Published online Mar 6, 2022. doi: 10.12998/wjcc.v10.i7.2072
Peer-review started: July 15, 2021
First decision: September 5, 2021
Revised: September 24, 2021
Accepted: February 10, 2022
Article in press: February 10, 2022
Published online: March 6, 2022
Processing time: 229 Days and 22.6 Hours
The N-Myc downstream-regulated gene (NDRG) family is comprised of four members (NDRG1-4) involved in various important biological processes. However, there is no systematic evaluation of the prognostic of the NDRG family in hepatocellular carcinoma (HCC).
To analyze comprehensively the biological role of the NDRG family in HCC.
The NDRG family expression was explored using The Cancer Genome Atlas. DNA methylation interactive visualization database was used for methylation analysis of the NDRG family. The NDRG family genomic alteration was assessed using the cBioPortal. Single-sample Gene Set Enrichment Analysis was used to determine the degree of immune cell infiltration in tumors.
NDRG1 and NDRG3 were up-regulated in HCC, while NDRG2 was down-regulated. Consistent with expression patterns, high expression of NDRG1 and NDRG3 was associated with poor survival outcomes (P < 0.05). High expression of NDRG2 was associated with favorable survival (P < 0.005). An NDRG-based signature that statistically stratified the prognosis of the patients was constructed. The percentage of genetic alterations in the NDRG family varied from 0.3% to 11.0%, and the NDRG1 mutation rate was the highest. NDRG 1-3 expression was associated with various types of infiltrated immune cells. Gene ontology analysis revealed that organic acid catabolism was the most important biological process related to the NDRG family. Gene Set Enrichment Analysis showed that metabolic, proliferation, and immune-related gene sets were enriched during NDRG1 and NDRG3 high expression and NDRG2 low expression.
Overexpression of NDRG1 and NDRG3 and down-expression of NDRG2 are correlated with poor overall HCC prognosis. Our results may provide new insights into the indispensable role of NDRG1, 2, and 3 in the development of HCC and guide a promising new strategy for treating HCC.
Core Tip: The N-Myc downstream-regulated gene (NDRG) family comprised of four members (NDRG1-4) is involved in various important biological processes. However, there is no systematic evaluation of the prognostic of the NDRG family in hepatocellular carcinoma (HCC). We aim to analyze systematically the role of the NDRG family in HCC by various bioinformatics tools. We found that NDRG1-3 may be the promising biomarker for the diagnosis and prognosis of HCC and may provide promising new targets and strategies for HCC treatment.
- Citation: Yin X, Yu H, He XK, Yan SX. Prognostic and biological role of the N-Myc downstream-regulated gene family in hepatocellular carcinoma. World J Clin Cases 2022; 10(7): 2072-2086
- URL: https://www.wjgnet.com/2307-8960/full/v10/i7/2072.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i7.2072
Liver cancer is currently a leading cause of cancer-related deaths globally, with almost 800000 deaths per year[1,2]. The incidence of liver cancer has increased rapidly over recent years[3,4]. Hepatocellular carcinoma (HCC) represents the major histological type of primary liver cancer (around 90%), placing a heavy burden on the healthcare system[3]. Despite significant advances in HCC management, the prognosis remains inferior[5,6]. High frequencies of postoperative recurrence and metastasis are considered responsible for most deaths. To date, the molecular characteristics of HCC remain poorly understood. Thus, it is extremely crucial to provide insights into the genetic events driving hepatocarcinogenesis and identify reliable biomarkers for HCC detection and treatment.
The N-Myc downstream-regulated gene (NDRG) family comprises NDRG1, NDRG2, NDRG3, and NDRG4, located on different chromosomes[7]. The five amino acids at the C-terminus of NDRG1-4 are evolutionary conserved. Sequence differences between NDRG1–4 are in the N- and C-terminal regions (except for five amino acid residues in the C-terminal)[8]. Increasing studies demonstrate that NDRG1-4 contribute to cell proliferation, differentiation, development, and immune response[9-11]. Recent studies focused on NDRG1-4 biological function in cancer progression and metastasis[12-14]. For instance, NDRG1 had been proved to act as a metastasis suppressor in multiple cancer types like glioma, prostate, and colorectal cancer[15-17]; this makes treatment against NDRG1 a plausible anti-metastatic therapy. NDRG2 played a tumor suppressor role in glioblastoma[18], liver cancer[19], and colorectal cancer[20]. NDRG3 expression was also elevated in colorectal cancer[21], laryngeal cancer[22], and lung cancer[23] and associated with worse overall survival.
Nevertheless, the prognostic role and potential biological functions of the NDRG family in HCC are far from fully demonstrated. The present study addressed these limitations by utilizing public databases and integrative bioinformatics analysis. The expression patterns and prognostic significance of the NDRG family in HCC were comprehensively analyzed. In addition, the relationship between the NDRG gene family expression and the immune infiltration level in HCC was also investigated. Finally, we explored the potential biological processes of the NDRG gene family involved in HCC by various gene enrichment analyses. This study aimed to analyze comprehensively expression patterns and biological functions of the NDRG family.
The gene and protein expression data of the NDRG family (NDRG1-4) were collected from Gene Expression Profiling Interactive Analysis (http:// gepia.cancer-pku.cn/index.html)[24], N-Myc database (www.oncomine.org), and Human Protein Atlas (www.proteinatlas.org)[25]. The RNA-seq data and related clinical information (age, gender, mutation, and survival information) of liver hepatocellular carcinoma (LIHC) were downloaded from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/)[26]. DNA Methylation Interactive Visualization Database was used for methylation analysis of the NDRG family[27]. The genomic alteration of the NDRG family was assessed using the cBioPortal[28].
To build the NDRG family-based signature, the multiply Cox model was used to screen candidate prognostic genes that are significantly associated with OS. Finally, the risk score formula was calculated using the regression coefficients from the multiply Cox model. The risk score of each patient was calculated by the formula of the prognostic score, and patients were divided into high and low-risk groups. The optimal cutoff value was obtained by the R package (OptimalCutpoints)[29]. Besides, the prognostic performances of the NDRG signature over 1-, 2-, and 3-year were compared by the receiver operating characteristics (ROC) curve (R package, survival ROC)[30].
The DNA methylation of CG sites in the gene promoter regions of NDRG1-4 among HCC tissues was obtained from the DNA Methylation Interactive Visualization Database[27]. The methylation level of NDRG1-4 between tumors and normal samples was compared by the Student’s t-test. The Spearman’s rank correlation between methylation levels and messenger RNA (mRNA) expressions of NDRG1-4 was calculated. The genomic alteration of NDRG was download and analyzed using cBioPortal[28]. The distribution and frequency of somatic mutation, amplification, and deletion data of NDRG1-4 were evaluated.
Single-sample Gene Set Enrichment Analysis (GSEA) was used to determine the degree of immune cell infiltration in tumors[31,32]. Immune cell subpopulation gene signatures were collected from lectures. Twenty-four immune cell of [B cells, T follicular helper (TFH), T helper (Th)1, Th2, Th17, Treg, Tγδ, T effector memory, T central memory, CD8+ T cells, mast cells, neutrophils, eosinophils, macrophages, plasmacytoid dendritic cells (DCs), inactivated DCs, activated DCs, CD56dim natural killer (NK) cells, and CD56bright NK cells] were identified from the previous lectures[33,34]. The Spearman’s rank correlation between levels of immune subpopulations and NDRG expression was calculated.
To explore the underlying biological function of NDRG1-3, two different bioinformatics ways were used. The correlation coefficient between NDRG1-3 and other genes was calculated, and the top 500 closely related genes were chosen for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis[35,36]. Annotation and visualization of results were performed by Clusterprolfe[37]. Besides, high expression (top 30%) and low expression (bottom 30%) groups of HCC patients were divided according to NDRG1-4 expression. The GSEA was performed between high- and low-NDRG groups.
Wilcoxon signed-rank test was adopted to compare NDRG1-4 expression between tumor and adjacent nontumorous tissues. Sample paired t-test was used to compare the differential expression of NDRG1-4 among paired samples. The univariate and multivariate Cox regression analyses were performed to evaluate and identify independent prognostic predictors for overall survival (OS). A P value < 0.05 was considered statistically significant, and all P values were two-tailed. All statistical analyses were performed by R software (version 3.6.0).
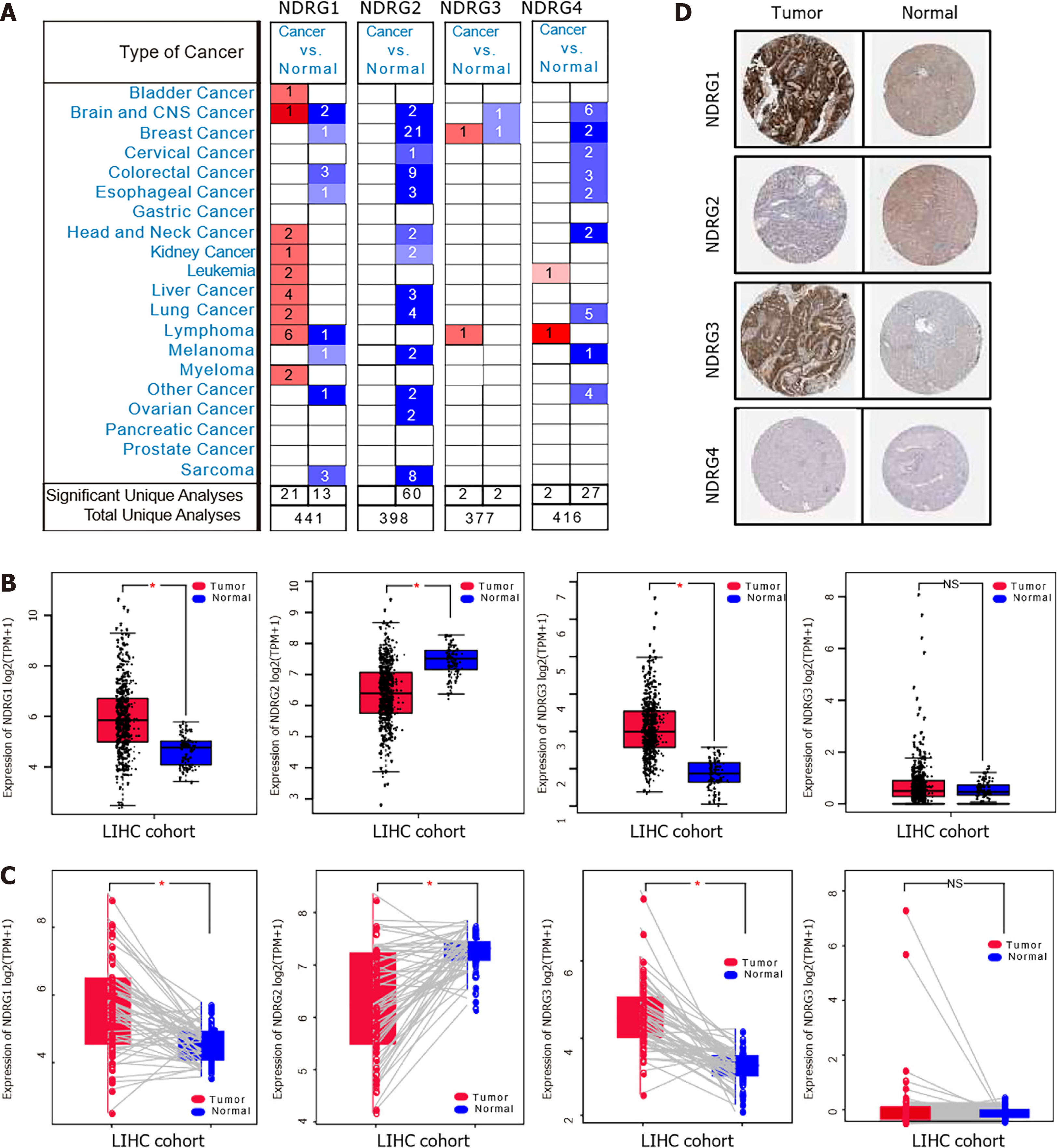
The mRNA expression patterns of NDRG1-4 were retrieved from Oncomine and TCGA databases, as shown in Figure 1A. NDRG1 was significantly up-regulated in 21 datasets (including lymphoma, liver cancer, and lung cancer) and down-regulated in 13 datasets (including sarcoma, colorectal cancer, and brain and CNS cancer). NDRG2 was significantly and consistently down-regulated in 60 datasets (including breast cancer, colorectal cancer, and sarcoma). NDRG3 was up-regulated in two datasets (breast cancer and lymphoma) and down-regulated in two datasets (brain cancer, CNS cancer, and breast cancer). The expression of NDRG4 was significantly increased in two datasets (lymphoma and leukemia) and decreased in 27 datasets (including brain and CNS cancer, lung cancer, and colorectal cancer). These data indicated apparent heterogeneity among NDRG1-4 expression patterns in different types of cancer. To validate further NDRG1-4 expression patterns in multiple cancer types, NDRG1-4 dysregulation in the TCGA pan-cancer cohort was analyzed. As shown in Supplementary Figure 1, in most cancer types, NDRG1 and NDRG3 were up-regulated, while NDRG2 and NDRG4 were down-regulated.
Next, we focus on NDRG1-4 dysregulation in HCC. The mRNA levels of NDRG1 and NDRG3 were significantly increased in tumor samples (Figure 1B). At the same time, NDRG2 expression was significantly decreased in HCC. No significant difference in NDRG4 expression was observed between tumor and normal samples. These patterns between tumor samples and their paired normal samples were also validated (Figure 1C). Noteworthy, immunohistochemistry shows that NDRG1 and NDRG3 protein levels were elevated, while NDRG2 protein was significantly reduced in the HCC (Figure 1D). Together, these data provided strong evidence that NDRG1 and NDRG3 may have an oncogenic role in HCC and NDRG2 may have an oncosuppressive role.
To assess the relevance of NDRG family genes in a clinical context, the NDRG gene family expression concerning different clinicopathological features was explored (Supplementary Figure 2). Higher expression of NDRG1 was associated with advanced-stage (III/IV), higher T stage (T3/T4), higher grade (G3/G4), and micro/macrovascular invasion. Lower expression of NDRG2 was associated with younger age (≤ 65), earlier stage (I/II), and lower grade (G1/G2). Higher expression of NDRG3 was associated with younger age (≤ 65), gender (female), advanced stage (III/IV), higher T stage (T3/T4), higher grade (G3/G4), and micro/macrovascular invasion. Higher NDRG4 expression was associated with gender (female) and no-hepatitis B virus infection.
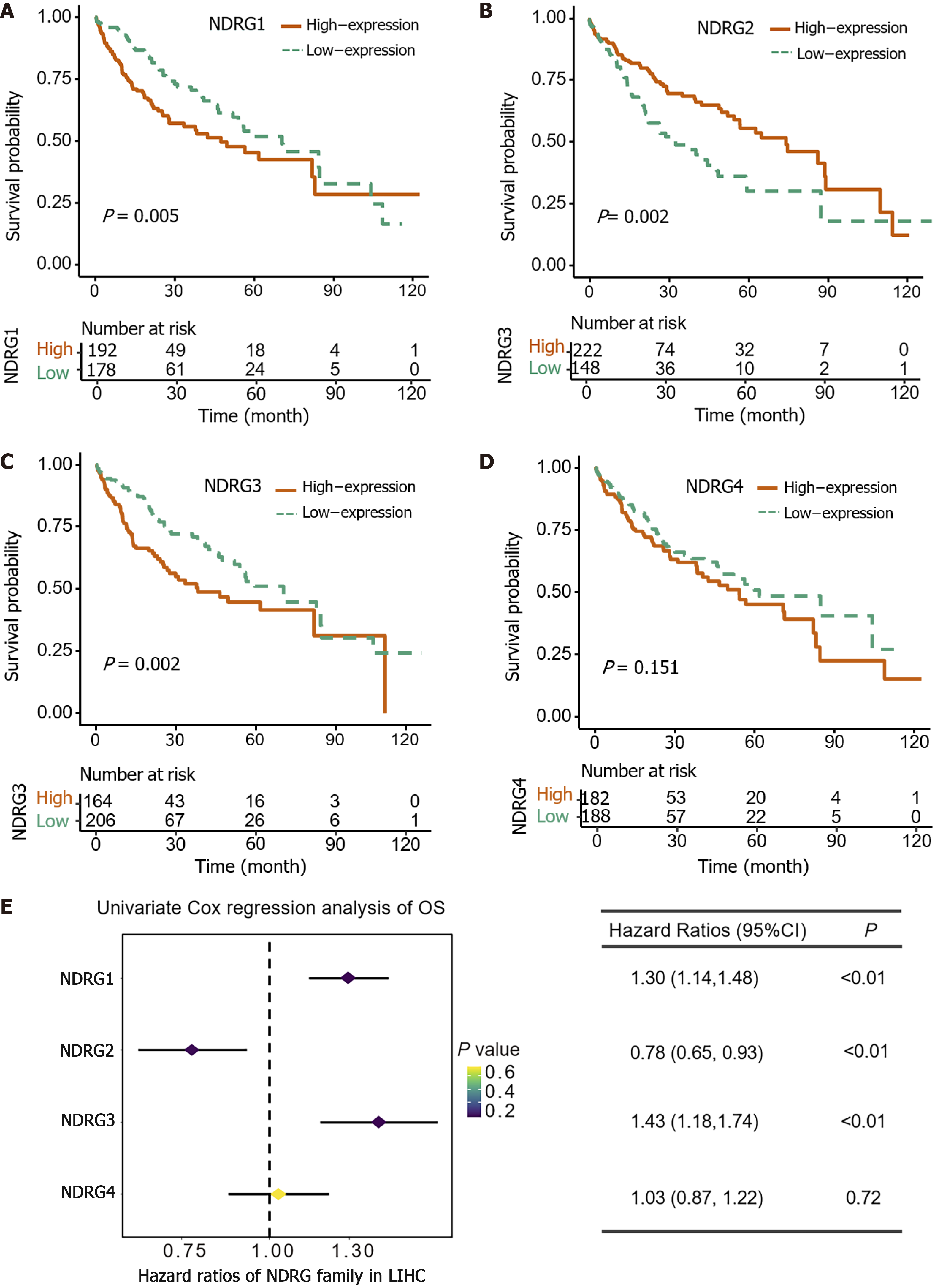
Furthermore, the prognostic role of the NDRG genes family in the LIHC cohort was evaluated. Consistent with expression patterns, higher expression of NDRG1 and NDRG3 was associated with worse survival outcomes (P < 0.05) (Figure 2A and C). Higher NDRG2 expression was associated with better survival outcomes (p<0.005) (Figure 2B). No significant relationship was found between NDRG4 and survival outcome (Figure 2D). In the univariate Cox’s regression analysis of OS, NDRG1 and NDRG3 were significantly associated with worse OS [hazard ratio (HR) = 1.30, 95% confidence interval (CI): 1.14-1.48, P < 0.01; HR = 1.43, 95%CI: 1.18-1.74, P < 0.01, Figure 2E], while NDRG2 showed a significant association with better OS (HR = 0.78, 95%CI: 0.65-0.93, P < 0.01, Figure 2E). NDRG4 had no significant correlation with OS (HR = 1.03, 95%CI: 0.87-1.22, P = 0.72, Figure 2E).
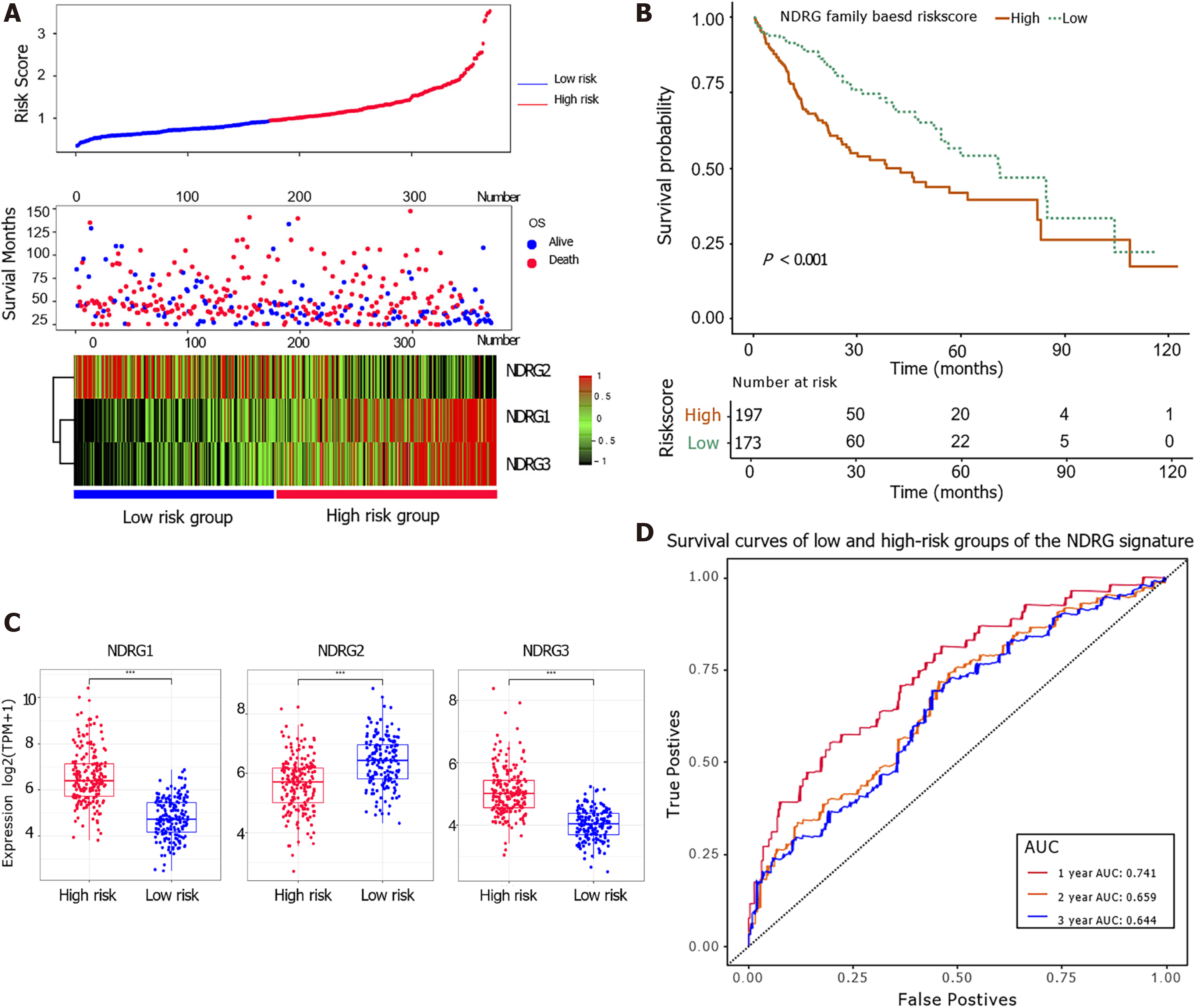
The multivariable Cox proportional hazards regression model was used to construct prognostic signatures to assess the prognosis risk of HCC patients based on their NDRG gene expression. Three prognostic genes of the NDRG family (NDRG1, NDRG2, and NDRG3) were included in the prognostic score. The following equation constructed a signature risk score, risk score = 0.33 × NDRG1expr – 0.16 × NDRG2expr + 0.34 × NDRG3expr. Patients' risk scores in the TCGA cohort were calculated, and patients were assigned into high-risk and low-risk groups based on their scores. Patients with high risk had shorter survival and higher NDRG1 and NDRG3 expression (Figure 3A). Kaplan-Meier analysis showed that patients with a higher score were associated with worse OS (Figure 3B). NDRG1 and NDRG3 were up-regulated in the high-risk group, while NDRG2 was down-regulated in the low-risk group (Figure 3C). The ROC analysis further indicated that the NDRG-based signature could statistically stratify patients' prognoses (Figure 3D). The area under the curve of 1-year survival in the total cohort was 0.741, indicating the good prognostic performance of the NDRG family-based signature.
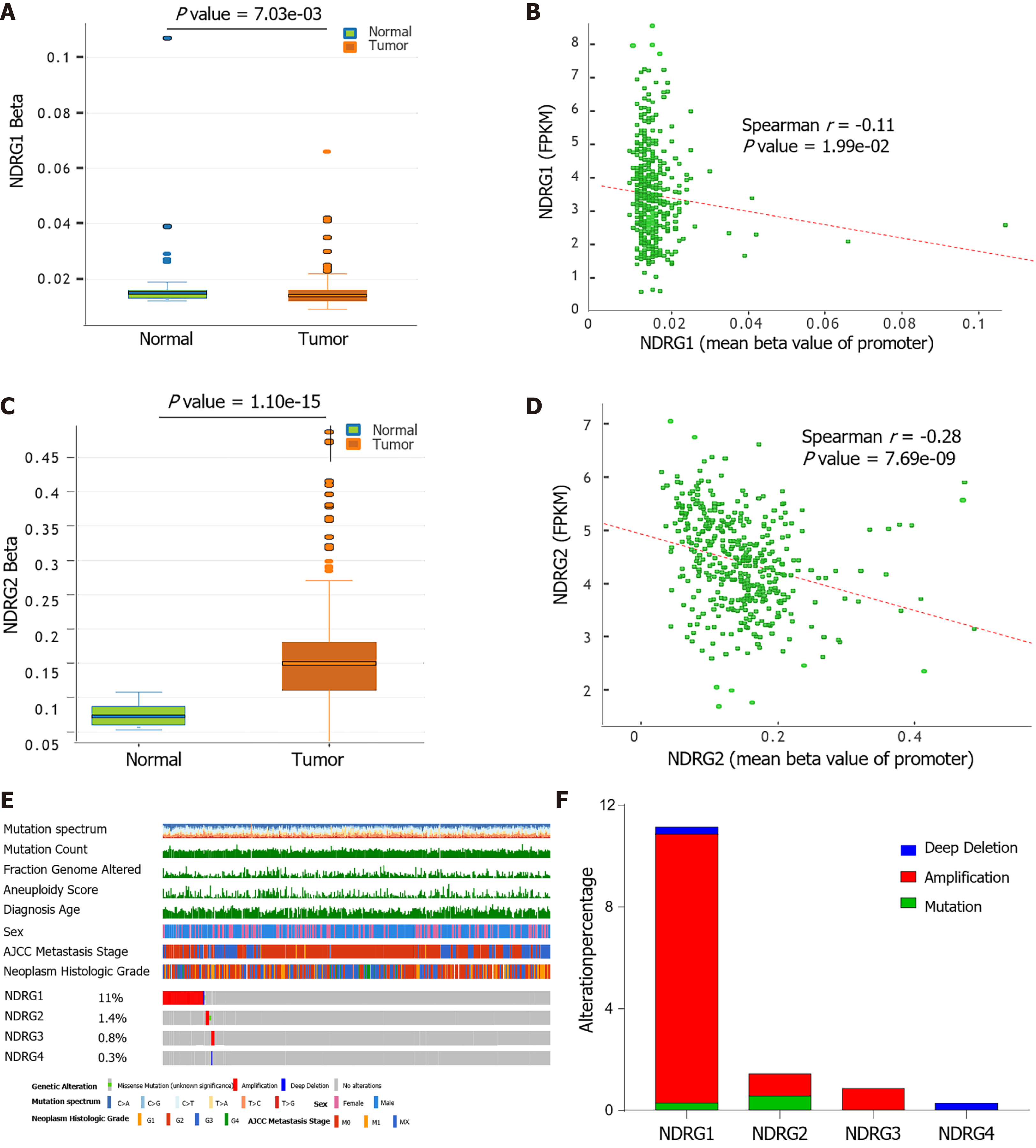
DNA methylation mediates transcriptional silencing of genes, and hypomethylation leads to overexpression (REF). To determine the methylation status of the NDRG genes, the methylation level of NDRG1-4 genes in HCC tissues was further explored. The promoter of NDRG1 was hypomethylated in HCC tissues compared to healthy tissues (Figure 4A). NDRG1 hypomethylation was significantly linked to the increased NDRG1 expression (Figure 4B). While higher NDRG2 methylation level was observed in tumors, and NDRG2 hypermethylation resulted in decreased levels of NDRG2 protein (Figure 4C and D). There was no significant difference between NDRG3 and NDRG4 methylation levels. Moreover, NDRG somatic mutations were also investigated by the cBioPortal website in the LIHC-TCGA cohort. As shown in Figure 4E, the percentage of genetic alterations of the NDRG family varied from 0.3% to 11% (missense mutation, deletion, and amplification) in the OncoPrints. The percentages of genetic mutation in NDRG1, NDRG2, and NDRG3 were 11%, 1.4%, 0.8%, and 0.3%, respectively. The distribution indicated that amplification accounts for most mutations of NDRG1 and NDRG3 (Figure 4F).
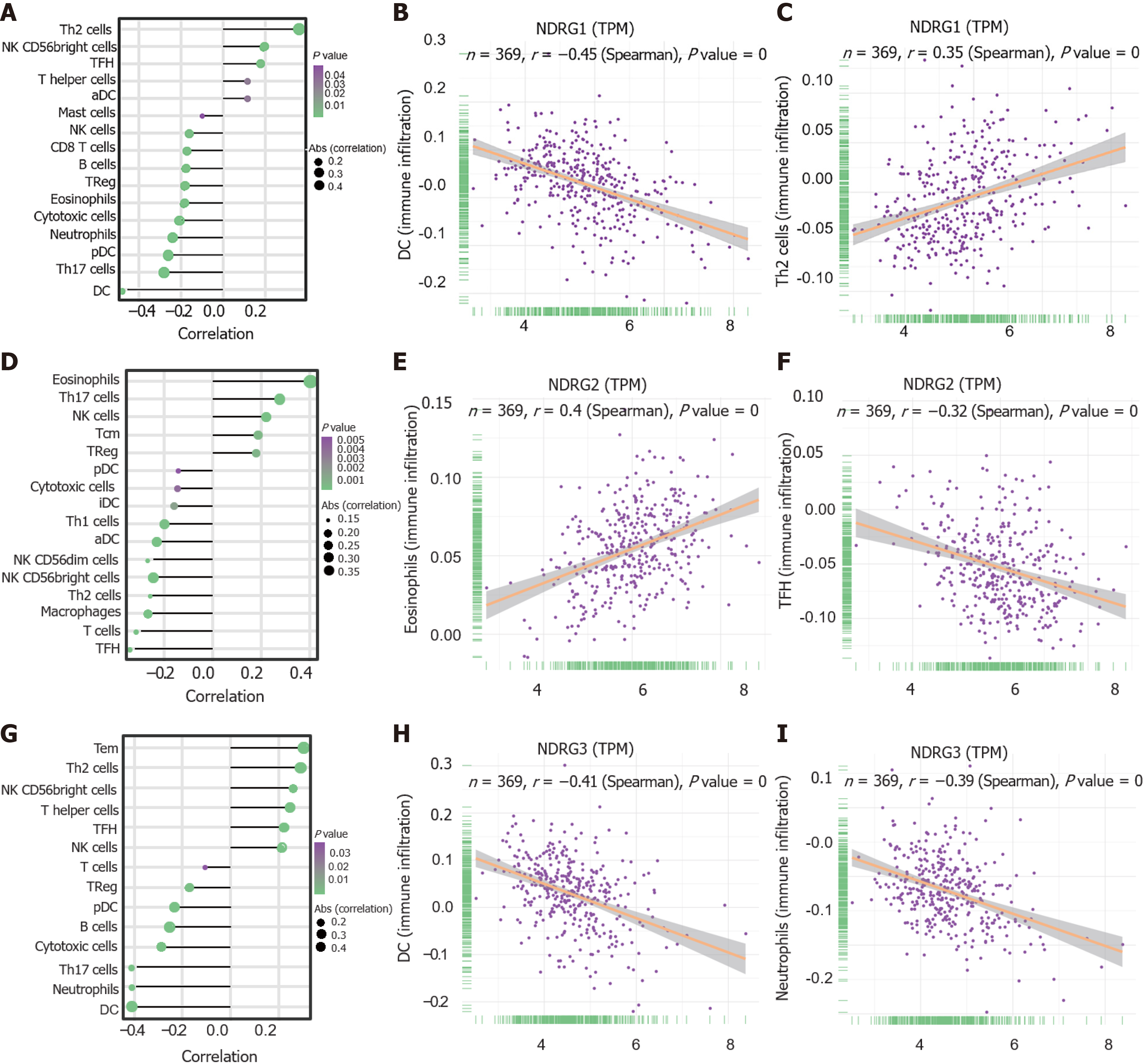
Given that the increasing evidence showed the associations among infiltrating immune cells and cancer prognosis levels, the correlation between tumor-infiltrating immune cells and NDRG members was further explored. Generally, several types of immune cells significantly correlated to NDRG family members (Figure 5A, D and G). DCs showed the strongest negative correlation with NDRG1 expression (r = -0.45, P < 0.001; Figure 5B). Th2 cells were positively associated with NDRG1 expression (r = 0.35, P < 0.001; Figure 5C). Eosinophils cells were positively associated with NDRG2 expression (r = 0.4, P < 0.001; Figure 5E) and TFH cells were negatively associated with NDRG2 expression (r = -0.32, P < 0.001; Figure 5F). DC cells and neutrophils were negatively associated with NDRG3 expression (r = -0.41, P < 0.001; Figure 5H, r = -0.39, P < 0.001; Figure 5I).
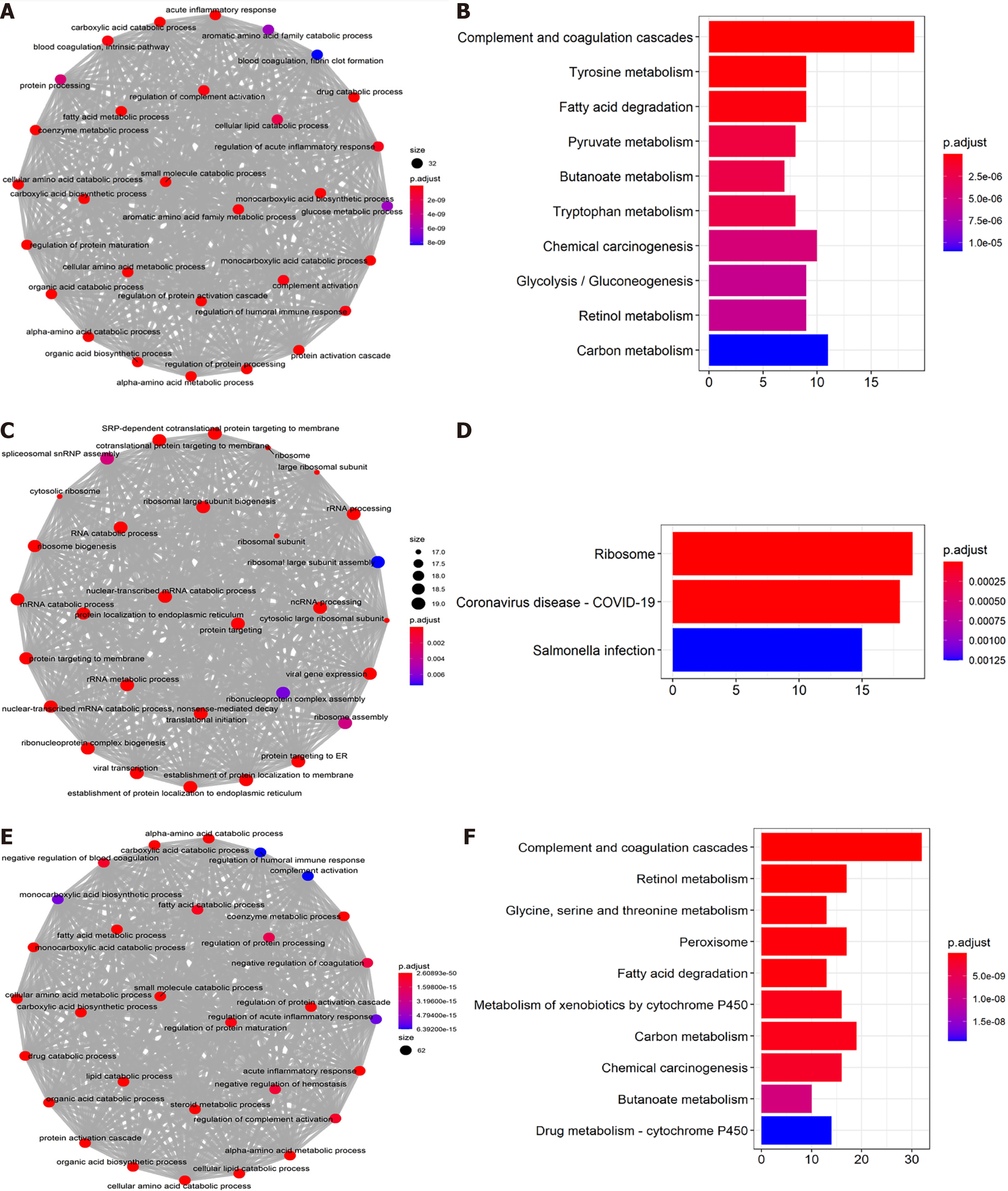
The first 500 closely co-expressed NDRG1-3 genes were annotated by GO and cluster Profiler's KEGG pathway enrichment analysis. Organic acid catabolic process (GO: 0046395), carboxylic acid catabolic process (GO: 0046395), and small molecule catabolic process (GO: 0044282) were the top three significantly enriched biological processes (BP) for NDRG1 (Figure 6A). Complement and coagulation cascades, tyrosine metabolism, and fatty acid degradation were the top three significant KEGG pathways for NDRG1 (Figure 6B). Co-translational protein targeting to the membrane (GO: 0006613), nuclear-transcribed mRNA catabolic process (GO: 0000956), and protein targeting to endoplasmic reticulum (GO: 0045047) were the top three significantly enriched BP for NDRG2 (Figure 6C). The ribosome, coronavirus disease 2019, and salmonella infection were the top three significant KEGG pathways for NDRG2 (Figure 6D). Fatty acid catabolic process (GO: 0009062), carboxylic acid catabolic process (GO: 0046395), and small molecule catabolic process (GO: 0044282) were the top three significantly enriched BP for NDRG3 (Figure 6E). Complement and coagulation cascades, retinol metabolism, and glycine, serine, and threonine metabolism were the top three significant KEGG pathways for NDRG3 (Figure 6F).
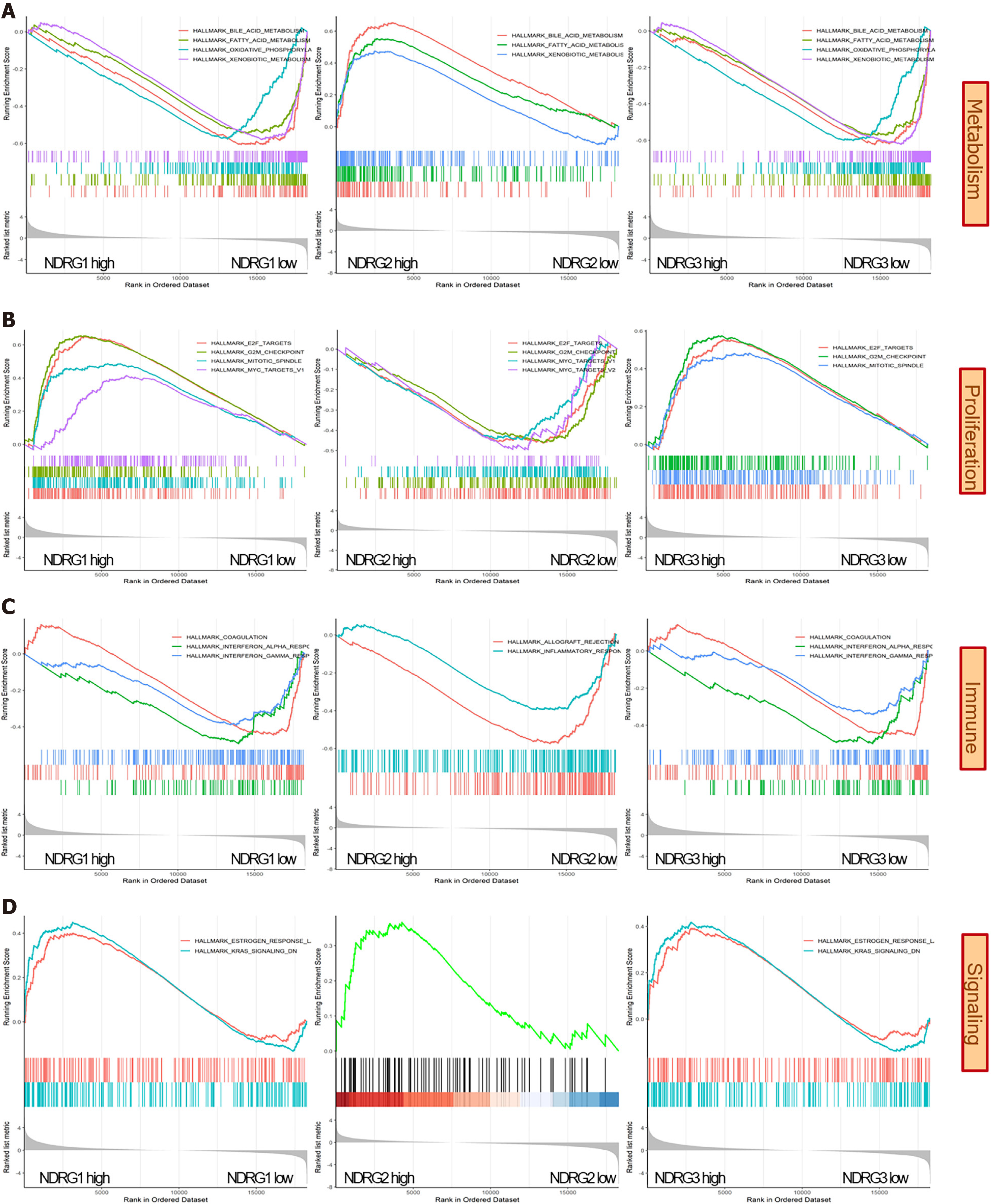
After GO and KEGG enrichment analysis, the potential biological roles of the NDRG family were further explored by the GSEA. Among 50 hallmark gene sets, metabolic (bile acid metabolism, fatty acid metabolism, and xenobiotic metabolism) and proliferation (G2-M checkpoint and E2 transcription factor targets) gene sets were enriched when the NDRG1 and NDRG3 expression are high and NDRG2 expression is low (Figure 7A and B). Immune-related sets (interferon response and coagulation) were enriched in low NDRG1 and NDRG3 expression, while gene sets (inflammation and allograft rejection) were enriched in low NDRG2 expression (Figure 7C). Signaling sets (estrogen response and Kirsten rat sarcoma viral oncogene homolog signaling down) were enriched in high NDRG1 and NDRG3 expression, and signaling sets (androgen response) were enriched in high NDRG2 expression (Figure 7D).
Previous studies have demonstrated that NDRG1-4 genes are differentially expressed in various organs and involved in essential processes, such as cell proliferation, apoptosis, and differentiation[38]. To our knowledge, expression patterns, prognostic significance, and biological roles of NDRG1-4 in HCC have not been comprehensively characterized.
NDRG1 is the most investigated gene among the NDRG gene family; it is involved in tumor growth, metastasis, and angiogenesis[39-43]. Cheng et al[40] reported that NDRG1 is up-regulated in HCC, and its expression is associated with a poor prognosis. Furthermore, high NDRG1 expression is correlated with metastasis and recurrence in HCC[41-43]. In the current study, we demonstrated NDRG1 was up-regulated in HCC, and high NDRG1 expression has a poor prognostic effect for overall survival, consistent with the previous study. Furthermore, high hypomethylation of the NDRG1 gene promoter leads to low NDRG1 mRNA expression in HCC tissue. The frequency of genetic alteration in NDRG1 was highest among the NDRG family, with amplifications being the most common alteration type. Amplification of the NDRG1 gene might be one reason for high NDRG1 expression. NDRG1 expression was positively associated with Th2 cells, NK CD56 bright cells, and TFH cells. Negative correlations between NDRG1 and DC, Th17, and plasmacytoid DC were also noticed. It is suggested that NDRG1 could alter the degree of tumor immune cell infiltration and was involved in the tumor microenvironment. Gene enrichment analysis of NDRG1 showed that NDRG1 has a wide range of effects on metabolism, proliferation, and immunity. The current study's findings could provide the impetus for future research to explore NDRG1 function on immunology and metabolism.
Like NDRG1, NDRG3 was elevated in HCC and associated with an unfavorable prognosis. According to Shi et al[44], high NDRG3 expression could facilitate HCC metastasis via activation of WNT/β-catenin signaling. High NDRG3 expression was associated with low infiltrating levels of DC cells and neutrophils. Enrichment analysis indicated that NDRG3 is involved in metabolism, proliferation, estrogen response, and Kirsten rat sarcoma viral oncogene homolog signaling. Unlike NDRG1 and NDRG3, NDRG2 was significantly down-regulated in HCC, and elevated NDRG2 expression was associated with a favorable prognosis. Previous studies supported that high expression of NDRG2 was negatively correlated with the malignant phenotype of HCC[19,45]. Guo et al[46] reported that NDRG2 could inhibit lactate dehydrogenase A expression, thus inhibiting the Warburg effect and HCC malignant progression. Promoter hypermethylation contributed to reducing NDRG2 mRNA, consistent with a previous study showing that NDRG2 carried extensively methylated CpG sites[47]. NDRG2 expression was positively correlated with the abundance of eosinophils and negatively correlated with TFH abundance. Further studies are required to verify our findings. Gene enrichment analysis indicated that NDRG2 had the opposite role in HCC development. Although NDRG4 was reported to be a potential tumor suppressor and prognostic marker of colorectal and gastric cancers, NDRG4 role in HCC remains unknown[48]. NDRG4 expression showed no significant difference between tumor and non-tumor tissues in the current study. Besides, NDRG4 was not associated with HCC prognosis. We speculate that NDRG4 might not be involved in HCC development.
Furthermore, multivariate Cox regression analysis established a prognostic risk score model with three NDRG genes. There was a significant difference in patient survival between the low-risk and high-risk groups. This NDRG-based signature exhibited high prognostic accuracy in the TCGA cohort, as demonstrated by ROC curves. Consistently, our model could predict the 1-, 2-, and 3-year survival probabilities for HCC patients. Taken together, our findings revealed the potential value of NDRG genes in predicting prognosis.
We acknowledge several limitations in the current study. Our NDRG family-based signature might be subject to sampling bias, and further studies with larger samples are required to validate our signature’s performance. Besides, biological experiments are warranted to validate their functions in liver cancer.
In summary, NDRG1 and NDRG3 were significantly highly expressed in HCC, while NDRG2 was significantly down-regulated in HCC. High expression of NDRG1 and NDRG3 and low NDRG2 expression were significantly associated with worse OS. Additionally, an NDRG-based prognostic score was built as a prognostic biomarker for OS of HCC patients. Compared with NDRG1 and NDRG3, NDRG2 had a distinct biological role in metabolism, which provided insights for further investigation of the NDRG family as potential targets in HCC.
The N-Myc downstream regulatory gene (NDRG) family consists of four members (NDRG1-4), which participate in various important biological processes. However, the prognosis of NDRG family in hepatocellular carcinoma (HCC) has not been systematically evaluated yet.
The study revealed prognostic and potential biological role of the NDRG1, NDRG2, and NDRG3 in HCC. A prognostic risk score model with three NDRG family genes was established through multivariate Cox regression analysis and exhibited high prognostic accuracy.
To analyze comprehensively the prognostic and biological role of the NDRG family in HCC.
The expression of the NDRG family was downloaded from a public database. The related clinical information of HCC was downloaded from The Cancer Genome Atlas. The methylation analysis, genomic changes, and prognostic value of NDRG family were evaluated by online tools. Single sample gene set enrichment analysis was used to determine the degree of immune cell infiltration in tumors.
The expression of NDRG1 and NDRG3 in HCC was significantly higher than that in non-cancerous tissues, while the expression of NDRG2 was down regulated in HCC. The high expression of NDRG1 and NDRG3 and the low expression of NDRG2 were associated with the poor prognosis of HCC. The mutation rate of NDRG1 gene was the highest. The expression of NDRG1-3 is related to various types of filtered immune cells. Gene Ontology analysis showed that organic acid catabolism is the most important biological process related to NDRG family. Gene Set Enrichment Analysis showed that the gene sets related to metabolism, proliferation, and immunity were enriched in the high expression of NDRG1 and NDRF3 and the low expression of NDRG2.
The high expression of NDRG1 and NDRG3 and the low expression of NDRG2 were significantly correlated with poor overall survival in HCC. In addition, we also established a prognostic score based on NDRG as a prognostic biomarker of overall survival in patients with HCC. Compared with NDRG1 and NDRG3, NDRG2 has a unique biological role in metabolism, which provides insights for the further study of NDRG family as a potential target of HCC.
To provide preliminary suggestions for the clinical study of HCC and insights for the further study of the NDRG family as a potential target of HCC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Emran TB, Vishnoi S S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Yu HG
| 1. | Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1933] [Cited by in RCA: 1871] [Article Influence: 207.9] [Reference Citation Analysis (4)] |
| 2. | Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 778] [Article Influence: 155.6] [Reference Citation Analysis (0)] |
| 3. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15305] [Article Influence: 3061.0] [Reference Citation Analysis (4)] |
| 4. | Sagnelli E, Macera M, Russo A, Coppola N, Sagnelli C. Epidemiological and etiological variations in hepatocellular carcinoma. Infection. 2020;48:7-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 5. | Altekruse SF, Henley SJ, Cucinelli JE, McGlynn KA. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am J Gastroenterol. 2014;109:542-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 355] [Article Influence: 32.3] [Reference Citation Analysis (1)] |
| 6. | Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, Zhu AX, Murad MH, Marrero JA. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2107] [Cited by in RCA: 3023] [Article Influence: 431.9] [Reference Citation Analysis (3)] |
| 7. | Zhao W, Tang R, Huang Y, Wang W, Zhou Z, Gu S, Dai J, Ying K, Xie Y, Mao Y. Cloning and expression pattern of the human NDRG3 gene. Biochim Biophys Acta. 2001;1519:134-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Melotte V, Qu X, Ongenaert M, van Criekinge W, de Bruïne AP, Baldwin HS, van Engeland M. The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J. 2010;24:4153-4166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 233] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 9. | Chen B, Nelson DM, Sadovsky Y. N-myc down-regulated gene 1 modulates the response of term human trophoblasts to hypoxic injury. J Biol Chem. 2006;281:2764-2772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 116] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 10. | Taketomi Y, Sunaga K, Tanaka S, Nakamura M, Arata S, Okuda T, Moon TC, Chang HW, Sugimoto Y, Kokame K, Miyata T, Murakami M, Kudo I. Impaired mast cell maturation and degranulation and attenuated allergic responses in Ndrg1-deficient mice. J Immunol. 2007;178:7042-7053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Takahashi K, Yamada M, Ohata H, Honda K. Ndrg2 promotes neurite outgrowth of NGF-differentiated PC12 cells. Neurosci Lett. 2005;388:157-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Yao L, Zhang J, Liu X. NDRG2: a Myc-repressed gene involved in cancer and cell stress. Acta Biochim Biophys Sin (Shanghai). 2008;40:625-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 94] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 13. | Ellen TP, Ke Q, Zhang P, Costa M. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis. 2008;29:2-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 173] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 14. | Jing JS, Li H, Wang SC, Ma JM, Yu LQ, Zhou H. NDRG3 overexpression is associated with a poor prognosis in patients with hepatocellular carcinoma. Biosci Rep. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Strzelczyk B, Szulc A, Rzepko R, Kitowska A, Skokowski J, Szutowicz A, Pawelczyk T. Identification of high-risk stage II colorectal tumors by combined analysis of the NDRG1 gene expression and the depth of tumor invasion. Ann Surg Oncol. 2009;16:1287-1294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 16. | Liu W, Iiizumi-Gairani M, Okuda H, Kobayashi A, Watabe M, Pai SK, Pandey PR, Xing F, Fukuda K, Modur V, Hirota S, Suzuki K, Chiba T, Endo M, Sugai T, Watabe K. KAI1 gene is engaged in NDRG1 gene-mediated metastasis suppression through the ATF3-NFkappaB complex in human prostate cancer. J Biol Chem. 2011;286:18949-18959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 79] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 17. | Schonkeren SL, Massen M, van der Horst R, Koch A, Vaes N, Melotte V. Nervous NDRGs: the N-myc downstream-regulated gene family in the central and peripheral nervous system. Neurogenetics. 2019;20:173-186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 18. | Deng Y, Yao L, Chau L, Ng SS, Peng Y, Liu X, Au WS, Wang J, Li F, Ji S, Han H, Nie X, Li Q, Kung HF, Leung SY, Lin MC. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer. 2003;106:342-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 188] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 19. | Pan T, Zhang M, Zhang F, Yan G, Ru Y, Wang Q, Zhang Y, Wei X, Xu X, Shen L, Zhang J, Wu K, Yao L, Li X. NDRG2 overexpression suppresses hepatoma cells survival during metabolic stress through disturbing the activation of fatty acid oxidation. Biochem Biophys Res Commun. 2017;483:860-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Kim YJ, Yoon SY, Kim JT, Choi SC, Lim JS, Kim JH, Song EY, Lee HG, Choi I, Kim JW. NDRG2 suppresses cell proliferation through down-regulation of AP-1 activity in human colon carcinoma cells. Int J Cancer. 2009;124:7-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 21. | Li T, Sun R, Lu M, Chang J, Meng X, Wu H. NDRG3 facilitates colorectal cancer metastasis through activating Src phosphorylation. Onco Targets Ther. 2018;11:2843-2852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Ma J, Liu S, Zhang W, Zhang F, Wang S, Wu L, Yan R, Wang C, Zha Z, Sun J. High expression of NDRG3 associates with positive lymph node metastasis and unfavourable overall survival in laryngeal squamous cell carcinoma. Pathology. 2016;48:691-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Luo X, Hou N, Chen X, Xu Z, Xu J, Wang L, Yang S, Liu S, Xu L, Chen Y, Xiong L, Wang J, Fan W. High expression of NDRG3 associates with unfavorable overall survival in non-small cell lung cancer. Cancer Biomark. 2018;21:461-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7078] [Article Influence: 884.8] [Reference Citation Analysis (0)] |
| 25. | Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1668] [Cited by in RCA: 1892] [Article Influence: 135.1] [Reference Citation Analysis (0)] |
| 26. | Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med. 2016;375:1109-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 969] [Cited by in RCA: 1090] [Article Influence: 121.1] [Reference Citation Analysis (0)] |
| 27. | Ding W, Chen J, Feng G, Chen G, Wu J, Guo Y, Ni X, Shi T. DNMIVD: DNA methylation interactive visualization database. Nucleic Acids Res. 2020;48:D856-D862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 28. | Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9144] [Cited by in RCA: 12771] [Article Influence: 982.4] [Reference Citation Analysis (0)] |
| 29. | Lopez-Raton M, Rodriguez-Alvarez MX, Cadarso-Suárez C, Gude-Sampedro F. OptimalCutpoints: An R Package for Selecting Optimal Cutpoints in Diagnostic Tests. J Stat Softw. 61:1-36. [RCA] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 303] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 30. | Kamarudin AN, Cox T, Kolamunnage-Dona R. Time-dependent ROC curve analysis in medical research: current methods and applications. BMC Med Res Methodol. 2017;17:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 535] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 31. | Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545-15550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27252] [Cited by in RCA: 37415] [Article Influence: 1870.8] [Reference Citation Analysis (0)] |
| 32. | Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Fröhling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1822] [Cited by in RCA: 2690] [Article Influence: 168.1] [Reference Citation Analysis (0)] |
| 33. | Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 2952] [Article Influence: 246.0] [Reference Citation Analysis (0)] |
| 34. | Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, Thomson E, Monteiro R, Guimaraes P, Walters B, Wiser J, Butte AJ. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 579] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 35. | Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18868] [Cited by in RCA: 24696] [Article Influence: 987.8] [Reference Citation Analysis (0)] |
| 36. | Gene Ontology Consortium. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440-D444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 530] [Cited by in RCA: 525] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 37. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 22157] [Article Influence: 1704.4] [Reference Citation Analysis (0)] |
| 38. | Geleta B, Makonnen E, Abay SM. N-myc Downstream Regulated Gene (NDRG): Role in Cancer Metastasis Suppression and as Drug Target in Cancer Therapeutics. J Cancer Sci Ther. 8:154-159. [DOI] [Full Text] |
| 39. | Bae DH, Jansson PJ, Huang ML, Kovacevic Z, Kalinowski D, Lee CS, Sahni S, Richardson DR. The role of NDRG1 in the pathology and potential treatment of human cancers. J Clin Pathol. 2013;66:911-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 40. | Cheng J, Xie HY, Xu X, Wu J, Wei X, Su R, Zhang W, Lv Z, Zheng S, Zhou L. NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer Lett. 2011;310:35-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 41. | Broggini T, Wüstner M, Harms C, Stange L, Blaes J, Thomé C, Harms U, Mueller S, Weiler M, Wick W, Vajkoczy P, Czabanka M. NDRG1 overexpressing gliomas are characterized by reduced tumor vascularization and resistance to antiangiogenic treatment. Cancer Lett. 2016;380:568-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 42. | Sharma A, Mendonca J, Ying J, Kim HS, Verdone JE, Zarif JC, Carducci M, Hammers H, Pienta KJ, Kachhap S. The prostate metastasis suppressor gene NDRG1 differentially regulates cell motility and invasion. Mol Oncol. 2017;11:655-669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 43. | Kovacevic Z, Richardson DR. The metastasis suppressor, Ndrg-1: a new ally in the fight against cancer. Carcinogenesis. 2006;27:2355-2366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 162] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 44. | Shi J, Zheng H, Yuan L. High NDRG3 expression facilitates HCC metastasis by promoting nuclear translocation of β-catenin. BMB Rep. 2019;52:451-456. [PubMed] |
| 45. | Zheng J, Li Y, Yang J, Liu Q, Shi M, Zhang R, Shi H, Ren Q, Ma J, Guo H, Tao Y, Xue Y, Jiang N, Yao L, Liu W. NDRG2 inhibits hepatocellular carcinoma adhesion, migration and invasion by regulating CD24 expression. BMC Cancer. 2011;11 251:1-251:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 46. | Guo Y, Li X, Sun X, Wang J, Yang X, Zhou X, Liu X, Liu W, Yuan J, Yao L, Shen L. Combined Aberrant Expression of NDRG2 and LDHA Predicts Hepatocellular Carcinoma Prognosis and Mediates the Anti-tumor Effect of Gemcitabine. Int J Biol Sci. 2019;15:1771-1786. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 47. | Lee DC, Kang YK, Kim WH, Jang YJ, Kim DJ, Park IY, Sohn BH, Sohn HA, Lee HG, Lim JS, Kim JW, Song EY, Kim DM, Lee MN, Oh GT, Kim SJ, Park KC, Yoo HS, Choi JY, Yeom YI. Functional and clinical evidence for NDRG2 as a candidate suppressor of liver cancer metastasis. Cancer Res. 2008;68:4210-4220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 113] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 48. | Melotte V, Lentjes MH, van den Bosch SM, Hellebrekers DM, de Hoon JP, Wouters KA, Daenen KL, Partouns-Hendriks IE, Stessels F, Louwagie J, Smits KM, Weijenberg MP, Sanduleanu S, Khalid-de Bakker CA, Oort FA, Meijer GA, Jonkers DM, Herman JG, de Bruïne AP, van Engeland M. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009;101:916-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 167] [Article Influence: 10.4] [Reference Citation Analysis (0)] |