E, Ali F, Bishop NJ, Arundel P, Högler W, Balasubramanian M. Phenotypic variability in patients with osteogenesis imperfecta caused by BMP1 mutations. Am J Med Genet A. 2016;170:3150-3156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 47. | Zhang Y, Chen B, Li D, Zhou X, Chen Z. LncRNA NEAT1/miR-29b-3p/BMP1 axis promotes osteogenic differentiation in human bone marrow-derived mesenchymal stem cells. Pathol Res Pract. 2019;215:525-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 48. | Cabral WA, Ishikawa M, Garten M, Makareeva EN, Sargent BM, Weis M, Barnes AM, Webb EA, Shaw NJ, Ala-Kokko L, Lacbawan FL, Högler W, Leikin S, Blank PS, Zimmerberg J, Eyre DR, Yamada Y, Marini JC. Absence of the ER Cation Channel TMEM38B/TRIC-B Disrupts Intracellular Calcium Homeostasis and Dysregulates Collagen Synthesis in Recessive Osteogenesis Imperfecta. PLoS Genet. 2016;12:e1006156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 49. | Leoni L, Tonelli F, Besio R, Gioia R, Moccia F, Rossi A, Forlino A. Knocking out TMEM38B in human foetal osteoblasts hFOB 1.19 by CRISPR/Cas9: A model for recessive OI type XIV. PLoS One. 2021;16:e0257254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 50. | Webb EA, Balasubramanian M, Fratzl-Zelman N, Cabral WA, Titheradge H, Alsaedi A, Saraff V, Vogt J, Cole T, Stewart S, Crabtree NJ, Sargent BM, Gamsjaeger S, Paschalis EP, Roschger P, Klaushofer K, Shaw NJ, Marini JC, Högler W. Phenotypic Spectrum in Osteogenesis Imperfecta Due to Mutations in TMEM38B: Unraveling a Complex Cellular Defect. J Clin Endocrinol Metab. 2017;102:2019-2028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 51. | Ramzan K, Alotaibi M, Huma R, Afzal S. Detection of a Recurrent TMEM38B Gene Deletion Associated with Recessive Osteogenesis Imperfecta. Discoveries (Craiova). 2021;9:e124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 52. | Shaheen R, Alazami AM, Alshammari MJ, Faqeih E, Alhashmi N, Mousa N, Alsinani A, Ansari S, Alzahrani F, Al-Owain M, Alzayed ZS, Alkuraya FS. Study of autosomal recessive osteogenesis imperfecta in Arabia reveals a novel locus defined by TMEM38B mutation. J Med Genet. 2012;49:630-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 53. | Volodarsky M, Markus B, Cohen I, Staretz-Chacham O, Flusser H, Landau D, Shelef I, Langer Y, Birk OS. A deletion mutation in TMEM38B associated with autosomal recessive osteogenesis imperfecta. Hum Mutat. 2013;34:582-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 54. | Lv F, Xu XJ, Wang JY, Liu Y, Asan, Wang JW, Song LJ, Song YW, Jiang Y, Wang O, Xia WB, Xing XP, Li M. Two novel mutations in TMEM38B result in rare autosomal recessive osteogenesis imperfecta. J Hum Genet. 2016;61:539-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 55. | Laine CM, Joeng KS, Campeau PM, Kiviranta R, Tarkkonen K, Grover M, Lu JT, Pekkinen M, Wessman M, Heino TJ, Nieminen-Pihala V, Aronen M, Laine T, Kröger H, Cole WG, Lehesjoki AE, Nevarez L, Krakow D, Curry CJ, Cohn DH, Gibbs RA, Lee BH, Mäkitie O. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N Engl J Med. 2013;368:1809-1816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 273] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 56. | Lu Y, Dai Y, Wang Y, Zhai N, Zhang J, Liu J, Yin X, Li T, Ren X, Han J. Complex heterozygous WNT1 mutation in severe recessive osteogenesis imperfecta of a Chinese patient. Intractable Rare Dis Res. 2018;7:19-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 57. | Zhang B, Li R, Wang W, Zhou X, Luo B, Zhu Z, Zhang X, Ding A. The role of WNT1 mutant variant (WNT1) (c.677C>T) in osteogenesis imperfecta. Ann Hum Genet. 2020;84:447-455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 58. | Mäkitie O, Zillikens MC. Early-Onset Osteoporosis. Calcif Tissue Int. 2022;110:546-561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 59. | Nampoothiri S, Guillemyn B, Elcioglu N, Jagadeesh S, Yesodharan D, Suresh B, Turan S, Symoens S, Malfait F. Ptosis as a unique hallmark for autosomal recessive WNT1-associated osteogenesis imperfecta. Am J Med Genet A. 2019;179:908-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 60. | Pyott SM, Tran TT, Leistritz DF, Pepin MG, Mendelsohn NJ, Temme RT, Fernandez BA, Elsayed SM, Elsobky E, Verma I, Nair S, Turner EH, Smith JD, Jarvik GP, Byers PH. WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am J Hum Genet. 2013;92:590-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 169] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 61. | Keller RB, Tran TT, Pyott SM, Pepin MG, Savarirayan R, McGillivray G, Nickerson DA, Bamshad MJ, Byers PH. Monoallelic and biallelic CREB3L1 variant causes mild and severe osteogenesis imperfecta, respectively. Genet Med. 2018;20:411-419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 62. | Lu CL, Ortmeier S, Brudvig J, Moretti T, Cain J, Boyadjiev SA, Weimer JM, Kim J. Collagen has a unique SEC24 preference for efficient export from the endoplasmic reticulum. Traffic. 2022;23:81-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 63. | Guillemyn B, Kayserili H, Demuynck L, Sips P, De Paepe A, Syx D, Coucke PJ, Malfait F, Symoens S. A homozygous pathogenic missense variant broadens the phenotypic and mutational spectrum of CREB3L1-related osteogenesis imperfecta. Hum Mol Genet. 2019;28:1801-1809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 64. | Cayami FK, Maugeri A, Treurniet S, Setijowati ED, Teunissen BP, Eekhoff EMW, Pals G, Faradz SM, Micha D. The first family with adult osteogenesis imperfecta caused by a novel homozygous mutation in CREB3L1. Mol Genet Genomic Med. 2019;7:e823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 65. | Lindahl K, Åström E, Dragomir A, Symoens S, Coucke P, Larsson S, Paschalis E, Roschger P, Gamsjaeger S, Klaushofer K, Fratzl-Zelman N, Kindmark A. Homozygosity for CREB3L1 premature stop codon in first case of recessive osteogenesis imperfecta associated with OASIS-deficiency to survive infancy. Bone. 2018;114:268-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 66. | Lima MLS, Medeiros CACX, Guerra GCB, Santos R, Bader M, Pirih FQ, Araújo Júnior RF, Chan AB, Cruz LJ, Brito GAC, Leitão RFC, Silveira EJDD, Garcia VB, Martins AA, Araújo AA. AT1 and AT2 Receptor Knockout Changed Osteonectin and Bone Density in Mice in Periodontal Inflammation Experimental Model. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 67. | Rosset EM, Bradshaw AD. SPARC/osteonectin in mineralized tissue. Matrix Biol. 2016;52-54:78-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 208] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 68. | Durkin A, DeVile C, Arundel P, Bull M, Walsh J, Bishop NJ, Hupin E, Parekh S, Nadarajah R, Offiah AC, Calder A, Brock J, Baker D, Balasubramanian M. Expanding the phenotype of SPARC-related osteogenesis imperfecta: clinical findings in two patients with pathogenic variants in SPARC and literature review. J Med Genet. 2022;59:810-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 69. | Hayat A, Hussain S, Bilal M, Kausar M, Almuzzaini B, Abbas S, Tanveer A, Khan A, Siddiqi S, Foo JN, Ahmad F, Khan F, Khan B, Anees M, Mäkitie O, Alfadhel M, Ahmad W, Umair M. Biallelic variants in four genes underlying recessive osteogenesis imperfecta. Eur J Med Genet. 2020;63:103954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 70. | Mendoza-Londono R, Fahiminiya S, Majewski J; Care4Rare Canada Consortium, Tétreault M, Nadaf J, Kannu P, Sochett E, Howard A, Stimec J, Dupuis L, Roschger P, Klaushofer K, Palomo T, Ouellet J, Al-Jallad H, Mort JS, Moffatt P, Boudko S, Bächinger HP, Rauch F. Recessive osteogenesis imperfecta caused by missense mutations in SPARC. Am J Hum Genet. 2015;96:979-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 94] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 71. | Lin HH, Lo YL, Wang WC, Huang KY, I KY, Chang GW. Overexpression of FAM46A, a Non-canonical Poly(A) Polymerase, Promotes Hemin-Induced Hemoglobinization in K562 Cells. Front Cell Dev Biol. 2020;8:414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 72. | Watanabe T, Yamamoto T, Tsukano K, Hirano S, Horikawa A, Michiue T. Fam46a regulates BMP-dependent pre-placodal ectoderm differentiation in Xenopus. Development. 2018;145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 73. | Doyard M, Bacrot S, Huber C, Di Rocco M, Goldenberg A, Aglan MS, Brunelle P, Temtamy S, Michot C, Otaify GA, Haudry C, Castanet M, Leroux J, Bonnefont JP, Munnich A, Baujat G, Lapunzina P, Monnot S, Ruiz-Perez VL, Cormier-Daire V. FAM46A mutations are responsible for autosomal recessive osteogenesis imperfecta. J Med Genet. 2018;55:278-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 74. | Caengprasath N, Theerapanon T, Porntaveetus T, Shotelersuk V. MBTPS2, a membrane bound protease, underlying several distinct skin and bone disorders. J Transl Med. 2021;19:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 75. | Lindert U, Cabral WA, Ausavarat S, Tongkobpetch S, Ludin K, Barnes AM, Yeetong P, Weis M, Krabichler B, Srichomthong C, Makareeva EN, Janecke AR, Leikin S, Röthlisberger B, Rohrbach M, Kennerknecht I, Eyre DR, Suphapeetiporn K, Giunta C, Marini JC, Shotelersuk V. MBTPS2 mutations cause defective regulated intramembrane proteolysis in X-linked osteogenesis imperfecta. Nat Commun. 2016;7:11920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 76. | Chen C, Xu C, Li H, Jia M, Tang S. Novel mutation in MBTPS2 causes keratosis follicularis spinulosa decalvans in a large Chinese family. Int J Dermatol. 2019;58:493-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 77. | Naiki M, Mizuno S, Yamada K, Yamada Y, Kimura R, Oshiro M, Okamoto N, Makita Y, Seishima M, Wakamatsu N. MBTPS2 mutation causes BRESEK/BRESHECK syndrome. Am J Med Genet A. 2012;158A:97-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 78. | Oeffner F, Fischer G, Happle R, König A, Betz RC, Bornholdt D, Neidel U, Boente Mdel C, Redler S, Romero-Gomez J, Salhi A, Vera-Casaño A, Weirich C, Grzeschik KH. IFAP syndrome is caused by deficiency in MBTPS2, an intramembrane zinc metalloprotease essential for cholesterol homeostasis and ER stress response. Am J Hum Genet. 2009;84:459-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 79. | Moosa S, Yamamoto GL, Garbes L, Keupp K, Beleza-Meireles A, Moreno CA, Valadares ER, de Sousa SB, Maia S, Saraiva J, Honjo RS, Kim CA, Cabral de Menezes H, Lausch E, Lorini PV, Lamounier A Jr, Carniero TCB, Giunta C, Rohrbach M, Janner M, Semler O, Beleggia F, Li Y, Yigit G, Reintjes N, Altmüller J, Nürnberg P, Cavalcanti DP, Zabel B, Warman ML, Bertola DR, Wollnik B, Netzer C. Autosomal-Recessive Mutations in MESD Cause Osteogenesis Imperfecta. Am J Hum Genet. 2019;105:836-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 80. | Stürznickel J, Jähn-Rickert K, Zustin J, Hennig F, Delsmann MM, Schoner K, Rehder H, Kreczy A, Schinke T, Amling M, Kornak U, Oheim R. Compound Heterozygous Frameshift Mutations in MESD Cause a Lethal Syndrome Suggestive of Osteogenesis Imperfecta Type XX. J Bone Miner Res. 2021;36:1077-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 81. | Efthymiou S, Herman I, Rahman F, Anwar N, Maroofian R, Yip J, Mitani T, Calame DG, Hunter JV, Sutton VR, Yilmaz Gulec E, Duan R, Fatih JM, Marafi D, Pehlivan D, Jhangiani SN, Gibbs RA, Posey JE; SYNAPS Study Group, Maqbool S, Lupski JR, Houlden H. Two novel bi-allelic KDELR2 missense variants cause osteogenesis imperfecta with neurodevelopmental features. Am J Med Genet A. 2021;185:2241-2249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 82. | van Dijk FS, Semler O, Etich J, Köhler A, Jimenez-Estrada JA, Bravenboer N, Claeys L, Riesebos E, Gegic S, Piersma SR, Jimenez CR, Waisfisz Q, Flores CL, Nevado J, Harsevoort AJ, Janus GJM, Franken AAM, van der Sar AM, Meijers-Heijboer H, Heath KE, Lapunzina P, Nikkels PGJ, Santen GWE, Nüchel J, Plomann M, Wagener R, Rehberg M, Hoyer-Kuhn H, Eekhoff EMW, Pals G, Mörgelin M, Newstead S, Wilson BT, Ruiz-Perez VL, Maugeri A, Netzer C, Zaucke F, Micha D. Interaction between KDELR2 and HSP47 as a Key Determinant in Osteogenesis Imperfecta Caused by Bi-allelic Variants in KDELR2. Am J Hum Genet. 2020;107:989-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 83. | Dubail J, Brunelle P, Baujat G, Huber C, Doyard M, Michot C, Chavassieux P, Khairouni A, Topouchian V, Monnot S, Koumakis E, Cormier-Daire V. Homozygous Loss-of-Function Mutations in CCDC134 Are Responsible for a Severe Form of Osteogenesis Imperfecta. J Bone Miner Res. 2020;35:1470-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 84. | Ali TM, Linnenkamp BDW, Yamamoto GL, Honjo RS, Cabral de Menezes Filho H, Kim CA, Bertola DR. The recurrent homozygous translation start site variant in CCDC134 in an individual with severe osteogenesis imperfecta of non-Morrocan ancestry. Am J Med Genet A. 2022;188:1545-1549. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 85. | Holick MF, Shirvani A, Charoenngam N. Fetal Fractures in an Infant with Maternal Ehlers-Danlos Syndrome, CCDC134 Pathogenic Mutation and a Negative Genetic Test for Osteogenesis Imperfecta. Children (Basel). 2021;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 86. | Zhytnik L, Simm K, Salumets A, Peters M, Märtson A, Maasalu K. Reproductive options for families at risk of Osteogenesis Imperfecta: a review. Orphanet J Rare Dis. 2020;15:128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 87. | Zhytnik L, Maasalu K, Duy BH, Pashenko A, Khmyzov S, Reimann E, Prans E, Kõks S, Märtson A. De novo and inherited pathogenic variants in collagen-related osteogenesis imperfecta. Mol Genet Genomic Med. 2019;7:e559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 88. | Mei Y, Zhang H, Zhang Z. Comparing Clinical and Genetic Characteristics of De Novo and Inherited COL1A1/COL1A2 Variants in a Large Chinese Cohort of Osteogenesis Imperfecta. Front Endocrinol (Lausanne). 2022;13:935905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 89. | Pyott SM, Pepin MG, Schwarze U, Yang K, Smith G, Byers PH. Recurrence of perinatal lethal osteogenesis imperfecta in sibships: parsing the risk between parental mosaicism for dominant mutations and autosomal recessive inheritance. Genet Med. 2011;13:125-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 90. | Mathijssen IB, van Maarle MC, Kleiss IIM, Redeker EJW, Ten Kate LP, Henneman L, Meijers-Heijboer H. With expanded carrier screening, founder populations run the risk of being overlooked. J Community Genet. 2017;8:327-333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 91. | Cabral WA, Barnes AM, Adeyemo A, Cushing K, Chitayat D, Porter FD, Panny SR, Gulamali-Majid F, Tishkoff SA, Rebbeck TR, Gueye SM, Bailey-Wilson JE, Brody LC, Rotimi CN, Marini JC. A founder mutation in LEPRE1 carried by 1.5% of West Africans and 0.4% of African Americans causes lethal recessive osteogenesis imperfecta. Genet Med. 2012;14:543-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 92. | Essawi O, Symoens S, Fannana M, Darwish M, Farraj M, Willaert A, Essawi T, Callewaert B, De Paepe A, Malfait F, Coucke PJ. Genetic analysis of osteogenesis imperfecta in the Palestinian population: molecular screening of 49 affected families. Mol Genet Genomic Med. 2018;6:15-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 93. | Schwarze U, Cundy T, Pyott SM, Christiansen HE, Hegde MR, Bank RA, Pals G, Ankala A, Conneely K, Seaver L, Yandow SM, Raney E, Babovic-Vuksanovic D, Stoler J, Ben-Neriah Z, Segel R, Lieberman S, Siderius L, Al-Aqeel A, Hannibal M, Hudgins L, McPherson E, Clemens M, Sussman MD, Steiner RD, Mahan J, Smith R, Anyane-Yeboa K, Wynn J, Chong K, Uster T, Aftimos S, Sutton VR, Davis EC, Kim LS, Weis MA, Eyre D, Byers PH. Mutations in FKBP10, which result in Bruck syndrome and recessive forms of osteogenesis imperfecta, inhibit the hydroxylation of telopeptide lysines in bone collagen. Hum Mol Genet. 2013;22:1-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 107] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 94. | Vorster A, Beighton P, Chetty M, Ganie Y, Henderson B, Honey E, Maré P, Thompson D, Fieggen K, Viljoen D, Ramesar R. Osteogenesis imperfecta type 3 in South Africa: Causative mutations in FKBP10. S Afr Med J. 2017;107:457-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 95. | Zhu W, Yan K, Chen X, Zhao W, Wu Y, Tang H, Chen M, Wu J, Wang P, Zhang R, Shen Y, Zhang D. A Founder Pathogenic Variant of PPIB Unique to Chinese Population Causes Osteogenesis Imperfecta IX. Front Genet. 2021;12:717294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 96. | Hu J, Li LJ, Zheng WB, Zhao DC, Wang O, Jiang Y, Xing XP, Li M, Xia W. A novel mutation in PLS3 causes extremely rare X-linked osteogenesis imperfecta. Mol Genet Genomic Med. 2020;8:e1525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 97. | Garibaldi N, Besio R, Dalgleish R, Villani S, Barnes AM, Marini JC, Forlino A. Dissecting the phenotypic variability of osteogenesis imperfecta. Dis Model Mech. 2022;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 98. | Yang L, Liu B, Dong X, Wu J, Sun C, Xi L, Cheng R, Wu B, Wang H, Tong S, Wang D, Luo F. Clinical severity prediction in children with osteogenesis imperfecta caused by COL1A1/2 defects. Osteoporos Int. 2022;33:1373-1384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 99. | Zhang J, Lai Z, Shi L, Tian Y, Luo A, Xu Z, Ma X, Wang S. Repeated superovulation increases the risk of osteoporosis and cardiovascular diseases by accelerating ovarian aging in mice. Aging (Albany NY). 2018;10:1089-1102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 100. | De Rycke M, Berckmoes V. Preimplantation Genetic Testing for Monogenic Disorders. Genes (Basel). 2020;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 101. | Zanetti BF, Braga DPAF, Azevedo MC, Setti AS, Figueira RCS, Iaconelli A Jr, Borges E Jr. Preimplantation genetic testing for monogenic diseases: a Brazilian IVF centre experience. JBRA Assist Reprod. 2019;23:99-105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 102. | Bonanni G, Trevisan V, Zollino M, De Santis M, Romanzi F, Lanzone A, Bevilacqua E. Case Report: Challenges of Non-Invasive Prenatal Testing (NIPT): A Case Report of Confined Placental Mosaicism and Clinical Considerations. Front Genet. 2022;13:881284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 103. | Gorduza EV, Popescu R, Caba L, Ivanov I, Martiniuc V, Nedelea F, Militaru M, Socolov DG. Prenatal diagnosis of 21 trisomy by quantification of methylated fetal DNA in maternal blood: study on 10 pregnancies. Rev Rom Med Lab. 2013;21:275-284. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 104. | Gug C, Mozos I, Ratiu A, Tudor A, Gorduza EV, Caba L, Gug M, Cojocariu C, Furau C, Furau G, Vaida MA, Stoicanescu D. Genetic Counseling and Management: The First Study to Report NIPT Findings in a Romanian Population. Medicina (Kaunas). 2022;58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 105. | Liehr T. False-positives and false-negatives in non-invasive prenatal testing (NIPT): what can we learn from a meta-analyses on > 750,000 tests? Mol Cytogenet. 2022;15:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 106. | Shaw J, Scotchman E, Chandler N, Chitty LS. PREIMPLANTATION GENETIC TESTING: Non-invasive prenatal testing for aneuploidy, copy-number variants and single-gene disorders. Reproduction. 2020;160:A1-A11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 107. | Deguchi M, Tsuji S, Katsura D, Kasahara K, Kimura F, Murakami T. Current Overview of Osteogenesis Imperfecta. Medicina (Kaunas). 2021;57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 108. | Gug C, Caba L, Mozos I, Stoian D, Atasie D, Gug M, Gorduza EV. Rare splicing mutation in COL1A1 gene identified by whole exomes sequencing in a patient with osteogenesis imperfecta type I followed by prenatal diagnosis: A case report and review of the literature. Gene. 2020;741:144565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 109. | Milks KS, Hill LM, Hosseinzadeh K. Evaluating skeletal dysplasias on prenatal ultrasound: an emphasis on predicting lethality. Pediatr Radiol. 2017;47:134-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 110. | Gilligan LA, Calvo-Garcia MA, Weaver KN, Kline-Fath BM. Fetal magnetic resonance imaging of skeletal dysplasias. Pediatr Radiol. 2020;50:224-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 111. | Nelson DB, Dashe JS, McIntire DD, Twickler DM. Fetal skeletal dysplasias: sonographic indices associated with adverse outcomes. J Ultrasound Med. 2014;33:1085-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 112. | Waratani M, Ito F, Tanaka Y, Mabuchi A, Mori T, Kitawaki J. Prenatal diagnosis of fetal skeletal dysplasia using 3-dimensional computed tomography: a prospective study. BMC Musculoskelet Disord. 2020;21:662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 113. | Bellur S, Jain M, Cuthbertson D, Krakow D, Shapiro JR, Steiner RD, Smith PA, Bober MB, Hart T, Krischer J, Mullins M, Byers PH, Pepin M, Durigova M, Glorieux FH, Rauch F, Sutton VR, Lee B; Members of the BBD Consortium, Nagamani SC. Cesarean delivery is not associated with decreased at-birth fracture rates in osteogenesis imperfecta. Genet Med. 2016;18:570-576. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 114. | Chamunyonga F, Masendeke KL, Mateveke B. Osteogenesis imperfecta and pregnancy: a case report. J Med Case Rep. 2019;13:363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 115. | Cozzolino M, Perelli F, Maggio L, Coccia ME, Quaranta M, Gizzo S, Mecacci F. Management of osteogenesis imperfecta type I in pregnancy; a review of literature applied to clinical practice. Arch Gynecol Obstet. 2016;293:1153-1159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 116. | Ralston SH, Gaston MS. Management of Osteogenesis Imperfecta. Front Endocrinol (Lausanne). 2019;10:924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 117. | Folkestad L, Hald JD, Canudas-Romo V, Gram J, Hermann AP, Langdahl B, Abrahamsen B, Brixen K. Mortality and Causes of Death in Patients With Osteogenesis Imperfecta: A Register-Based Nationwide Cohort Study. J Bone Miner Res. 2016;31:2159-2166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 118. | Balasubramanian M, Verschueren A, Kleevens S, Luyckx I, Perik M, Schirwani S, Mortier G, Morisaki H, Rodrigus I, Van Laer L, Verstraeten A, Loeys B. Aortic aneurysm/dissection and osteogenesis imperfecta: Four new families and review of the literature. Bone. 2019;121:191-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 119. | Jensen PR, Andersen TL, Chavassieux P, Roux JP, Delaisse JM. Bisphosphonates impair the onset of bone formation at remodeling sites. Bone. 2021;145:115850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 120. | Bains JS, Carter EM, Citron KP, Boskey AL, Shapiro JR, Steiner RD, Smith PA, Bober MB, Hart T, Cuthbertson D, Krischer J, Byers PH, Pepin M, Durigova M, Glorieux FH, Rauch F, Sliepka JM, Sutton VR, Lee B; Members of the BBD Consortium, Nagamani SC, Raggio CL. A Multicenter Observational Cohort Study to Evaluate the Effects of Bisphosphonate Exposure on Bone Mineral Density and Other Health Outcomes in Osteogenesis Imperfecta. JBMR Plus. 2019;3:e10118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 121. | Bradbury LA, Barlow S, Geoghegan F, Hannon RA, Stuckey SL, Wass JA, Russell RG, Brown MA, Duncan EL. Risedronate in adults with osteogenesis imperfecta type I: increased bone mineral density and decreased bone turnover, but high fracture rate persists. Osteoporos Int. 2012;23:285-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 122. | Dwan K, Phillipi CA, Steiner RD, Basel D. Bisphosphonate therapy for osteogenesis imperfecta. Cochrane Database Syst Rev. 2016;10:CD005088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 117] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 123. | Ying ZM, Hu B, Yan SG. Oral Bisphosphonate Therapy for Osteogenesis Imperfecta: A Systematic Review and Meta-Analysis of Six Randomized Placebo-Controlled Trials. Orthop Surg. 2020;12:1293-1303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 124. | Ma S, Goh EL, Jin A, Bhattacharya R, Boughton OR, Patel B, Karunaratne A, Vo NT, Atwood R, Cobb JP, Hansen U, Abel RL. Long-term effects of bisphosphonate therapy: perforations, microcracks and mechanical properties. Sci Rep. 2017;7:43399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 125. | Green SB, Pappas AL. Effects of maternal bisphosphonate use on fetal and neonatal outcomes. Am J Health Syst Pharm. 2014;71:2029-2036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 126. | Botor M, Fus-Kujawa A, Uroczynska M, Stepien KL, Galicka A, Gawron K, Sieron AL. Osteogenesis Imperfecta: Current and Prospective Therapies. Biomolecules. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 127. | Hoyer-Kuhn H, Rehberg M, Netzer C, Schoenau E, Semler O. Individualized treatment with denosumab in children with osteogenesis imperfecta - follow up of a trial cohort. Orphanet J Rare Dis. 2019;14:219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 128. | Lee LR, Holman AE, Li X, Vasiljevski ER, O'Donohue AK, Cheng TL, Little DG, Schindeler A, Biggin A, Munns CF. Combination treatment with growth hormone and zoledronic acid in a mouse model of Osteogenesis imperfecta. Bone. 2022;159:116378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 129. | Rossi V, Lee B, Marom R. Osteogenesis imperfecta: advancements in genetics and treatment. Curr Opin Pediatr. 2019;31:708-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 130. | Orwoll ES, Shapiro J, Veith S, Wang Y, Lapidus J, Vanek C, Reeder JL, Keaveny TM, Lee DC, Mullins MA, Nagamani SC, Lee B. Evaluation of teriparatide treatment in adults with osteogenesis imperfecta. J Clin Invest. 2014;124:491-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 131. | Vahle JL, Sato M, Long GG, Young JK, Francis PC, Engelhardt JA, Westmore MS, Linda Y, Nold JB. Skeletal changes in rats given daily subcutaneous injections of recombinant human parathyroid hormone (1-34) for 2 years and relevance to human safety. Toxicol Pathol. 2002;30:312-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 506] [Cited by in RCA: 454] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 132. | Glorieux FH, Devogelaer JP, Durigova M, Goemaere S, Hemsley S, Jakob F, Junker U, Ruckle J, Seefried L, Winkle PJ. BPS804 Anti-Sclerostin Antibody in Adults With Moderate Osteogenesis Imperfecta: Results of a Randomized Phase 2a Trial. J Bone Miner Res. 2017;32:1496-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 133. | Uehara M, Nakamura Y, Nakano M, Miyazaki A, Suzuki T, Takahashi J. Efficacy of romosozumab for osteoporosis in a patient with osteogenesis imperfecta: A case report. Mod Rheumatol Case Rep. 2022;6:128-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 134. | Lv F, Cai X, Yang W, Gao L, Chen L, Wu J, Ji L. Denosumab or romosozumab therapy and risk of cardiovascular events in patients with primary osteoporosis: Systematic review and meta- analysis. Bone. 2020;130:115121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 135. | Song IW, Nagamani SC, Nguyen D, Grafe I, Sutton VR, Gannon FH, Munivez E, Jiang MM, Tran A, Wallace M, Esposito P, Musaad S, Strudthoff E, McGuire S, Thornton M, Shenava V, Rosenfeld S, Huang S, Shypailo R, Orwoll E, Lee B. Targeting TGF-β for treatment of osteogenesis imperfecta. J Clin Invest. 2022;132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 56] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 136. | Chen S, Grover M, Sibai T, Black J, Rianon N, Rajagopal A, Munivez E, Bertin T, Dawson B, Chen Y, Jiang MM, Lee B, Yang T, Bae Y. Losartan increases bone mass and accelerates chondrocyte hypertrophy in developing skeleton. Mol Genet Metab. 2015;115:53-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 137. | Götherström C, David AL, Walther-Jallow L, Åström E, Westgren M. Mesenchymal Stem Cell Therapy for Osteogenesis Imperfecta. Clin Obstet Gynecol. 2021;64:898-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 138. | Horwitz EM, Gordon PL, Koo WK, Marx JC, Neel MD, McNall RY, Muul L, Hofmann T. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proc Natl Acad Sci U S A. 2002;99:8932-8937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1315] [Cited by in RCA: 1202] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 139. | Schindeler A, Lee LR, O'Donohue AK, Ginn SL, Munns CF. Curative Cell and Gene Therapy for Osteogenesis Imperfecta. J Bone Miner Res. 2022;37:826-836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 140. | Jung H, Rim YA, Park N, Nam Y, Ju JH. Restoration of Osteogenesis by CRISPR/Cas9 Genome Editing of the Mutated COL1A1 Gene in Osteogenesis Imperfecta. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 141. | Duran I, Zieba J, Csukasi F, Martin JH, Wachtell D, Barad M, Dawson B, Fafilek B, Jacobsen CM, Ambrose CG, Cohn DH, Krejci P, Lee BH, Krakow D. 4-PBA Treatment Improves Bone Phenotypes in the Aga2 Mouse Model of Osteogenesis Imperfecta. J Bone Miner Res. 2022;37:675-686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 142. | Scheiber AL, Wilkinson KJ, Suzuki A, Enomoto-Iwamoto M, Kaito T, Cheah KS, Iwamoto M, Leikin S, Otsuru S. 4PBA reduces growth deficiency in osteogenesis imperfecta by enhancing transition of hypertrophic chondrocytes to osteoblasts. JCI Insight. 2022;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |