Published online Feb 26, 2022. doi: 10.12998/wjcc.v10.i6.1889
Peer-review started: July 15, 2021
First decision: November 11, 2021
Revised: November 16, 2021
Accepted: January 11, 2022
Article in press: January 11, 2022
Published online: February 26, 2022
Processing time: 223 Days and 15.6 Hours
This case report describes a novel genotypic and phenotypic presentation of Alazami-Yuan syndrome, and contributes to the current knowledge on the condition.
We report an 11-year-old boy with Alazami-Yuan syndrome. The main clinical manifestations were rapid development of puberty, typical facial features of Cornelia de Lange syndrome, and normal intelligence. Peripheral blood DNA samples obtained from the patient and his parents were sequenced using high-throughput whole-exosome sequencing, which was verified by Sanger sequencing. The results showed that there was a compound heterozygous mutation of c.1052delT and c.76A>T in the TATA-Box Binding Protein Associated Factor 6 (TAF6) gene. The mutation of c.1052delT was from his mother and the mutation of c.76A>T was from his father.
This study extends the mutation spectrum of the TAF6 gene, and provides a molecular basis for the etiological diagnosis of Alazami-Yuan syndrome and genetic consultation for the family.
Core Tip: We report an 11-year-old boy with Alazami-Yuan syndrome. The main clinical manifestations were rapid development of puberty, typical facial features of Cornelia de Lange syndrome, and normal intelligence. DNA sequencing test showed that there was a compound heterozygous mutation of c.1052delT and c.76A>T in the TATA-Box Binding Protein Associated Factor 6 (TAF6) gene. This study extends the mutation spectrum of the TAF6 gene, and provides a molecular basis for the etiological diagnosis of Alazami-Yuan syndrome and genetic consultation for the family.
- Citation: Lin SZ, Feng JH, Sun LP, Ma HW, Wang WQ, Li JY. Novel compound heterozygous variants in the TAF6 gene in a patient with Alazami-Yuan syndrome: A case report. World J Clin Cases 2022; 10(6): 1889-1895
- URL: https://www.wjgnet.com/2307-8960/full/v10/i6/1889.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i6.1889
Alazami-Yuan syndrome is an autosomal recessive genetic disease caused by mutation of the TATA-Box Binding Protein Associated Factor 6 (TAF6) gene. Its clinical features are similar to those of Cornelia de Lange syndrome (CdLS). Typical features include short stature, mental retardation, arched eyebrows, conjoined eyebrows, protruding bridge of the nose, nose tilted forward, and a thin upper lip[1,2]. There are differences in the gene mutation site and genetic mode between Alazami-Yuan syndrome and CdLS. CdLS is caused by mutations in Nipped-B-like protein (NIPBL), structural maintenance of chromosomes 1A (SMC1A), SMC3, RAD21, and histone deacetylase 8, and the genetic mode is autosomal dominant inheritance and X-linked dominant inheritance[3].
In this case study, two new mutations of the TAF6 gene were found by high-throughput whole-exosome sequencing in an 11-year-old patient with rapid development of puberty and special facial features.
An 11-year-old male patient was referred to our clinic due to testicular enlargement and rapid growth in height.
The patient presented with testicular enlargement without obvious cause, no pubic hair, no spermatorrhea, and a small amount of beard hair for 6 mo. Peripheral blood DNA samples obtained from the patient and his parents were sequenced using high-throughput whole-exosome sequencing, which was verified by Sanger sequencing.
The patient was the 2.1 kg (< -3sd), 46 cm (< -1sd), and the product of a 36 wk pre
His non-consanguineous parents were clinically normal. His father and mother were 170 and 151 cm in height, respectively. There was no family history of genetic or infectious diseases.
On physical examination at his visit at 11 years of age, his weight was 52.5 kg and length was 146.1 cm. The patient had a clear mind, good spirit, normal hair, and no yellow coloring or bleeding spots on the skin. He had arched eyebrows, protruding bridge of the nose, forward leaning nostrils, a thin upper lip, a small amount of beard, normal jaw, and an inconspicuous laryngeal knot. Both pupils were equal in size and were sensitive to light. Breath sounds in both lungs were clear, and dry and moist rales were not heard. Heart sounds were strong and regular, the heart rate was 90 bpm, and no pathological murmur was found in each valve area. The abdomen was soft, no tenderness and rebound pain was observed, the liver and spleen were unpalpable. The big toes on both feet were widened and the limbs were normal. Limb muscle tension was normal. Physiological reflexes were present, and pathological reflexes were not found. Bilateral testes were symmetrical, about 8-10 mL in size, without pubic hair.
The patient’s liver function, kidney function, electrolytes, blood glucose and blood lipids were normal. Insulin-like growth factor was normal. Karyotype analysis of cultured cells revealed a karyotype of 46XY. Sex hormone levels were as follows: estradiol 25 pg/mL, (adult male reference value: < 20-47 pg/mL), follicle-stimulating hormone 6.62 mIU/mL (adult male reference value: 1.27-19.26 mIU/mL), luteinizing hormone 3.20 mIU/mL (adult male reference value: 1.24-8.62 mIU/mL), and testosterone 1.75 ng/mL (adult male reference value: 1.75-7.81 ng/mL). Total 25 hydroxy vitamin D was 13.79 ng/mL (reference value < 20 ng/mL vitamin D deficiency). Fasting insulin was 23.1 mU/L (reference value: 2.3-11.8 mU/L). Thyroid function was evaluated as follows: Triiodothyronine 6.84 pmol/L (reference value: 2.63-5.71 pmol/L), thyroxine 12.10 pmol/L (reference value: 9.01-19.05 pmol/L), and thyroid-stimulating hormone 1.9145 μIU/mL (reference value: 0.30-4.80 μIU/mL).
The patient underwent a skeletal examination, and the results showed that the bone age was 13 years. Magnetic resonance imaging of the pituitary gland was normal. Slight lateral curvature of the thoracic spine was observed.
Informed consent was obtained from the parents on behalf of the proband for whole-exome sequencing, mitochondrial sequencing, and the publication of photographs. DNA was obtained from peripheral blood samples from the patient and his parents. Sequencing and analyses were performed by the Beijing Mygenostics (Beijing, China), which is a high-tech biotechnology company providing life science instruments, reagents and technical services. The second generation sequencer Illumina NextSeqTM 500 (Illumina, San Diego, CA, United States) was used to sequence the captured region at two ends, with a reading length of 150 bp. After sequencing the target region, the splices and low-quality data in the sequencing data were removed. Using Burrows-Wheeler Aligner software to compare with the reference genome (hg19 version), the data on sequencing depth, homogeneity, and probe specificity were analyzed. Genome Analysis Toolkit software was used to detect the polymorphic sites in the comparison data of each sample, and statistical analyses of the data on single nucleotide polymorphisms (SNPs) and insertion deletion mutations (indels) were conducted. The SNPs and indels were screened using the database of SNPs, (http://www.ncbi.nlm.nih.gov/SNP), 1000 human genome (http://www.internationalgenome.org), and the Exome Aggregation Consortium database (http://exac.broadinstitute.org). Application of the human gene mutation database (HGMD, http://www.hgmd.cf.ac.uk) and the human Online Mendelian genetic database (OMIM, http://omim.org) confirmed the reported pathogenic gene locus. The effects of variation on protein structure and pathogenicity were predicted by Rev, Polyphen-2, and Sift. The American College of Medical Genetics and Genomics (ACMG) sequence variation interpretation standards and guidelines[4] were used for a comprehensive evaluation of the pathogenicity of mutation sites.
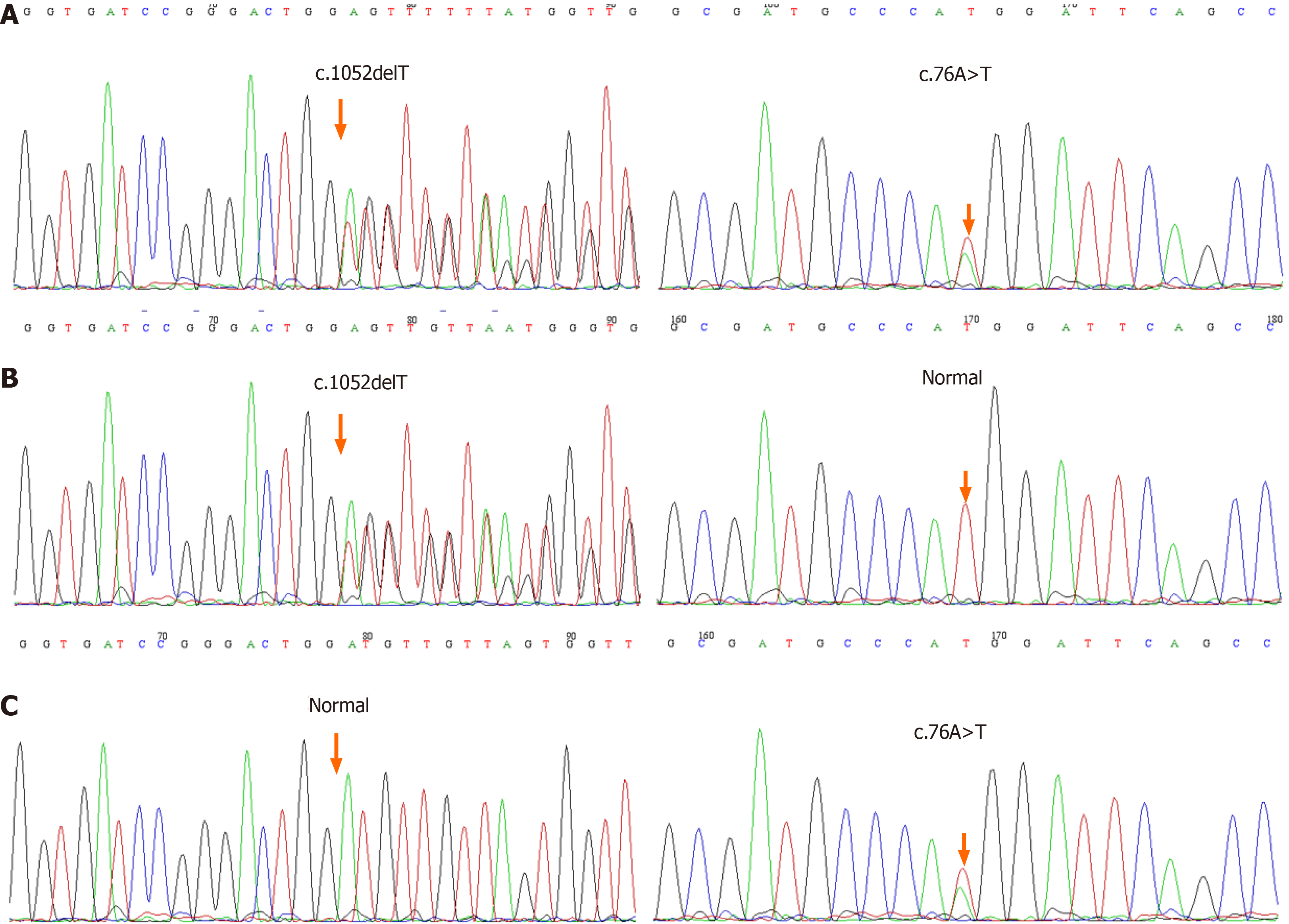
Whole-exome sequencing showed that there was complex heterozygous variation of the TAF6 gene in this patient, one of which was an unreported frameshift mutation c.1052delT (p.I351Tfs*40), which may lead to the loss of gene function; the frequency of the variation in the normal population database is unknown, and is a low-frequency variation; the results of protein function prediction are unknown, and are not reported in the HGMD database. According to Sanger sequencing, the variation originated from the child’s mother, and the paternal gene was wild-type (Figure 1). According to ACMG guidelines, the mutation was suspected to be pathogenic.
The other was a missense mutation c.76A>T(p.M26L), which has not been reported. This missense mutation showed 76 nucleotide deficiency changes from adenine to thymine, resulting in the 26 amino acids changing from methionine to leucine. The frequency of the mutation in the normal population database is 0.0014, and is a low-frequency mutation; the results of protein function prediction are unknown, and are not reported in the HGMD database. According to Sanger sequencing, the variation originated from the child’s father, and the maternal gene was wild-type (Figure 1). According to ACMG guidelines, the clinical significance of the variation is unknown.
Sanger sequencing showed that there was a compound heterozygous mutation of c.1052delT and c.76A>T in the TAF6 gene.
According to the clinical manifestations, laboratory tests, and gene sequencing results, the clinical phenotype of the patient was Alazami-Yuan syndrome. The boy’s weight was above the normal range, and he was given exercise and diet guidance. The patient’s total 25 hydroxy vitamin D level was low, and vitamin D 2000 U was administered once a day for 3 mo, and calcium carbonate 500 mg once a day for 3 mo. To improve the final height of the child, 3.75 mg of diphereline was injected once every 28 d, and 10 U recombinant human growth hormone was injected sub
After 4 mo of treatment, the child’s height increased by 3.6 cm, his weight decreased by 0.7 kg, and the vitamin D level returned to normal. During treatment, skin at the injection site was good, fasting blood glucose and nail function were normal, and there was no eyelid edema, headache, or other adverse reactions.
The TAF6 gene is located on chromosome 7q22.1, which is involved in the initiation and activation of RNA transcription and is closely related to human cell viability[5]. The mutation of TAF6 gene can lead to Alazami-Yuan syndrome. In published cases, 5 patients from two families have been reported. Their parents were consanguineous, and the mutation types were homozygous mutations, with mutation locations at c.136c>T and c.212t>C[1,2]. The clinical manifestations of these patients were similar, with short stature, mental retardation, and typical facial features of CdLS. We report a case of compound heterozygous mutation in which the parents were non-consanguineous and the mutation location was c.1052delT and c.76A>T. The main clinical manifestations were rapid puberty and special body surface characteristics, including arched eyebrows, protruding nose bridge, forward leaning nose, thin upper lip and widened big toes on both feet. The child had normal intelligence and was born small for gestational age, but had no history of feeding difficulties. Growth and development before puberty were basically normal.
CdLS (OMIM: 122470) is a type of multiple congenital dysplasia. Patients usually have physiological, cognitive and behavioral characteristics[6]. According to previous case reports, CdLS1 caused by gene mutation of NIPBL accounts for about 50% to 60% of CdLS cases[7]. A large number of individuals with typical CdLS carry a mosaic NIPBL variation[8]. Although individuals with typical CdLS phenotypes are likely to have mutations in NIPBL, individuals with one of the other pathogenic CdLS genes can also meet the standard for typical CdLS[9-14]. Typical CdLS has a unique craniofacial appearance and growth pattern, as well as limb deformities. However, not all CdLS patients show typical phenotypes, and there are differences in the manifestations of the disease itself, from mild to severe, and the degree of facial and limb involvement is also different[15]. In this report, although the patient had typical facial features of CdLS, he had a unique clinical phenotype and gene mutation sites, which has practical significance for in-depth research and clinical guidance.
The patient first attended hospital due to enlarged testicles for 6 mo. His bone age was 2 years older than his actual age, and puberty developed rapidly. In order to improve the final height of this patient, he was treated with the combination of diphereline and recombinant human growth hormone. The patient's special facial features were similar to those of CdLS which attracted our attention at his first visit, but the patient had no mental retardation or language deficiency. In order to determine the cause of the disease, we used high-throughput whole-exome sequencing and identified a compound heterozygous mutation of the TAF6 gene. Most patients with CdLS have new mutations, and the risk of their parents having another CdLS child is low. In this case, the two mutated genes were from the father and mother, respectively. The probability of the parents having a child with Alazami-Yuan syndrome was 25%, and the probability of carrying the pathogenic gene in a subsequent child was 50%. It is suggested that prenatal consultation and diagnosis should be carried out if the child's mother has subsequent pregnancies.
Herein, the rapid development of puberty and older bone age were defined for the first time in the TAF6-related phenotype. We suggest that TAF6 should be considered in individuals with rapid development of puberty and CdLS-overlapping features. Furthermore, our patient was found to be a compound heterozygote for two novel pathogenic variants in TAF6. Identification of a compound heterozygote should encourage clinicians to consider Alazami-Yuan syndrome in patients with similar clinical features and without a family history of consanguinity.
We would like to thank the child and his family members for agreeing to participate in this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Pediatrics
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: El-Shabrawi MH S-Editor: Liu JH L-Editor: Filipodia P-Editor: Liu JH
| 1. | Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah MH, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F, Kurdi W, Alfadhel M, Babay Z, Alsogheer M, Kaya N, Al-Hassnan ZN, Abdel-Salam GM, Al-Sannaa N, Al Mutairi F, El Khashab HY, Bohlega S, Jia X, Nguyen HC, Hammami R, Adly N, Mohamed JY, Abdulwahab F, Ibrahim N, Naim EA, Al-Younes B, Meyer BF, Hashem M, Shaheen R, Xiong Y, Abouelhoda M, Aldeeri AA, Monies DM, Alkuraya FS. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 355] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 2. | Yuan B, Pehlivan D, Karaca E, Patel N, Charng WL, Gambin T, Gonzaga-Jauregui C, Sutton VR, Yesil G, Bozdogan ST, Tos T, Koparir A, Koparir E, Beck CR, Gu S, Aslan H, Yuregir OO, Al Rubeaan K, Alnaqeb D, Alshammari MJ, Bayram Y, Atik MM, Aydin H, Geckinli BB, Seven M, Ulucan H, Fenercioglu E, Ozen M, Jhangiani S, Muzny DM, Boerwinkle E, Tuysuz B, Alkuraya FS, Gibbs RA, Lupski JR. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J Clin Invest. 2015;125:636-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 133] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 3. | Mannini L, Cucco F, Quarantotti V, Krantz ID, Musio A. Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Hum Mutat. 2013;34:1589-1596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 4. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19696] [Cited by in RCA: 22474] [Article Influence: 2247.4] [Reference Citation Analysis (0)] |
| 5. | Kamtchueng C, Stébenne MÉ, Delannoy A, Wilhelm E, Léger H, Benecke AG, Bell B. Alternative splicing of TAF6: downstream transcriptome impacts and upstream RNA splice control elements. PLoS One. 2014;9:e102399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 6. | Kline AD, Grados M, Sponseller P, Levy HP, Blagowidow N, Schoedel C, Rampolla J, Clemens DK, Krantz I, Kimball A, Pichard C, Tuchman D. Natural history of aging in Cornelia de Lange syndrome. Am J Med Genet C Semin Med Genet. 2007;145C:248-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 114] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 7. | Sirmaci A, Spiliopoulos M, Brancati F, Powell E, Duman D, Abrams A, Bademci G, Agolini E, Guo S, Konuk B, Kavaz A, Blanton S, Digilio MC, Dallapiccola B, Young J, Zuchner S, Tekin M. Mutations in ANKRD11 cause KBG syndrome, characterized by intellectual disability, skeletal malformations, and macrodontia. Am J Hum Genet. 2011;89:289-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 8. | Huisman SA, Redeker EJ, Maas SM, Mannens MM, Hennekam RC. High rate of mosaicism in individuals with Cornelia de Lange syndrome. J Med Genet. 2013;50:339-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 9. | Huisman S, Mulder PA, Redeker E, Bader I, Bisgaard AM, Brooks A, Cereda A, Cinca C, Clark D, Cormier-Daire V, Deardorff MA, Diderich K, Elting M, van Essen A, FitzPatrick D, Gervasini C, Gillessen-Kaesbach G, Girisha KM, Hilhorst-Hofstee Y, Hopman S, Horn D, Isrie M, Jansen S, Jespersgaard C, Kaiser FJ, Kaur M, Kleefstra T, Krantz ID, Lakeman P, Landlust A, Lessel D, Michot C, Moss J, Noon SE, Oliver C, Parenti I, Pie J, Ramos FJ, Rieubland C, Russo S, Selicorni A, Tümer Z, Vorstenbosch R, Wenger TL, van Balkom I, Piening S, Wierzba J, Hennekam RC. Phenotypes and genotypes in individuals with SMC1A variants. Am J Med Genet A. 2017;173:2108-2125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 10. | Deardorff MA, Kaur M, Yaeger D, et al Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007. 80(3): 485-94.. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 439] [Cited by in RCA: 402] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 11. | Parenti I, Gervasini C, Pozojevic J, Graul-Neumann L, Azzollini J, Braunholz D, Watrin E, Wendt KS, Cereda A, Cittaro D, Gillessen-Kaesbach G, Lazarevic D, Mariani M, Russo S, Werner R, Krawitz P, Larizza L, Selicorni A, Kaiser FJ. Broadening of cohesinopathies: exome sequencing identifies mutations in ANKRD11 in two patients with Cornelia de Lange-overlapping phenotype. Clin Genet. 2016;89:74-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 12. | Woods SA, Robinson HB, Kohler LJ, Agamanolis D, Sterbenz G, Khalifa M. Exome sequencing identifies a novel EP300 frame shift mutation in a patient with features that overlap Cornelia de Lange syndrome. Am J Med Genet A. 2014;164A:251-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 13. | Ansari M, Poke G, Ferry Q, Williamson K, Aldridge R, Meynert AM, Bengani H, Chan CY, Kayserili H, Avci S, Hennekam RC, Lampe AK, Redeker E, Homfray T, Ross A, Falkenberg Smeland M, Mansour S, Parker MJ, Cook JA, Splitt M, Fisher RB, Fryer A, Magee AC, Wilkie A, Barnicoat A, Brady AF, Cooper NS, Mercer C, Deshpande C, Bennett CP, Pilz DT, Ruddy D, Cilliers D, Johnson DS, Josifova D, Rosser E, Thompson EM, Wakeling E, Kinning E, Stewart F, Flinter F, Girisha KM, Cox H, Firth HV, Kingston H, Wee JS, Hurst JA, Clayton-Smith J, Tolmie J, Vogt J, Tatton-Brown K, Chandler K, Prescott K, Wilson L, Behnam M, McEntagart M, Davidson R, Lynch SA, Sisodiya S, Mehta SG, McKee SA, Mohammed S, Holden S, Park SM, Holder SE, Harrison V, McConnell V, Lam WK, Green AJ, Donnai D, Bitner-Glindzicz M, Donnelly DE, Nellåker C, Taylor MS, FitzPatrick DR. Genetic heterogeneity in Cornelia de Lange syndrome (CdLS) and CdLS-like phenotypes with observed and predicted levels of mosaicism. J Med Genet. 2014;51:659-668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 14. | Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2647] [Cited by in RCA: 2668] [Article Influence: 222.3] [Reference Citation Analysis (0)] |
| 15. | Kline AD, Moss JF, Selicorni A, Bisgaard AM, Deardorff MA, Gillett PM, Ishman SL, Kerr LM, Levin AV, Mulder PA, Ramos FJ, Wierzba J, Ajmone PF, Axtell D, Blagowidow N, Cereda A, Costantino A, Cormier-Daire V, FitzPatrick D, Grados M, Groves L, Guthrie W, Huisman S, Kaiser FJ, Koekkoek G, Levis M, Mariani M, McCleery JP, Menke LA, Metrena A, O'Connor J, Oliver C, Pie J, Piening S, Potter CJ, Quaglio AL, Redeker E, Richman D, Rigamonti C, Shi A, Tümer Z, Van Balkom IDC, Hennekam RC. Diagnosis and management of Cornelia de Lange syndrome: first international consensus statement. Nat Rev Genet. 2018;19:649-666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 243] [Article Influence: 34.7] [Reference Citation Analysis (0)] |