Published online Jan 6, 2023. doi: 10.12998/wjcc.v11.i1.92
Peer-review started: September 28, 2022
First decision: November 4, 2022
Revised: November 10, 2022
Accepted: December 21, 2022
Article in press: December 21, 2022
Published online: January 6, 2023
Processing time: 98 Days and 14.5 Hours
In recently diagnosed patients with thyroid cancer, papillary thyroid cancer (PTC), as the most common histological subtype, accounts for 90% of all cases. Although PTC is known as a relatively adolescent malignant disease, there still is a high possibility of recurrence in PTC patients with a poor prognosis. Therefore, new biomarkers are necessary to guide more effective stratification of PTC patients and personalize therapy to avoid overtreatment or inadequate treatment. Accumulating evidence demonstrates that microRNAs (miRNAs) have broad application prospects as diagnostic biomarkers in cancer.
To explore novel markers consisting of miRNA-associated signatures for PTC prognostication.
We obtained and analyzed the data of 497 PTC patients from The Cancer Genome Atlas. The patients were randomly assigned to either a training or testing cohort.
We discovered 237 differentially expressed miRNAs in tumorous thyroid tissues compared with normal tissues, which contained 172 up-regulated and 65 down-regulated miRNAs. The evaluation of differently expressed miRNAs was conducted using our risk score model. We then successfully generated a four-miRNA potential prognostic signature [risk score = (-0.001 × hsa-miR-181a-2-3p) + (0.003 × hsa-miR-138-5p) + (-0.018 × hsa-miR-424-3p) + (0.284 × hsa-miR-612)], which reliably distinguished patients from high and low risk with a significant difference in the overall survival (P < 0.01) and was effective in predicting the five-year disease survival rate with the area under the receiver operating characteristic curve of 0.937 and 0.812 in the training and testing cohorts, respectively. Additionally, there was a trend indicated that high-risk patients had shorter relapse-free survival, although statistical significance was not reached (P = 0.082) in our sequencing cohort.
Our results indicated a four-miRNA signature that has a robust predictive effect on the prognosis of PTC. Accordingly, we would recommend more radical therapy and closer follow-ups for high-risk groups.
Core Tip: Thyroid cancer is the most prevalent endocrine malignancy in the world, and its incidence is rapidly rising. In this paper, an efficient and accurate prognostic prediction model for thyroid cancer was constructed, which is valuable for future clinical studies.
- Citation: Yang F, Zhou YL. Identification of a four-miRNA signature predicts the prognosis of papillary thyroid cancer. World J Clin Cases 2023; 11(1): 92-103
- URL: https://www.wjgnet.com/2307-8960/full/v11/i1/92.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i1.92
Thyroid cancer is the most prevalent endocrine malignancy in the world, and its incidence is rapidly rising[1]. It ranks as the fifth most common cancer in female patients, and its incidence is approximately three times higher than in males in most regions and populations[2,3]. Differentiated thyroid cancer derives from follicular epithelial cells; the main subtypes comprise papillary thyroid cancer (PTC), follicular thyroid cancer, and Hurthle cell carcinoma[4]. The most common histological subtype of thyroid cancer is PTC, which accounts for 90% of newly diagnosed thyroid cancers and has the best prognosis among all subtypes[4]. PTC usually behaves like an indolent disease in most patients and can be well controlled or cured using the appropriate surgical procedure or with the help of radioiodine. However, the recurrence rate of PTC remains high, and the dedifferentiation of PTC would potentially lead to invasiveness and a poor prognosis[5].
Given that PTC is a heterogeneous disease, optimal treatment for PTC patients has long been a heated controversy. While more aggressive treatment of cancer can reduce the recurrence of disease and mortality rates, it will also give rise to treatment-related complications[6]. To alleviate this dilemma, we need to improve risk stratification to identify patients with worse outcomes more accurately. Therefore, introducing new biomarkers as an advanced method in improving the overall survival (OS) of PTC patients is favored.
MicroRNAs (miRNAs) are types of noncoding, single-stranded RNA molecules consisting of approximately 18 to 25 nucleotides. The specific binding of miRNAs to the complementary mRNA can either facilitate mRNA degradation or prevent mRNA translation into protein[7]. Previous reports have suggested that miRNAs are essential in the tumorigenesis and progression of PTC[8,9]. Accumulating evidence has also demonstrated that miRNAs have broad application prospects, such as diagnostic biomarkers and therapeutic targets in cancer[10]. However, the limitations of existing studies include inadequate sample quantitation and limited comprehensive analyses on many PTC samples. With the help of The Cancer Genome Atlas (TCGA) database, we could investigate cancer-specific signatures, which contain large-scale miRNA expression data and prognostic survival data.
In this study, we utilize data from the TCGA database to conduct a comprehensive analysis and screen out differentially expressed miRNAs. We then assess the prognostic value of these miRNAs using a risk score model. A panel of four miRNAs is generated as a prognostic signature, which is then tested in PTC patients. Such a practical tool has satisfying potential in stratifying PTC patients and individualized therapy to avoid overtreatment or inadequate treatment.
Level three miRNASeq datasets of 507 PTC and 58 normal samples and the corresponding clinical data of PTC patients were extracted from the TCGA database (http://cancergenome.nih.gov) on December 12, 2019[11]. The inclusion criteria of the studied samples were as follows: (1) The data contained both miRNA sequencing and clinical information; (2) the sample had prognosis information; and (3) the histological typing was PTC. A total of 497 PTC samples met our criteria and were selected for further analysis. The entire set was randomly separated into a training cohort (249 cases) and a test cohort (248 cases). The detailed baseline characteristics of the entire set are listed in Supplementary Table 1.
An analysis of the global miRNA expression profile detected 2202 miRNAs. Then, the miRNA expression profiles were standardized using the R package of edgeR[12]. EdgeR was also utilized to sift out the differentially expressed miRNAs according to the following criteria: (1) Fold change (FC) > 2 for up- or down-regulation; and (2) false discovery rate (FDR) < 0.05. Based on the analysis of the differentially expressed miRNA, a volcano plot was produced with label colors that were determined by the filtering criteria.
First, a univariate cox regression analysis was used to sift out each differently expressed miRNA that was related to patients’ OS. Subsequently, these differently expressed miRNAs with P < 0.05 were selected for the least absolute shrinkage and selection operator analysis (LASSO). The LASSO analysis created a more refined model by constructing a penalty function. Finally, we established general multivariate stepwise Cox regression models to identify which of the significant miRNAs was an independent predictor of prognosis.
MiRNAs that had a notable association with OS in the multivariate Cox regression analysis were utilized to construct the miRNA signature, which was used to estimate the prognostic risk score for each patient. The miRNA signature was built using the coefficients obtained from the Cox regression analysis. The standards were as follows: Risk score = (-0.001 × hsa-miR-181a-2-3p) + (0.003 × hsa-miR-138-5p) + (-0.018 × hsa-miR-424-3p) + (0.284 × hsa-miR-612). Subsequently, according to the same median risk score as the cutoff point, patients in both the training and testing cohorts were divided into the low-risk and high-risk groups. Next, the area under the curve (AUC) of the time-dependent ROC analysis was analyzed to reveal the predictive effect of the miRNA-based classifier and prognostic model. To make the prognostic miRNA signature more convenient in clinical practice, we also constructed a prognostic nomogram. Furthermore, a calibration curve was carried out to assess the consistency of model prediction and the actual outcome.
The miRDB (http://www.mirdb.org/miRDB/) and TargetScan (http://www.targetscan.org/) were used for the prediction of target genes of four miRNAs. Furthermore, overlapping target genes from these two online analysis databases were analyzed using the functional enrichment analysis tool FunRich[13]. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were used to analyze the function of these target genes. These target genes of four miRNAs are listed in Supplementary Table 2.
The Mann-Whitney U test and the χ2 test were used to analyze the associations of continuous and categorical variables between the training and testing cohorts, respectively. Survival analyses were compared using log-rank tests, while the Kaplan-Meier method was adopted to plot the survival curves. P < 0.05 was considered statistically significant. All statistical analyses were performed using SPSS version 22.0 (Chicago, IL, United States) and R version 3.6.1.
We extracted and investigated the data of 497 PTC patients from the TCGA database. The patients were randomly assigned to either the training or testing cohort. Table 1 demonstrates the detailed clinical characteristics (age, sex, vital status, stages, and T/N/M classification) of both cohorts, finding no significant differences between the two groups (P > 0.05).
| Variable | Train cohort | Test cohort | P value |
| (n = 249) | (n = 248) | ||
| Age | 0.968 | ||
| < 45 | 112 | 112 | |
| ≥ 45 | 137 | 138 | |
| Sex | 0.064 | ||
| Male | 75 | 57 | |
| Female | 174 | 193 | |
| Vital status | 0.617 | ||
| Death | 9 | 7 | |
| Alive | 240 | 241 | |
| Stages | 0.768 | ||
| I | 140 | 138 | |
| II | 28 | 23 | |
| III | 53 | 61 | |
| IV | 28 | 26 | |
| T classification | 0.302 | ||
| T1 | 67 | 76 | |
| T2 | 92 | 74 | |
| T3 | 78 | 85 | |
| T4 | 12 | 11 | |
| Tx | 0 | 2 | |
| N classification | 0.166 | ||
| N0 | 107 | 121 | |
| N1 | 120 | 99 | |
| Nx | 22 | 28 | |
| M classification | 0.584 | ||
| M0 | 139 | 139 | |
| M1 | 3 | 6 | |
| Mx | 107 | 103 |
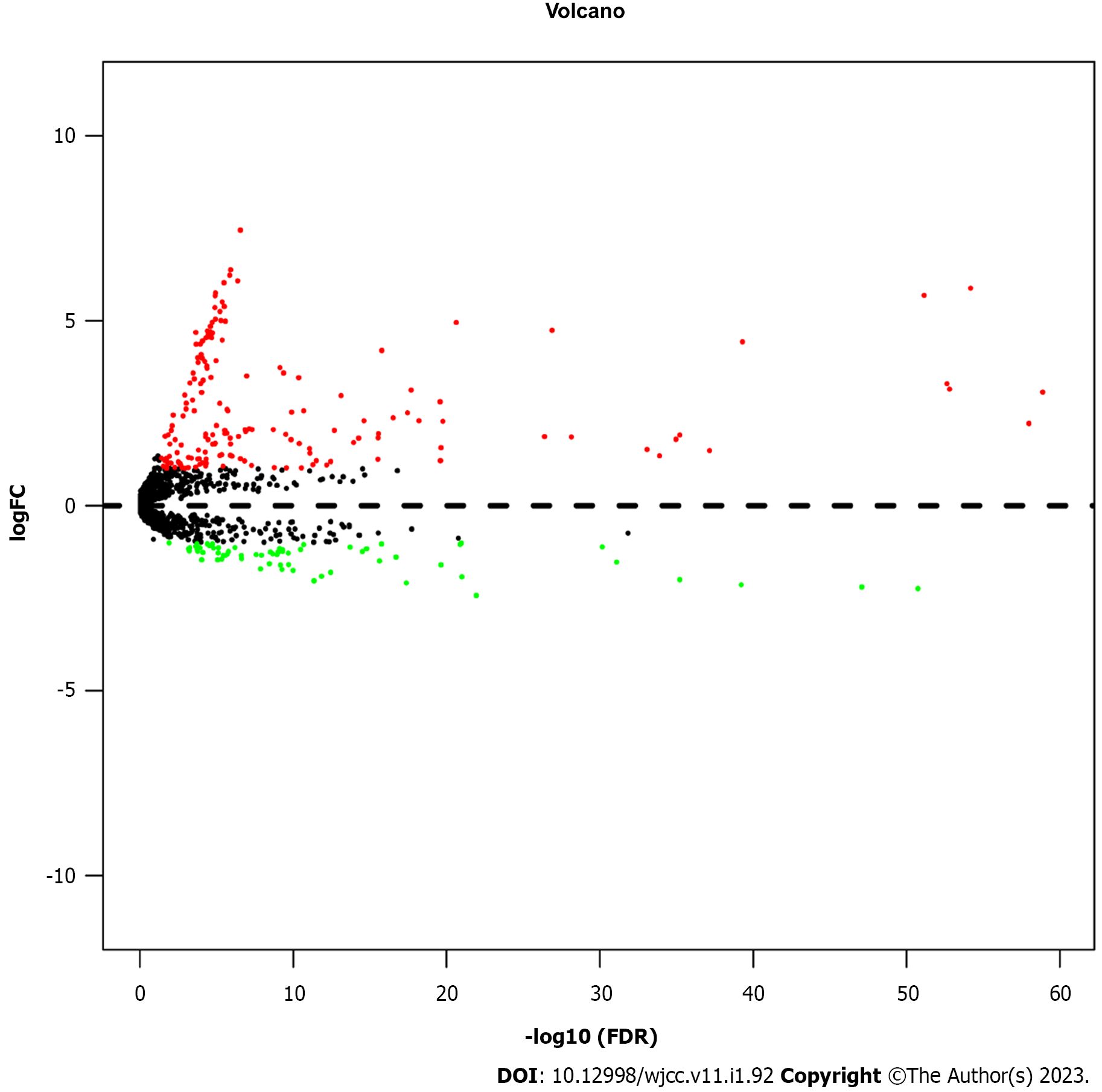
To select significantly differentially expressed miRNAs in the 497 PTC tissues and 59 adjacent normal thyroid tissues (Figure 1), we performed a volcano plot to evaluate miRNA expression variation with the standard of FC > 2 or < 0.5, as well as FDR < 0.05. A total of 237 miRNAs were differentially expressed in the cancerous tissues compared with the normal tissues, including 172 up-regulated miRNAs and 65 down-regulated miRNAs. These 237 significantly differently expressed miRNAs were identified as potential prognostic biomarkers for PTC. To screen out the OS-related miRNAs, a univariate cox regression analysis was performed for these significantly differentially expressed miRNAs in the training cohort. Subsequently, we found that 13 miRNAs were distinctly associated with the OS of PTC (Table 2).
| miRNA | P value | HR | Lower 95%CI | Upper 95%CI |
| hsa-miR-138-5p | 0.0001 | 1.0024 | 1.0011 | 1.0036 |
| hsa-miR-1179 | 0.0006 | 1.0120 | 1.0050 | 1.0191 |
| hsa-miR-138-1-3p | 0.0009 | 1.0280 | 1.0112 | 1.0451 |
| hsa-miR-612 | 0.0064 | 1.3720 | 1.0929 | 1.7222 |
| hsa-miR-7-2-3p | 0.0077 | 1.0109 | 1.0028 | 1.0191 |
| hsa-miR-146b-3p | 0.0165 | 0.9999 | 0.9998 | 0.9999 |
| hsa-miR-181a-2-3p | 0.0177 | 0.9999 | 0.9998 | 0.9999 |
| hsa-miR-146b-5p | 0.0224 | 0.9999 | 0.9999 | 0.9999 |
| hsa-miR-5682 | 0.0308 | 1.8605 | 1.0587 | 3.2696 |
| hsa-miR-31-5p | 0.0320 | 0.9952 | 0.9908 | 0.9995 |
| hsa-miR-424-3p | 0.0393 | 0.9790 | 0.9594 | 0.9989 |
| hsa-miR-6842-3p | 0.0437 | 0.9457 | 0.8959 | 0.9984 |
| hsa-miR-4709-3p | 0.0471 | 0.9702 | 0.9416 | 0.9996 |
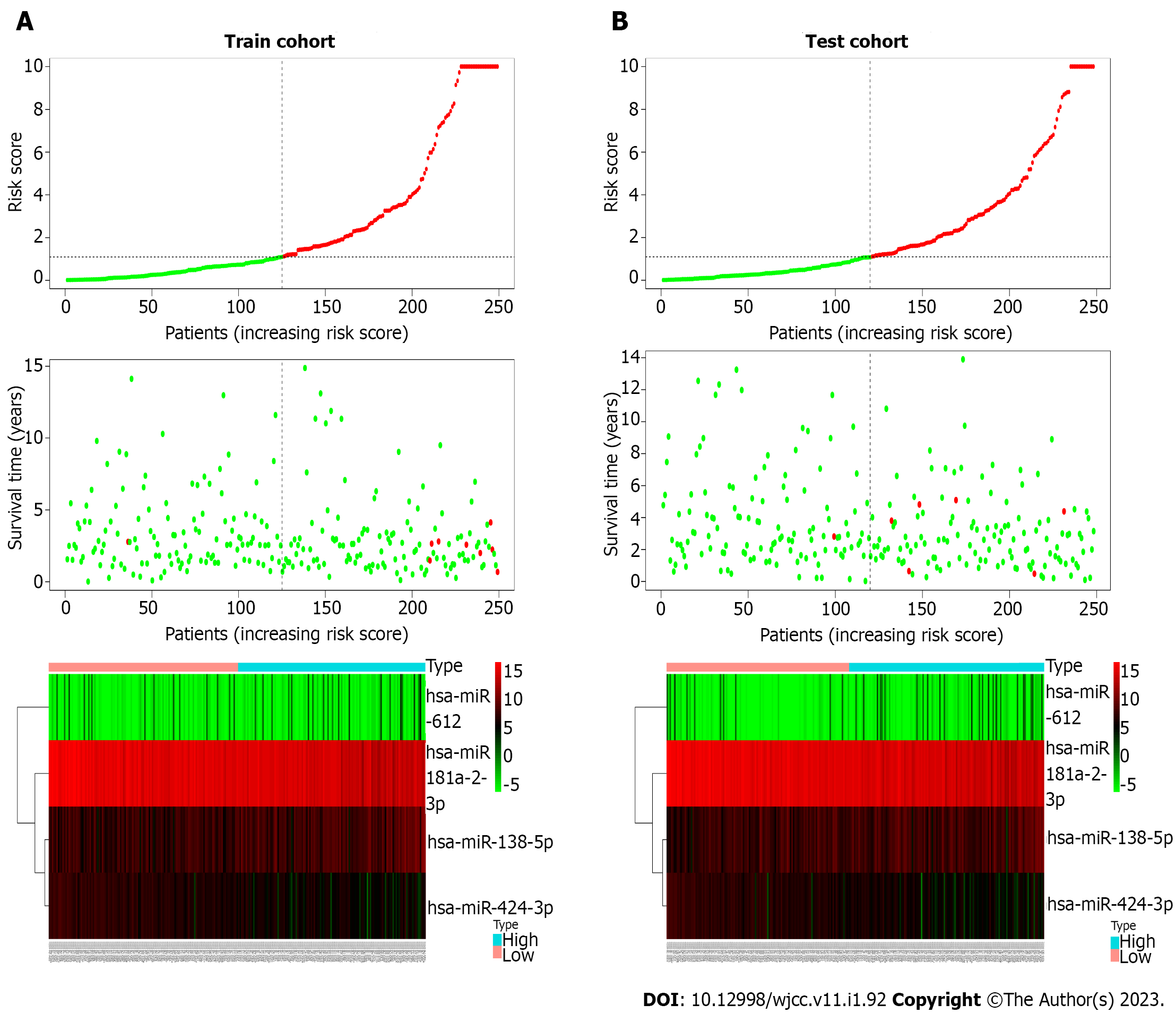
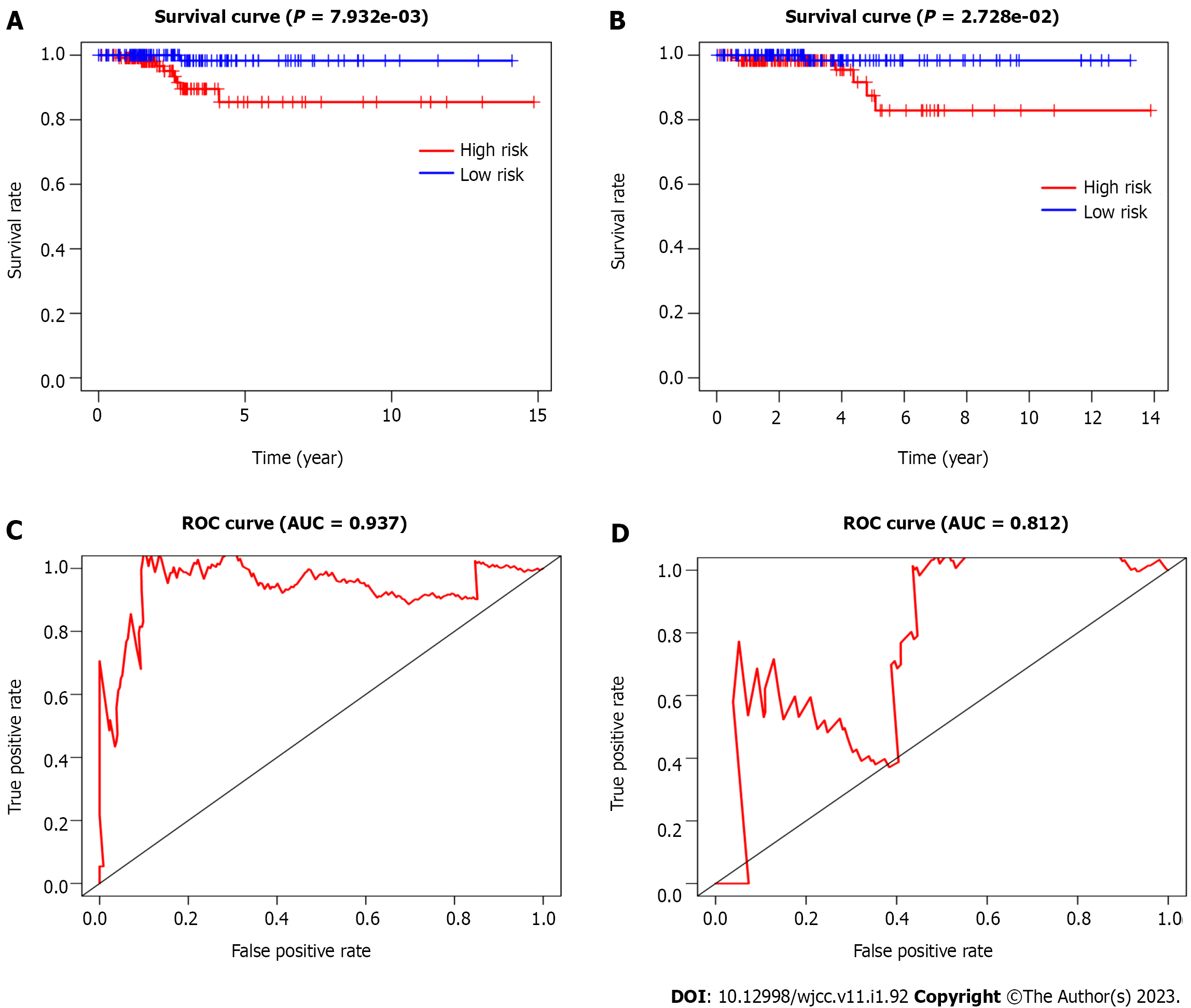
To construct the miRNA prognostic signature, these 13 miRNAs were further selected into the LASSO and multivariate cox regression analyses. The lambda value was set using lambda min, which is the value of lambda, to obtain the minimum mean cross-validated error. Four miRNAs with non-zero coefficients were defined. We finally managed to establish four miRNAs that had an independent prognostic effect for PTC in the training cohort based on the LASSO and Cox regression models (Table 3). To clearly reveal the weight of each weighting coefficient of miRNAs, a forest figure is presented in Supplementary Figure 1. We then chose the same median risk score in the above two independent cohorts as the cutoff point, classifying patients into the low-risk and high-risk groups. Figure 2 shows the distribution of the miRNA-based risk score, OS, and four miRNAs expression profiles of the training and testing cohorts. The Kaplan-Meier survival analysis indicated a much worse prognosis (P < 0.001) in the high-risk group (Figure 3A and C).
| miRNA | Coefficient | P value | HR | Lower 95%CI | Upper 95%CI |
| hsa-miR-181a-2-3p | -0.001 | 0.0800 | 1.000 | 1.000 | 1.000 |
| hsa-miR-138-5p | 0.003 | 0.0001 | 1.003 | 1.001 | 1.004 |
| hsa-miR-424-3p | -0.018 | 0.0590 | 0.982 | 0.964 | 1.001 |
| hsa-miR-612 | 0.284 | 0.0242 | 1.329 | 1.038 | 1.702 |
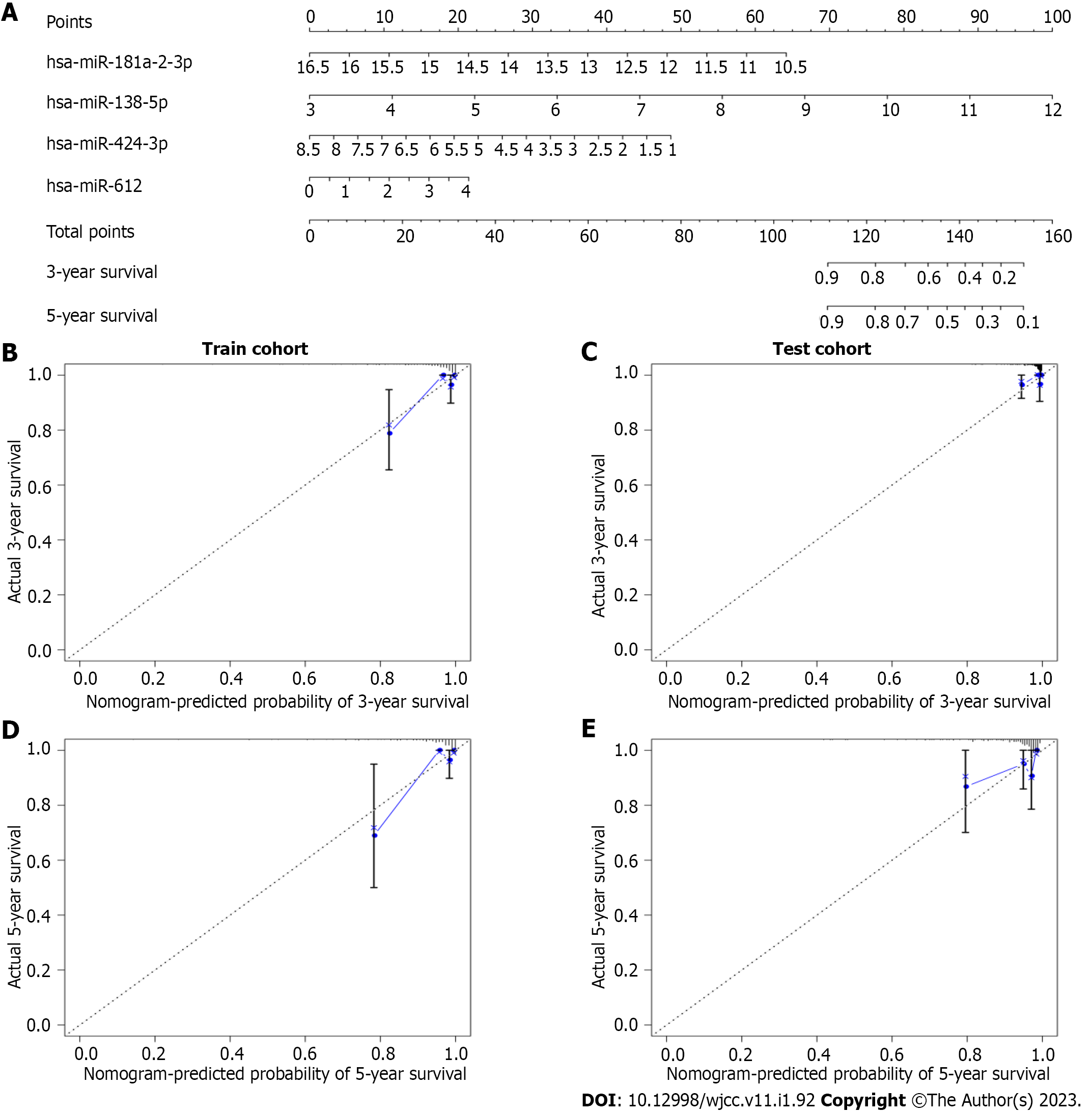
Next, we conducted a time-dependent ROC curve analysis to assess the performance of the four-miRNA signature in predicting the PTC prognosis. The AUC values of the four-miRNA signature at five years were 0.937 and 0.812 in the training and testing cohorts, respectively (Figure 3B and D). To make the prognostic miRNA signatures more convenient in clinical practice, a four-miRNA-based nomogram was established (Figure 4A). Furthermore, calibration plots of the four-miRNA-based prognostic model showed compactness in the training and testing cohorts for the 3- and 5-year survival rates, respectively, which indicated good calibration ability (Figure 4B-E).
The predictive effect of the four-miRNA signature on OS considering the clinicopathological features was evaluated using the univariate and multivariate Cox regression analyses. The multivariate Cox regression analysis showed that the four-miRNA signature was an independent prognostic factor associated with the OS of PTC patients (Table 4).
| Univariate analysis | Multivariate analysis | |||
| HR (95%CI) | P | HR (95%CI) | P | |
| Age (< 45 vs ≥ 45) | 0.013 (0.000-0.756) | 0.036 | - | - |
| Stage (I/II vs III/IV) | 0.141 (0.045-0.439) | 0.001 | - | - |
| Sex | 2.029 (0.734-5.613) | 0.173 | - | - |
| Four miRNA signature (high risk vs low risk) | 8.625 (1.956-38.03) | 0.004 | 6.186 (1.405-27.24) | 0.016 |
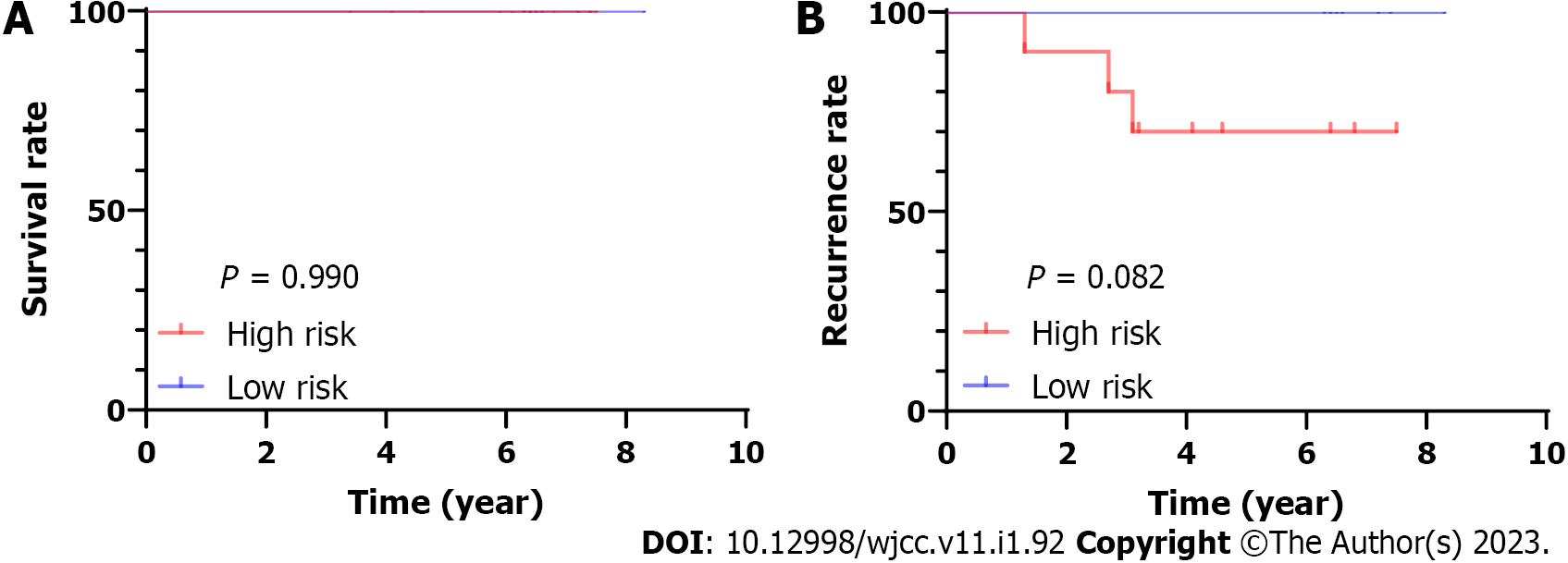
We performed whole-transcriptome sequencing of 19 paired-PTC tissue samples[14]. We obtained the expression of these four miRNAs and then calculated their risk score based on the four-miRNA signature in 19 PTC samples. Then, we chose the median risk score as the cutoff point, classifying patients into the low-risk and high-risk groups. We found that there was no significant difference in OS time between the high-risk and low-risk groups (P = 0.99). There was a trend indicated that high-risk patients had shorter relapse-free survival, although statistical significance was not reached. (P = 0.082, Figure 5).
The target genes of the selected four miRNAs, hsa-miR-181a-2-3p, hsa-miR-138-5p, hsa-miR-424-3p, and hsa-miR-612, were predicted by TargetScan and miRDB. Then, the biological roles of the overlapped target genes were assessed using an enrichment analysis. The GO biological processes were mainly enriched in the regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolism; transport; regulation of gene expression; epigenetics; and cell adhesion (Table 5). In addition, the KEGG pathways were remarkably enriched in the integrin family cell surface interactions, beta1 integrin cell surface interactions, and proteoglycan and syndecan-mediated signaling events (Table 6).
| Term | Count | Fold enrichment | P value |
| Regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 291 | 1.228 | < 0.001 |
| Transport | 127 | 1.247 | 0.005 |
| Regulation of gene expression, epigenetic | 12 | 2.171 | 0.008 |
| Cell adhesion | 12 | 2.653 | 0.001 |
| Term | Count | Fold enrichment | P value |
| Integrin family cell surface interactions | 149 | 1.265 | < 0.001 |
| Beta1 integrin cell surface interactions | 147 | 1.273 | < 0.001 |
| Proteoglycan syndecan-mediated signaling events | 146 | 1.267 | < 0.001 |
| PAR1-mediated thrombin signaling events | 145 | 1.306 | < 0.001 |
| Thrombin/PAR pathway | 145 | 1.305 | < 0.001 |
| VEGF and VEGFR signaling network | 145 | 1.301 | < 0.001 |
| Alpha9 beta1 integrin signaling events | 145 | 1.300 | < 0.001 |
| ErbB receptor signaling network | 145 | 1.294 | < 0.001 |
| S1P pathway | 145 | 1.294 | < 0.001 |
| TRAIL signaling pathway | 145 | 1.277 | < 0.001 |
In the era of precision medicine, molecular biomarker-guided treatment and more accurate patient survival prediction are demanding a prompt solution. These efforts have been enormously successful in the field of PTC. With the increasing use of thyroid fine-needle aspiration cytology, the classification of indeterminate nodules is becoming more accurate. However, approximately 20% of nodules lack specific characteristics in cytology[15]. ThyroSeq v3 Genomic Classifier is a tool that includes a variety of thyroid cancer-related point mutations, gene fusions, copy number variations, and gene expression alterations to achieve both high sensitivity and specificity in differentiating benign and malignant thyroid nodules sampled by FNA biopsy[16]. According to the latest research report, the ThyroSeq v3 demonstrated 94% sensitivity and 82% specificity of thyroid nodules with Bethesda III and IV cytology[17]. Moreover, a BRAF V600E mutation was reported in many studies associated with poorer clinicopathological outcomes of PTC[18]. Our previous studies showed that the co-existence of BRAF V600E and TERT promoter mutations was related to high-risk clinicopathological features of PTC[19].
With the advancement of sequencing technology and bioinformatics, we can obtain comprehensive DNA epigenetics, mRNA expression profiles, noncoding RNAs, and proteomics data. In the future, more molecular biomarker-based approaches will be developed based on different molecular data. As miRNAs are relatively stable and easily detected, they are promising biomarkers in the clinic[20-22]. In 2018, Liu et al[23] constructed a two-miRNA (hsa-miR-181a-2-3p and hsa-miR-138-1-3p) signature for a PTC prognosis assessment. The AUC value of the two-miRNA signature was 0.784. Additionally, Xiong et al[24] constructed another four-miRNA signature (hsa-miR-6843, hsa-miR-6730, hsa-miR-196a-2, and hsa-miR-206) as a potential prognostic biomarker in PTC. The AUC values of the four-miRNA signature were 0.886 and 0.882 in the training and testing cohorts, respectively. In this study, we analyzed large-scale miRNA sequencing data in the TCGA database and selected a total of 2203 miRNAs to provide a more comprehensive analysis. Consequently, we confirmed a four-miRNA signature that had a strong prognostic effect on the prognosis of PTC with relatively higher credibility. Although there was no statistical difference, we found a trend for shorter survival times in the high-risk group compared with the low-risk group in the sequencing cohort. This may be related to the inadequate sample size and short follow-up period.
It is widely recognized that miRNAs are pivotal regulators of gene expression in complex cellular processes, including cancer cell proliferation, metastasis, migration, and apoptosis[25]. The four miRNAs found in this study may be potential oncogene or tumor suppressor genes in PTC and independent predictors of PTC prognosis. Among these four miRNAs, hsa-miR-181a-2-3p has been reported as a potential prognostic indicator of PTC. However, its specific function and molecular mechanism in PTC have not been reported[23], and the biological functions of the remaining miRNAs remain unclear.
He et al[26] analyzed the miRNA expression of peripheral blood between healthy people and lung cancer patients and found a significant difference in the hsa-miR-138-5p expression in two groups. Hsa-miR-424-3p may be a key miRNA for liver cancer, which was involved in telomere maintenance via telomerase, protein sumoylation, the histone mRNA metabolic process, and angiotensin maturation[27]. However, many studies suggest that the down-regulation of hsa-miR-612 was involved in a variety of signal pathways that affect the pathophysiology of gastric cancer, liver cancer, and ovarian cancer. However, its relationship with PTC remains unclear[28-30]. As a result, it is necessary to conduct additional in vitro and in vivo experiments investigations using these four miRNAs.
In conclusion, this study indicated that the four-miRNA signature could accurately predict the prognosis of PTC. More intensive treatment and closer follow-ups for high-risk PTC patients are recommended.
Papillary thyroid cancer is a highly heterogeneous disease and therefore molecular markers need to be established to predict its prognosis.
The present study aims to explore novel markers consisting of microRNA (miRNA)-associated signatures for papillary thyroid cancer (PTC) prognostication.
To establish a practical tool has satisfying potential in stratifying PTC patients and individualized therapy to avoid overtreatment or inadequate treatment.
In this study, a panel of four miRNAs is generated as a prognostic signature, which is then tested in PTC patients.
The panel of four miRNAs could reliably distinguished PTC patients from high and low risk with a significant difference in the overall survival.
More intensive treatment and closer follow-ups for high-risk PTC patients are recommended.
Our prognostic signature contributed to individualized therapy to avoid overtreatment or inadequate treatment.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Hu YL, China; Kai K, Japan S-Editor: Chen YL L-Editor: A P-Editor: Chen YL
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13167] [Article Influence: 1881.0] [Reference Citation Analysis (4)] |
| 2. | Kilfoy BA, Zheng T, Holford TR, Han X, Ward MH, Sjodin A, Zhang Y, Bai Y, Zhu C, Guo GL, Rothman N. International patterns and trends in thyroid cancer incidence, 1973-2002. Cancer Causes Control. 2009;20:525-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 525] [Cited by in RCA: 495] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 3. | Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11065] [Cited by in RCA: 12187] [Article Influence: 1523.4] [Reference Citation Analysis (3)] |
| 4. | Wiltshire JJ, Drake TM, Uttley L, Balasubramanian SP. Systematic Review of Trends in the Incidence Rates of Thyroid Cancer. Thyroid. 2016;26:1541-1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 269] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 5. | McHenry CR, Stulberg JJ. Prophylactic central compartment neck dissection for papillary thyroid cancer. Surg Clin North Am. 2014;94:529-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Morris LG, Sikora AG, Tosteson TD, Davies L. The increasing incidence of thyroid cancer: the influence of access to care. Thyroid. 2013;23:885-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 384] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 7. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8847] [Cited by in RCA: 9285] [Article Influence: 464.3] [Reference Citation Analysis (0)] |
| 8. | Lee JC, Gundara JS, Glover A, Serpell J, Sidhu SB. MicroRNA expression profiles in the management of papillary thyroid cancer. Oncologist. 2014;19:1141-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Aragon Han P, Weng CH, Khawaja HT, Nagarajan N, Schneider EB, Umbricht CB, Witwer KW, Zeiger MA. MicroRNA Expression and Association with Clinicopathologic Features in Papillary Thyroid Cancer: A Systematic Review. Thyroid. 2015;25:1322-1329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 10. | Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 354] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 11. | Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, Chakravarty D, Daian F, Gao Q, Bailey MH, Liang WW, Foltz SM, Shmulevich I, Ding L, Heins Z, Ochoa A, Gross B, Gao J, Zhang H, Kundra R, Kandoth C, Bahceci I, Dervishi L, Dogrusoz U, Zhou W, Shen H, Laird PW, Way GP, Greene CS, Liang H, Xiao Y, Wang C, Iavarone A, Berger AH, Bivona TG, Lazar AJ, Hammer GD, Giordano T, Kwong LN, McArthur G, Huang C, Tward AD, Frederick MJ, McCormick F, Meyerson M; Cancer Genome Atlas Research Network, Van Allen EM, Cherniack AD, Ciriello G, Sander C, Schultz N. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018;173:321-337.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2333] [Cited by in RCA: 2120] [Article Influence: 302.9] [Reference Citation Analysis (0)] |
| 12. | Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22632] [Cited by in RCA: 29137] [Article Influence: 1821.1] [Reference Citation Analysis (0)] |
| 13. | Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, Bacic A, Hill AF, Stroud DA, Ryan MT, Agbinya JI, Mariadason JM, Burgess AW, Mathivanan S. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597-2601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 968] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 14. | Wang QX, Wang OC, Chen ED, Cai YF, Li Q, Jin YX, Jin WX, Wang YH, Zheng ZC, Xue L, Zhang XH. Erratum to: A panel of four genes accurately differentiates benign from malignant thyroid nodules. J Exp Clin Cancer Res. 2017;36:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 15. | Bongiovanni M, Spitale A, Faquin WC, Mazzucchelli L, Baloch ZW. The Bethesda System for Reporting Thyroid Cytopathology: a meta-analysis. Acta Cytol. 2012;56:333-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 660] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 16. | Nikiforova MN, Mercurio S, Wald AI, Barbi de Moura M, Callenberg K, Santana-Santos L, Gooding WE, Yip L, Ferris RL, Nikiforov YE. Analytical performance of the ThyroSeq v3 genomic classifier for cancer diagnosis in thyroid nodules. Cancer. 2018;124:1682-1690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 280] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 17. | Steward DL, Carty SE, Sippel RS, Yang SP, Sosa JA, Sipos JA, Figge JJ, Mandel S, Haugen BR, Burman KD, Baloch ZW, Lloyd RV, Seethala RR, Gooding WE, Chiosea SI, Gomes-Lima C, Ferris RL, Folek JM, Khawaja RA, Kundra P, Loh KS, Marshall CB, Mayson S, McCoy KL, Nga ME, Ngiam KY, Nikiforova MN, Poehls JL, Ringel MD, Yang H, Yip L, Nikiforov YE. Performance of a Multigene Genomic Classifier in Thyroid Nodules With Indeterminate Cytology: A Prospective Blinded Multicenter Study. JAMA Oncol. 2019;5:204-212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 313] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 18. | Xing M, Westra WH, Tufano RP, Cohen Y, Rosenbaum E, Rhoden KJ, Carson KA, Vasko V, Larin A, Tallini G, Tolaney S, Holt EH, Hui P, Umbricht CB, Basaria S, Ewertz M, Tufaro AP, Califano JA, Ringel MD, Zeiger MA, Sidransky D, Ladenson PW. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J Clin Endocrinol Metab. 2005;90:6373-6379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 716] [Cited by in RCA: 725] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 19. | Jin L, Chen E, Dong S, Cai Y, Zhang X, Zhou Y, Zeng R, Yang F, Pan C, Liu Y, Wu W, Xing M, Wang O. BRAF and TERT promoter mutations in the aggressiveness of papillary thyroid carcinoma: a study of 653 patients. Oncotarget. 2016;7:18346-18355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 20. | Qiu J, Zhang W, Zang C, Liu X, Liu F, Ge R, Sun Y, Xia Q. Identification of key genes and miRNAs markers of papillary thyroid cancer. Biol Res. 2018;51:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 21. | Mazeh H, Deutch T, Karas A, Bogardus KA, Mizrahi I, Gur-Wahnon D, Ben-Dov IZ. Next-Generation Sequencing Identifies a Highly Accurate miRNA Panel That Distinguishes Well-Differentiated Thyroid Cancer from Benign Thyroid Nodules. Cancer Epidemiol Biomarkers Prev. 2018;27:858-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Nixon AM, Provatopoulou X, Kalogera E, Zografos GN, Gounaris A. Circulating thyroid cancer biomarkers: Current limitations and future prospects. Clin Endocrinol (Oxf). 2017;87:117-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 23. | Liu T, You X, Sui J, Shen B, Zhang Y, Zhang XM, Yang S, Yao YZ, Yang F, Yin LH, Pu YP, Liang GY. Prognostic value of a two-microRNA signature for papillary thyroid cancer and a bioinformatic analysis of their possible functions. J Cell Biochem. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Chengfeng X, Gengming C, Junjia Z, Yunxia L. MicroRNA signature predicts survival in papillary thyroid carcinoma. J Cell Biochem. 2019;120:17050-17058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Hill M, Tran N. MicroRNAs Regulating MicroRNAs in Cancer. Trends Cancer. 2018;4:465-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 26. | He Q, Fang Y, Lu F, Pan J, Wang L, Gong W, Fei F, Cui J, Zhong J, Hu R, Liang M, Fang L, Wang H, Yu M, Zhang ZF. Analysis of differential expression profile of miRNA in peripheral blood of patients with lung cancer. J Clin Lab Anal. 2019;33:e23003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 27. | Wang X, Gao J, Zhou B, Xie J, Zhou G, Chen Y. Identification of prognostic markers for hepatocellular carcinoma based on miRNA expression profiles. Life Sci. 2019;232:116596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 28. | Zhu Y, Zhang HL, Wang QY, Chen MJ, Liu LB. Overexpression of microRNA-612 Restrains the Growth, Invasion, and Tumorigenesis of Melanoma Cells by Targeting Espin. Mol Cells. 2018;41:119-126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 29. | Yu H, Xu Y, Zhang D, Liu G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem Biophys Res Commun. 2018;503:2095-2100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 30. | Wang L, Bo X, Zheng Q, Ge W, Liu Y, Li B. Paired box 8 suppresses tumor angiogenesis and metastasis in gastric cancer through repression of FOXM1 via induction of microRNA-612. J Exp Clin Cancer Res. 2018;37:159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |