Published online Nov 26, 2022. doi: 10.12998/wjcc.v10.i33.12077
Peer-review started: May 7, 2022
First decision: July 13, 2022
Revised: August 15, 2022
Accepted: October 11, 2022
Article in press: October 11, 2022
Published online: November 26, 2022
Processing time: 200 Days and 0.8 Hours
Every year, esophageal cancer is responsible for 509000 deaths and around 572000 new cases worldwide. Although esophageal cancer treatment options have advanced, patients still have a dismal 5-year survival rate.
To investigate the relationship between genes associated to platelets and the prognosis of esophageal cancer.
We searched differentially expressed genes for changes between 151 tumor tissues and 653 normal, healthy tissues using the “limma” package. To develop a prediction model of platelet-related genes, a univariate Cox regression analysis and least absolute shrinkage and selection operator Cox regression analysis were carried out. Based on a median risk score, patients were divided into high-risk and low-risk categories. A nomogram was created to predict the 1-, 2-, and 3-year overall survival (OS) of esophageal cancer patients using four platelet-related gene signatures, TNM stages, and pathological type. Additionally, the concor
The prognosis of esophageal cancer was associated to APOOL, EP300, PLA2G6, and VAMP7 according to univariate Cox regression analysis and least absolute shrinkage and selection operator regression analysis. Patients with esophageal cancer at high risk had substantially shorter OS than those with cancer at low risk, according to a Kaplan-Meier analysis (P < 0.05). TNM stage (hazard ratio: 2.187, 95% confidence interval: 1.242-3.852, P = 0.007) in both univariate and multivariate Cox regression and risk score were independently correlated with OS (hazard ratio: 2.451, 95% confidence interval: 1.599-3.756, P < 0.001).
A survival risk score model and independent prognostic variables for esophageal cancer have been developed using APOOL, EP300, PLA2G6, and VAMP7. OS for esophageal cancer might be predicted using the nomogram based on TNM stage, pathological type, and risk score. The nomogram demonstrated strong predictive ability, as shown by the concordance index, receiver operating characteristic curve, and calibration curve.
Core Tip: Esophageal cancer is one of the most prevalent cancers. Despite significant improvements in esophageal cancer therapy over the past several years, the survival rates of patients with the malignancy are still extremely low. Numerous studies have demonstrated the important role platelets play in the initiation and growth of tumors. The precise underlying biological processes of platelet-related genes in esophageal cancer are unknown. An efficient risk score model based on the platelet-related genes may accurately predict the survival of patients with esophageal cancer, helping to clarify the relationship between platelet-related genes and the prognosis of the disease.
- Citation: Du QC, Wang XY, Hu CK, Zhou L, Fu Z, Liu S, Wang J, Ma YY, Liu MY, Yu H. Integrative analysis of platelet-related genes for the prognosis of esophageal cancer. World J Clin Cases 2022; 10(33): 12077-12088
- URL: https://www.wjgnet.com/2307-8960/full/v10/i33/12077.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i33.12077
Esophageal cancer incidence is increasing globally, and western countries have a higher prevalence of adenocarcinomas. Over 95% of esophageal cancer patients in China are diagnosed with esophageal squamous cell carcinomas; males are more likely than females to have this kind of cancer[1]. The 5-year survival rate for esophageal cancer patients is a little less than 20% despite improvements in therapy[2]. Target therapy and immunotherapy have made significant advancements in the treatment of esophageal cancer, although only some patients may benefit from them. Therefore, biomarkers to guide therapy and predict prognosis are urgently needed for patients with esophageal cancer.
In recent years, several attempts have been undertaken to better forecast the biology of each individual in esophageal cancer and to find prognostic and predictive biomarkers. Despite this, individuals with esophageal cancer often have a dismal prognosis because there are currently no reliable biomarkers for prognosis prediction and early identification.
Platelets may help tumor cells escape the immune system if they abnormally concentrate in the peripheral circulation and survive for prolonged periods of time. Platelet-derived growth factors, such as platelet factor 4, thrombospondin, and vascular endothelial growth factor, aid in the adhesion, invasion, angiogenesis, and tumor development of hematogenous cancer[3]. These findings suggest that platelets may be crucial in the growth of tumors in a variety of cancer types. Platelet research is mostly focused on the relationship between platelet characteristics and the prognosis of malignant tumors[4,5]. It may be possible to find novel treatment targets and improve the prognosis of malignant tumors by better understanding platelet-related genes and the underlying processes.
The link between platelet-related genes and survival in esophageal cancer patients has never been addressed, and only a small number of studies have looked at this relationship. This study examined the expression patterns of platelet-related genes and their prognostic significance in esophageal cancer by bioinformatics analysis. Finally, using pathway enrichment analysis to uncover probable biological roles for the platelet-related gene signature, we further investigated its function.
Animals were not used in the study nor were there any human participants, data, or tissue. A public database served as the source for all the data.
The Cancer Genome Atlas website at https://portal.gdc.cancer.gov/ allows users to obtain clinical and gene expression data of patients with esophageal cancer. Gene expression levels in normal tissue samples were extracted using Genotype-Tissue Expression databases (https://xenabrowser.net/). Using the “limma” package in R, raw data is normalized before further data processing. With the term “platelet” as the target keyword, 369 genes with a connection to platelets were found in AmiGO 2 (http://amigo.geneontology.org/).
Using the “limma” package and the Wilcoxon signed-rank test in the R project, we found platelet-related genes that were differently expressed between esophageal cancer and the healthy control group. By utilizing log fold change > 1 and false discovery rates < 0.05 as criteria, differentially expressed genes (DEGs) were chosen. The univariate Cox regression tests were used to evaluate the prognostic implications of overall survival (OS) for esophageal cancer. The intersection of DEGs and prognostic genes yielded the DEGs associated with the prognosis of esophageal cancer. Additionally, prospective risk factors based on genes connected to platelets were developed using least absolute shrinkage and selection operator Cox regression using the package “glmnet.” The penalty regularization parameter lambda (λ) was established using ten-fold cross validation, and we selected the value where the partial likelihood deviance was the minimum in order to prevent overfitting effects in the model. We generated the risk score using the following formula: risk score[6] = Σgenes Cox coefficient × genes expression levels. Then, based on their median risk ratings, patients were divided into high-risk and low-risk groups.
Using the R function “prcomp” from the “stats” package, principal component analysis was performed on each set of data. Additionally, the R package “Rtsne” was utilized to create the t-distributed stochastic neighbor embedding approach in order to visualize clustering. The optimal cutoffs for the survival analysis of each gene were determined using the “surv cutpoint” function of the “survminer” R package. Finally, a software package called “survival operating characteristic (ROC)” was adopted to analyze the time-dependent ROC curve in order to evaluate the gene signature’s predictive ability for time-dependent cancer mortality.
We used the “ClusterProfiler” R package to distinguish between the functional pathways enriched by the DEGs in the high-risk and low-risk groups based on Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Additionally, results from expression analysis and functional annotation enrichment analysis were displayed using the GO Circle and GO Chord plot methods in the “GO plot” R package.
With the use of prognostic gene signatures and a combination of clinicopathological factors, we developed a nomogram to predict each person’s probability of survival. Performance of the nomogram was evaluated using calibration plots and the concordance index. The dotted 45-degree line on the calibration graph represented the ideal forecast, while the X- and Y-axes on the graph represented the nomogram-predicted progression and the observed outcome, respectively. The model’s performance was assessed using a bootstrapping approach, which could compensate for overly optimistic assumptions.
R (version 4.1.0, http://www.r-project.org/) was used to perform the statistical analysis. The gene expression in tumor tissues and healthy control tissues was compared using the Student’s t test. The Fisher’s exact test or the χ2test, as applicable, were used to compare proportions. The Kaplan-Meier method was also used to assess progression-free survival and OS using log-rank testing. Univariate and multivariate Cox proportional hazard regression analyses were used to identify OS independent factors. Less than 0.05 was considered statistically significant for two-tailed P values.
We recruited 653 healthy individuals as controls and 151 patients with esophageal cancer as the case group. The Cancer Genome Atlas database contained details on 151 esophageal cancer cases, whereas the Genotype-Tissue Expression database contained information on 653 healthy controls. All information came from freely accessible public databases. The clinical characteristics of 151 esophageal cancer patients are summarized in Table 1.
| Variables | Esophageal cancer | Percent | |
| Age in yr | |||
| ≤ 65 | 94 | 62.3 | |
| > 65 | 57 | 37.7 | |
| Sex | |||
| Male | 129 | 85.4 | |
| Female | 22 | 14.6 | |
| Stage | |||
| Ⅰ | 18 | 11.9 | |
| Ⅱ | 70 | 46.4 | |
| Ⅲ | 51 | 33.8 | |
| Ⅳ | 12 | 7.9 | |
| Grade | |||
| G1 | 16 | 10.6 | |
| G2 | 60 | 39.7 | |
| G3 | 41 | 27.2 | |
| Gx | 34 | 22.5 | |
| Alcohol consumption | |||
| Yes | 105 | 69.5 | |
| No | 43 | 28.5 | |
| Unknown | 3 | 2.0 | |
| Symptoms | |||
| Yes | 116 | 76.8 | |
| No | 10 | 6.6 | |
| Unknown | 25 | 16.6 | |
| Pathological type | |||
| Squamous cell carcinoma | 77 | 51.0 | |
| Adenocarcinoma | 74 | 49.0 | |
| Survival status | |||
| Alive | 93 | 61.6 | |
| Deceased | 58 | 38.4 | |
| Smoking-yrs | 38.5 (30.0-50.0) | ||
| BMI | 24.4 (21.2-27.4) | ||
| Mean follow-up time in mo | 13.4 (7.7-23.1) | ||
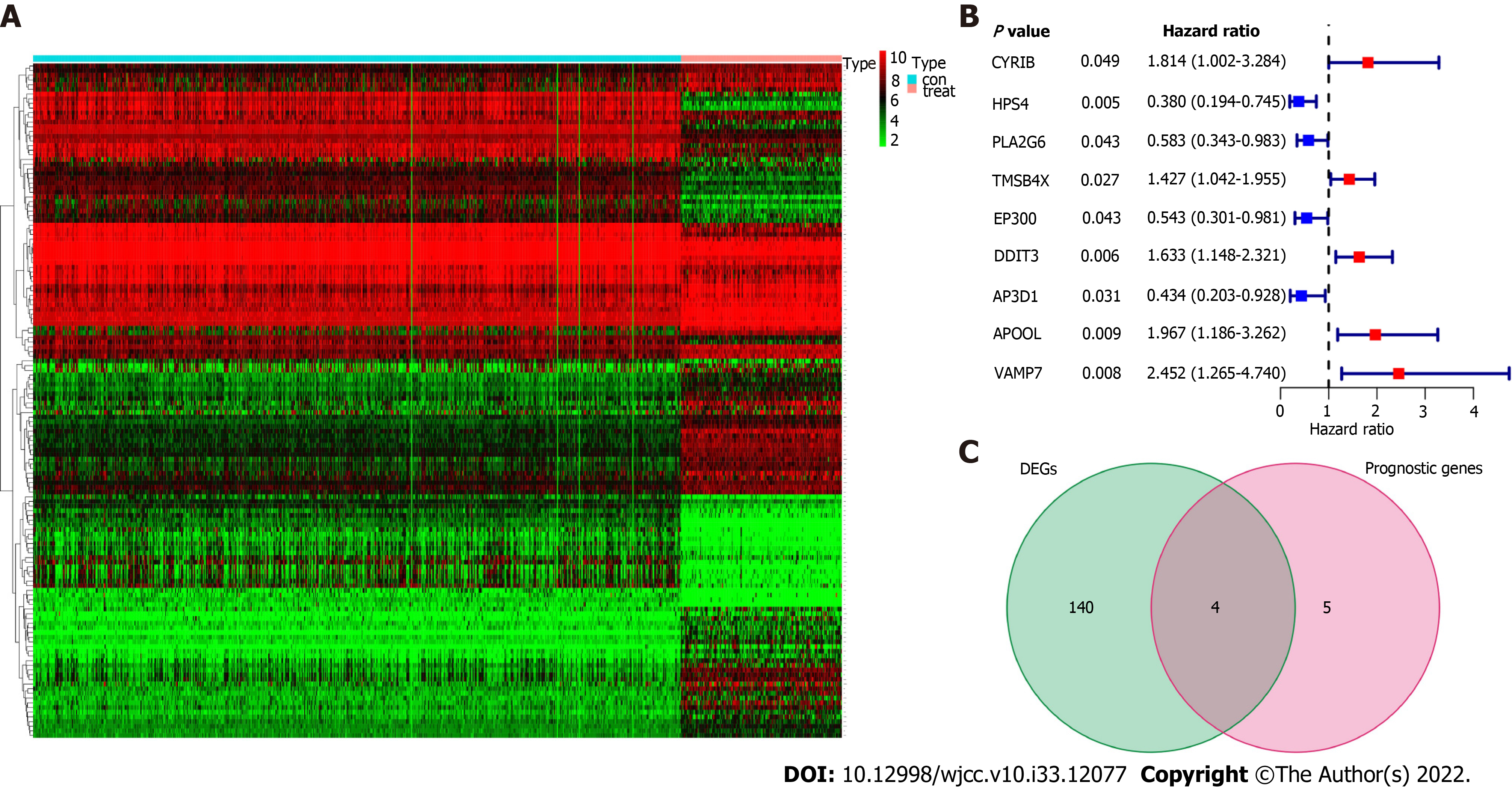
We discovered 144 DEGs when comparing the case group to the healthy control group, of which 77 genes were upregulated and 67 genes were downregulated (Figure 1A). Using univariate Cox proportional regression analysis for 369 platelet-related genes, we identified 9 platelet-related prognostic genes, including AP3D1, APOOL, CYRIB, DDIT3, EP300, HPS4, PLA2G6, TMSB4X, and VAMP7, as being substantially correlated with OS (Figure 1B, P < 0.05). We utilized a Venn diagram to discover the intersection between DEGs and platelet-related prognostic genes in order to further select platelet-related DEGs. The findings demonstrate that four genes, including APOOL, EP300, PLA2G6 and VAMP7, were connected to esophageal cancer prognosis (Figure 1C).
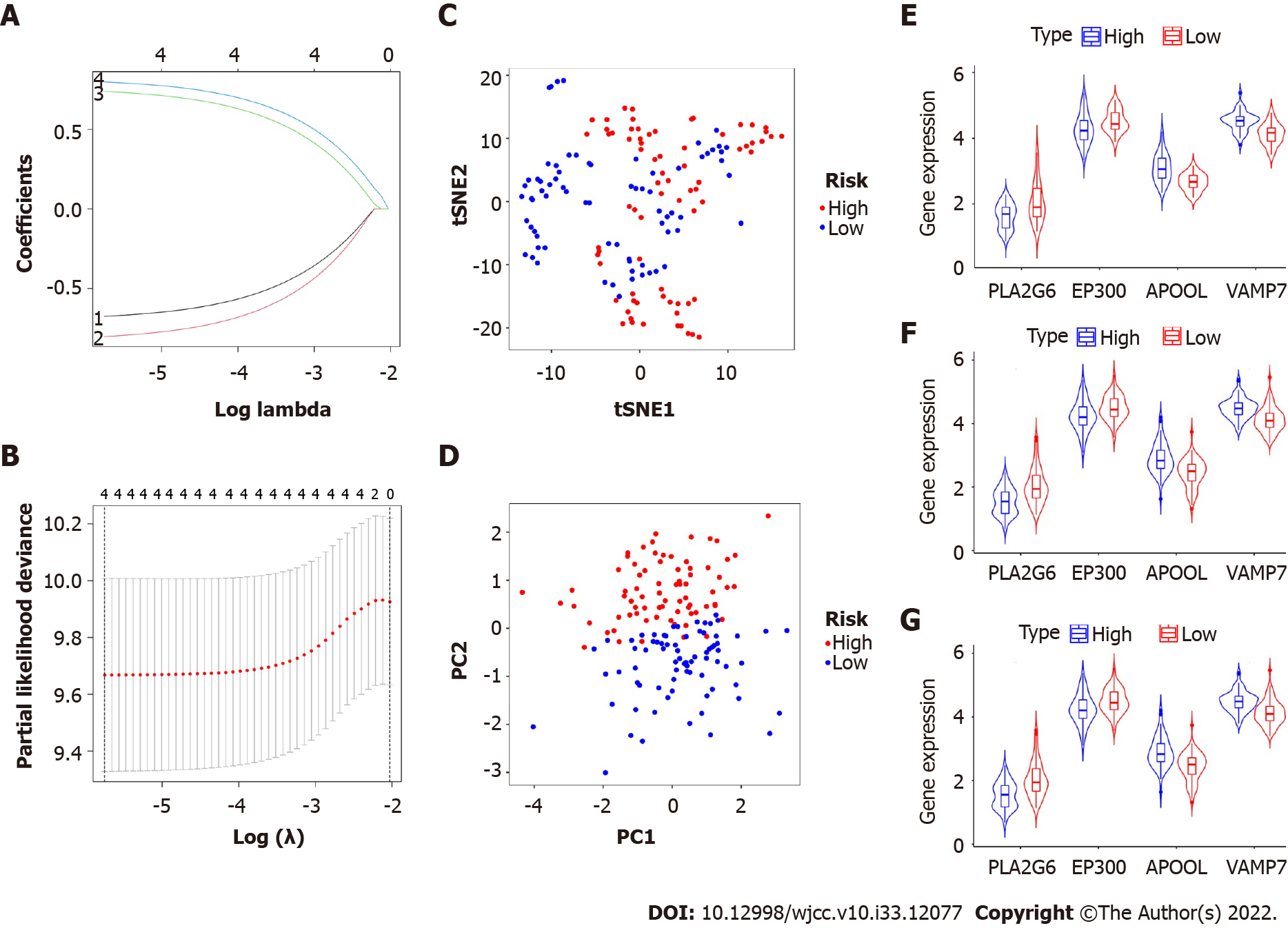
Additionally, we utilized the least absolute shrinkage and selection operator Cox analysis to choose the top four prognostic biomarkers, including APOOL, EP300, PLA2G6, and VAMP7, and we subsequently developed a classifier for predicting survival based on the four platelet-related prognostic genes (Figure 2A and B, Table 2). The formula used was: risk score = 0.739 × expression level of APOOL - 0.801 × expression level of EP300 - 0.674 × expression level of PLA2G6 + 0.798 × expression level of VAMP7. Using the aforementioned formula, we calculated the risk scores for each sample. The median risk score was then used to classify the patients into low-risk and high-risk categories. The distributions of the high- and low-risk groups were marginally dispersed, according to the t-distributed stochastic neighbor embedding analysis (Figure 2C). The expression of the four platelet-related genes was used to compare the differences between the low- and high-risk groupings using principal component analysis (Figure 2D). In conclusion, high-risk and low-risk groups can be easily distinguished by the predictive characteristics of platelet-related genes.
| Gene | Univariate Cox regression analysis | LASSO | ||
| HR | 95%CI | P | Coefficient | |
| PLA2G6 | 0.581 | 0.343-0.983 | 0.040 | -0.674 |
| EP300 | 0.543 | 0.301-0.981 | 0.040 | -0.801 |
| APOOL | 1.967 | 1.186-3.262 | 0.008 | 0.739 |
| VAMP7 | 2.452 | 1.269-4.740 | 0.008 | 0.798 |
To ascertain if our risk score model correlated with clinicopathological parameters, squamous cell carcinoma and adenocarcinoma were examined as two clinicopathological findings. A similar outcome was shown in subgroups of adenocarcinoma and squamous cell carcinoma as a same consequence of the evaluation between subgroups and the entire population (Figure 2E, F, and G). In contrast to PLA2G6 and EP300, which were strongly expressed in the low-risk group, APOOL and VAMP7 were substantially expressed in the high-risk group. Age, sex, grade, and stage were clinical factors that did not differ between the high- and low-risk groups in our research. However, there were still notable variances between the various pathological categories of the two groups. Patients with adenocarcinoma had a comparatively high-risk score (Table 3).
| Parameters | Risk model | χ2 | P | |
| High risk | Low risk | |||
| Age in yr | 0.321 | 0.571 | ||
| ≤ 65 | 45 | 49 | ||
| > 65 | 30 | 27 | ||
| Sex | 1.824 | 0.177 | ||
| Male | 67 | 62 | ||
| Female | 8 | 14 | ||
| Grade | 0.188 | 0.910 | ||
| G1-2 | 37 | 39 | ||
| G3 | 20 | 21 | ||
| Gx | 18 | 16 | ||
| Histological type | 4.134 | 0.042 | ||
| Squamous cell carcinoma | 32 | 45 | ||
| Adenocarcinoma | 43 | 31 | ||
| Stage | 2.416 | 0.120 | ||
| Ⅰ-Ⅱ | 39 | 49 | ||
| Ⅲ-Ⅳ | 36 | 27 | ||
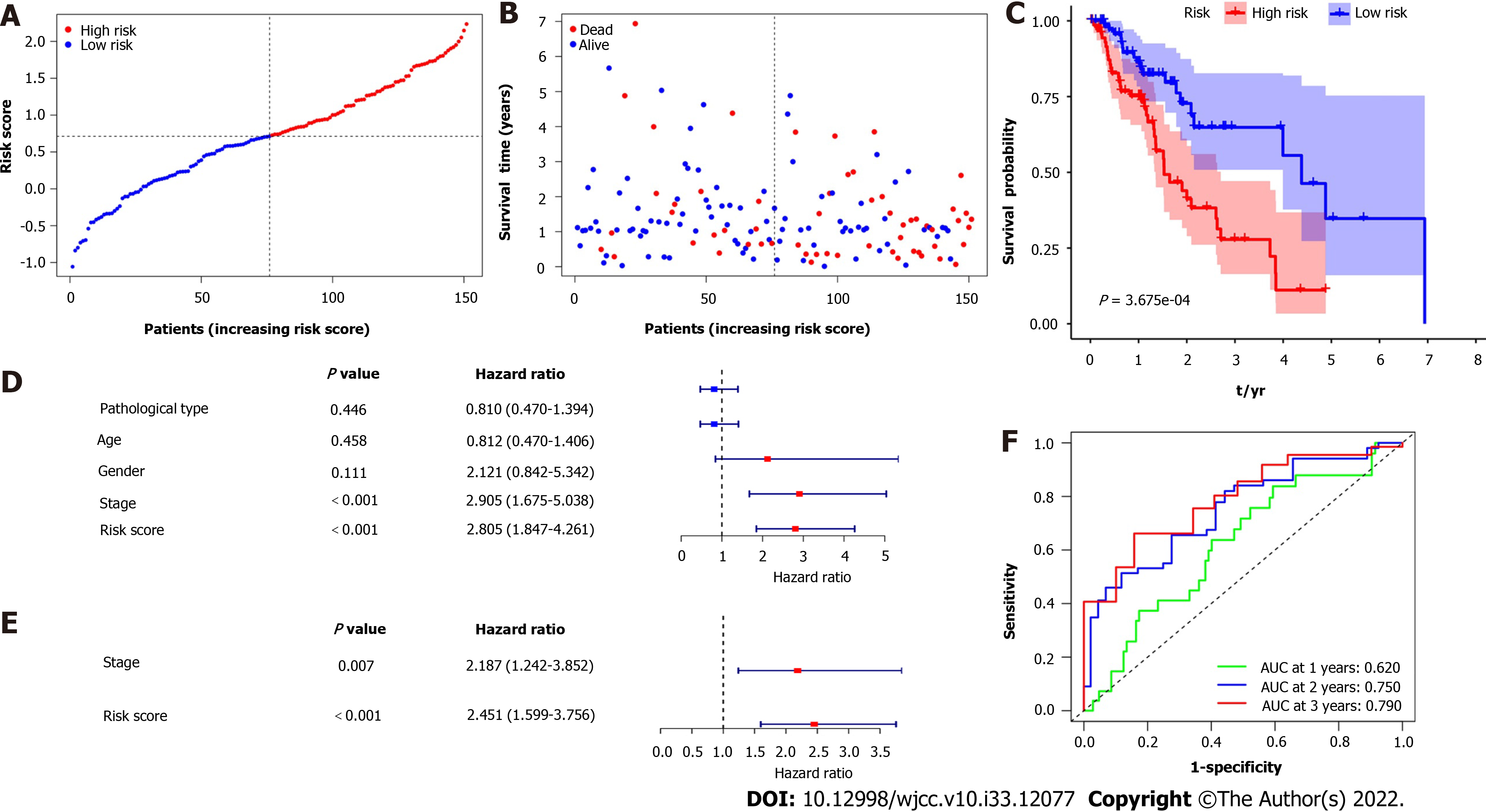
Figure 3A and B show that patient survival times decreased as risk ratings rose, and largely all of the patients who passed away belonged to the high-risk category. The prognostic significance of DEGs in esophageal cancer was assessed using Kaplan-Meier survival analysis (Figure 3C). Patient prognoses were considerably poorer in the high-risk group than in the low-risk group (P < 0.001). Additionally, the univariate Cox regression analysis was carried out to determine the prognostic significance of the features (Figure 3D). The stage and risk score were statistically significant (P < 0.001), while the individuals’ age, sex, and pathological type had no statistically significant impact on their survival. Furthermore, the multivariate analysis revealed that the stage and risk score were independent risk factors for OS (Figure 3E). The area under the curve (AUC) for survival after 1, 2, and 3 years using ROC analysis was 0.620, 0.750, and 0.790, respectively (Figure 3F). In other words, the platelet-related gene signature offered useful predictive value with clinical relevance for appropriately classifying OS patients.
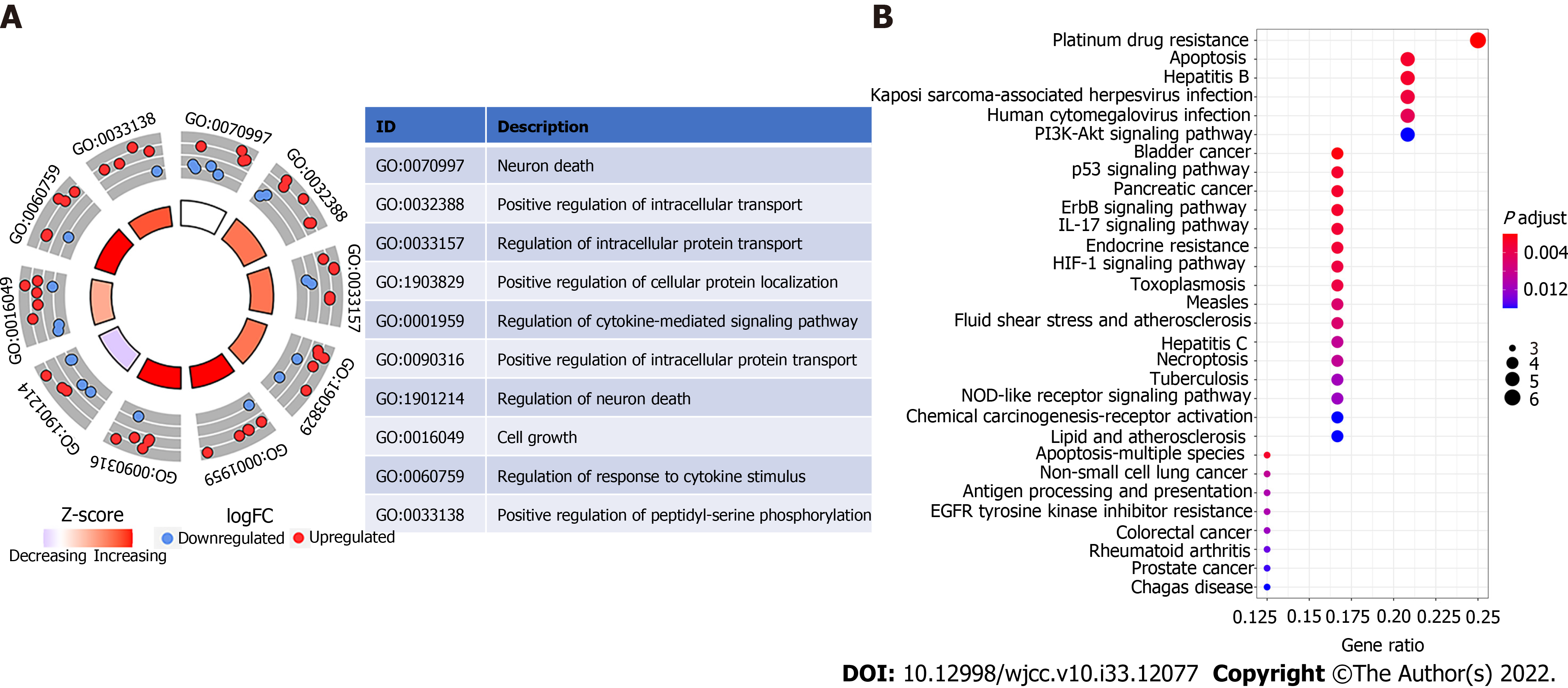
GO enrichment analysis revealed ten significantly enriched pathways, including those that control neuron death, cell growth, response to cytokine stimulus, positive regulation of phosphorylation, positive regulation of intracellular protein transport, positive regulation of cellular protein localization, positive regulation of intracellular transport, and regulation of cytokine-mediated signaling pathway (Figure 4A). “Neuron death” is a biological process word with the greatest richness (GO: 0070997). Platinum drug resistance was the most enriched and important route, according to KEGG analysis. Additionally, apoptosis was significantly enriched (Figure 4B).
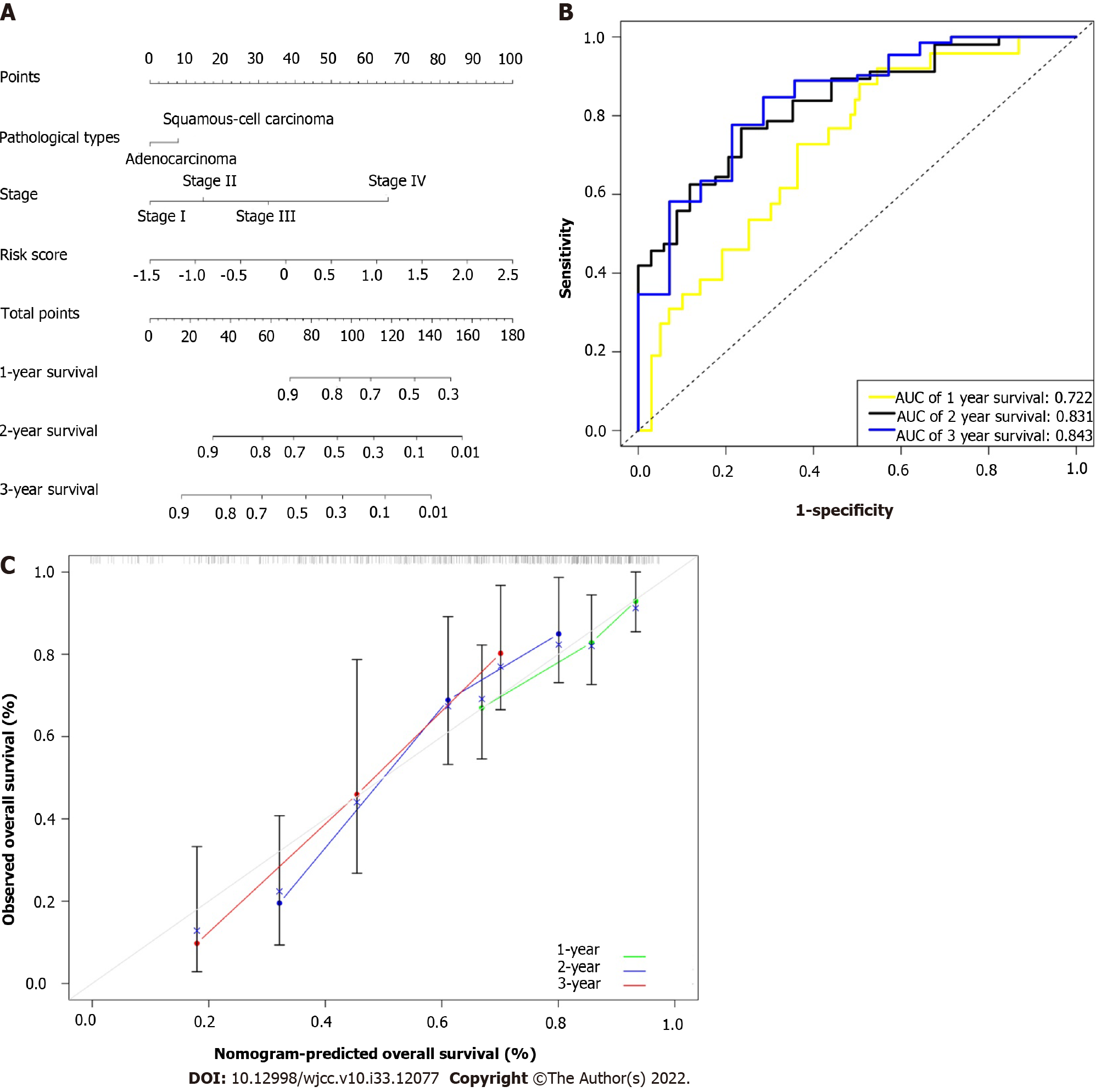
As previously mentioned, statistical analysis showed that the two independent prognostic indicators linked to OS were TNM stage and risk score. In order to eliminate pathological type interference, two pathological types were added to the nomogram. Finally, to predict the prognosis of esophageal cancer more accurately, we created a novel prognostic nomogram based on TNM stage, pathological type, and risk score (Figure 5A). The nomogram was used to evaluate each patient’s probability of survival, and the ROC curve was used to assess how well the nomogram predicted outcomes. In 1-, 2-, and 3-year intervals, this nomogram’s AUC was 0.722, 0.831, and 0.843, respectively (Figure 5B). The nomogram’s calibration curves for the probabilities of 1-, 2-, and 3-year survival demonstrated good agreement between prediction and observation (Figure 5C). The nomogram’s Harrell’s concordance index value was 0.729. After additional verification, our nomogram performed quite well.
Esophageal cancer is mostly an aging illness, peaking in incidence in the eighth decade of life, and the global elderly population is expanding quickly[7]. Even after full resection of esophageal cancer, the prognosis remains dismal despite advancements in multimodal treatments that integrate surgery, chemotherapy, radiation, and chemoradiotherapy[8]. The main treatment for early esophageal cancer was surgical resection; however, patients who had recurrence or progression of the cancer had a difficult time getting treatment[9]. Although tumor grade offers useful prognostic information, other trustworthy factors are required for more accurate prognosis prediction.
In terms of incidence, esophageal cancer is the eighth most common cancer worldwide, and it is the sixth most lethal[10]. The majority of those affected by this illness are elderly, and the average age of diagnosis is becoming older, peaking between 70-years-old and 75-years-old[11]. However, there were no age differences between the high-risk and low-risk esophageal cancer groups in our study.
Numerous forms of solid tumors typically exhibit platelet-related characteristics, which are essential to the development and growth of tumors[12,13]. Platelets help tumor cells grow, survive, and migrate[14]. To shield tumor cells from immune responses, platelets may form complexes with tumor cells[15]. Conversely, tumor cells are known to both activate platelets and cause platelet aggregation[12]. These mechanisms of tumor cell-platelet interactions, however, are still poorly understood. In order to further investigate the relationship between platelets and esophageal cancer, 369 platelet-related genes were included. By using univariate Cox and multivariate Cox analysis, four differentially expressed prognostic genes were identified, and a prognostic risk model based on a four platelet-related gene signature was established.
Four gene signatures have been discovered in this study utilizing univariate Cox regression analysis and the least absolute shrinkage and selection operator Cox regression model to predict OS for esophageal cancer. Additionally, 151 patients with esophageal cancer were divided into high- and low-risk groups based on the median risk model score using gene expression data. The prognosis between the high-risk group and the low-risk group differed significantly. As a result, the risk score method can accurately predict the results of esophageal cancer samples. GO and KEGG analyses were used to analyze biological processes and pathways in order to better investigate the mechanisms behind our risk model. Analyses of GO and KEGG data revealed that the majority of GO and KEGG enrichments were related to carcinogenesis and development. The risk model contained the four genes APOOL, EP300, PLA2G6 and VAMP7. While the expression levels of EP300 and PLA2G6 were positively connected with prognosis, the expression levels of APOOL and VAMP7 were adversely correlated with esophageal cancer.
According to Zhu et al[16], VAMP7 is implicated in the promotion of tumors since it is upregulated in high-risk individuals. Poor prognosis for esophageal squamous carcinoma was linked to EP300 epitopes as an oncogene[17]. The EP300 oncogene promotes tumor development, as was shown in earlier research in vitro using esophageal squamous cell carcinoma cell lines[17]. Studies on lung, colon, prostate, bladder, and breast cancer have also demonstrated this[18-20]. There are, however, few studies on APOOL and PLA2G6 in the growth and prognosis of tumors.
This study developed a nomogram for risk assessment based on multivariate Cox regression analysis to evaluate the OS of patients with esophageal cancer. The TNM stage and risk score are independent predictive factors for esophageal cancer, according to the univariate and multivariate Cox regression models. We further incorporated clinicopathological factors in addition to TNM stages and risk scores. The two histopathological types of primary esophageal cancer that are most frequently identified in clinics are squamous cell carcinoma and adenocarcinoma; they have the same potential risks, gene alterations, and treatments[21]. These two histopathological tumor types exhibit distinct behaviors in esophageal cancer, with squamous cell carcinoma exhibiting an earlier lymphatic migration and a poorer prognosis than adenocarcinoma[22]. Esophageal squamous cell carcinoma has a bad prognosis in terms of 5-year survival rates (15%–25%) since it is the most common subtype of esophageal cancer and has a higher occurrence in specific geographic areas[23]. The OS of esophageal cancer and the histopathological type were not significantly different in our study. But between the two pathological types, there was a statistically significant difference in the high-risk or low-risk groups. As a result, the nomogram also included pathological types.
In order to verify the nomogram’s effectiveness, its accuracy was assessed. The esophageal cancer cohort also demonstrated good calibration and discrimination; in particular, our high concordance index value (0.729) indicated the performance of the nomogram[24]. The nomogram had strong predictive power, as evidenced by the ROC curve, which had AUC values of 0.722 at 1 year, 0.831 at 2 years, and 0.843 at 3 years. The calibration curve’s observation and prediction exhibited a fair amount of agreement, indicating that the nomogram may be utilized to predict the 1-, 2-, and 3-year survival rates for the cohort with esophageal cancer[25]. Overall, the findings of the calibration curve, ROC curve, and concordance index revealed that our developed prediction nomogram had excellent prediction performance.
This study had a number of advantages. First, our study was the first to describe platelet-related genes and the prognosis of esophageal cancer. Four prognostic differential genes combined with histopathological types and TNM stages were used to construct a prognosis nomogram. The individualized prediction of this nomogram may reflect the survival rate of esophageal cancer candidates more correctly, making up for the inadequacy of earlier TNM stages. Second, our analysis differed from earlier research of similar studies. We choose more control group samples than in earlier research in order to exclude the interference of a small sample of the control group in the differential analysis. Finally, we developed a prognostic model that predicted 1-, 2-, and 3-year survival with high AUC utilizing a 4-gene signature, histopathological types, and TNM stages.
This study had a number of limitations. First, there was not enough esophageal cancer in our study’s sample to build prediction models based on adenocarcinoma and squamous cell carcinoma. Second, it was difficult to tell if the four DEGs differed between normal and malignant tissues due to a lack of additional histology specimens for external confirmation. Additionally, we utilized statistical analysis to find the related differential gene and GO and KEGG enrichment analysis to confirm the DEG’s pathway and enrichment function. There was no mechanism research to confirm these findings.
In conclusion, we constructed a novel esophageal cancer risk model based on four platelet-related genes. It was a great prognostic information tool that could be utilized to supplement the TNM staging system combined with histopathological types and risk scores.
The prognosis for esophageal cancer, one of the malignancies that responds least to cancer therapy, has not improved despite several breakthroughs in treatment. Improving patient outcomes depends on finding biomarkers and comprehending the molecular causes of esophageal cancer.
We wanted to create a risk score based on platelet-related gene signatures for prognosis prediction since the expression of platelet-related genes is strongly linked to patient prognosis.
To predict esophageal cancer prognosis, a risk model and nomogram constructed based on platelet-related gene signatures and clinical factors associated with prognosis could be utilized.
We constructed a trustworthy platelet-related gene signature to predict the prognosis of esophageal cancer using 151 samples of the disease. Then, an integrated nomogram for clinical practice was created utilizing a combined risk score, risk score, and TNM stage. The prognostic accuracy of the model was also supported by the receiver operating characteristic curve, concordance index, and related calibration curve.
The survival curve was created after constructing a prognostic risk model based on four platelet genes associated with prognosis. Patients in the high-risk group had a considerably lower life expectancy than those in the low-risk group, according to the Kaplan-Meier survival analysis. The risk score was an independent factor in predicting survival, according to results from both univariate and multivariate Cox regression analyses.
We identified a four-gene signature, constructed a risk score, and developed a prediction nomogram for patients with esophageal cancer based on the risk score, TNM staging, and histopathological type.
Identification and prediction of prognostic indicators are essential for esophageal cancer patients.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Surgery
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): E
P-Reviewer: Endo S, Japan; Micsik T, Hungary; Rasoulinejad A, Egypt S-Editor: Xing YX L-Editor: Filipodia P-Editor: Xing YX
| 1. | Yan MH, Hou XB, Cai BN, Qu BL, Dai XK, Liu F. Neoadjuvant chemoradiotherapy plus surgery in the treatment of potentially resectable thoracic esophageal squamous cell carcinoma. World J Clin Cases. 2020;8:6315-6321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (1)] |
| 2. | Lv J, Guo L, Liu JJ, Zhao HP, Zhang J, Wang JH. Alteration of the esophageal microbiota in Barrett's esophagus and esophageal adenocarcinoma. World J Gastroenterol. 2019;25:2149-2161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 85] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (2)] |
| 3. | Wang YQ, Zhi QJ, Wang XY, Yue DS, Li K, Jiang RC. Prognostic value of combined platelet, fibrinogen, neutrophil to lymphocyte ratio and platelet to lymphocyte ratio in patients with lung adenosquamous cancer. Oncol Lett. 2017;14:4331-4338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 4. | Li F, Wang L, Zhang Y, Feng W, Ju T, Liu Z, Wang Z, Du X. A Retrospective Study on Using a Novel Single Needle Cone Puncture Approach for the Iodine-125 Seed Brachytherapy in Treating Patients With Thoracic Malignancy. Front Oncol. 2021;11:640131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Han D, Zhang J, Zhao J, Lei T, Chen X, Zhang T, Wei H, Guan Y, Wang J, Zhang W, Zhao L, Yuan Z, Song Y, Liu N, Pang Q, Wang P. Platelet-to-lymphocyte ratio is an independent predictor of chemoradiotherapy-related esophageal fistula in esophageal cancer patients. Ann Transl Med. 2020;8:1163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Zhang T, Nie Y, Xia H, Zhang Y, Cai K, Chen X, Li H, Wang J. Identification of Immune-Related Prognostic Genes and LncRNAs Biomarkers Associated With Osteosarcoma Microenvironment. Front Oncol. 2020;10:1109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Cui MT, Liang ZW, Sun YZ, Wu J, Lu H, Wang WJ, Xu MD, Jiang M, Li W, Qian J, Duan WM. The prognostic roles of red blood cell-associated indicators in patients with resectable gastric cancers. Transl Cancer Res. 2020;9:2300-2311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Baba Y, Nomoto D, Okadome K, Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N, Baba H. Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma. Cancer Sci. 2020;111:3132-3141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 191] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 9. | Xu S, Zhou Y, Biekemitoufu H, Wang H, Li C, Zhang W, Ma Y. Expression of Twist, Slug and Snail in esophageal squamous cell carcinoma and their prognostic significance. Oncol Lett. 2021;21:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 10. | Kuwai T, Yamaguchi T, Imagawa H, Miura R, Sumida Y, Takasago T, Miyasako Y, Nishimura T, Iio S, Yamaguchi A, Kouno H, Kohno H, Ishaq S. Endoscopic submucosal dissection for early esophageal neoplasms using the stag beetle knife. World J Gastroenterol. 2018;24:1632-1640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 11. | Hirahara N, Tajima Y, Fujii Y, Kaji S, Yamamoto T, Hyakudomi R, Taniura T, Kawabata Y. Comprehensive Analysis of Red Blood Cell Distribution Width as a Preoperative Prognostic Predictor in Gastric Cancer. Anticancer Res. 2019;39:3121-3130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Huh G, Ryu JK, Chun JW, Kim JS, Park N, Cho IR, Paik WH, Lee SH, Kim YT. High platelet-to-lymphocyte ratio is associated with poor prognosis in patients with unresectable intrahepatic cholangiocarcinoma receiving gemcitabine plus cisplatin. BMC Cancer. 2020;20:907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Yang J, Xu H, Guo X, Zhang J, Ye X, Yang Y, Ma X. Pretreatment Inflammatory Indexes as Prognostic Predictors for Survival in Colorectal Cancer Patients Receiving Neoadjuvant Chemoradiotherapy. Sci Rep. 2018;8:3044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 14. | Bang KH, Na YG, Huh HW, Hwang SJ, Kim MS, Kim M, Lee HK, Cho CW. The Delivery Strategy of Paclitaxel Nanostructured Lipid Carrier Coated with Platelet Membrane. Cancers (Basel). 2019;11:807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 15. | Chen L, Zeng H, Yang J, Lu Y, Zhang D, Wang J, Kuang C, Zhu S, Wang M, Ma X. Survival and prognostic analysis of preoperative inflammatory markers in patients undergoing surgical resection for laryngeal squamous cell carcinoma. BMC Cancer. 2018;18:816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 16. | Zhu L, Dong L, Feng M, Yang F, Jiang W, Huang Z, Liu F, Wang L, Wang G, Li Q. Profiles of autophagy-related genes in esophageal adenocarcinoma. BMC Cancer. 2020;20:943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Bi Y, Kong P, Zhang L, Cui H, Xu X, Chang F, Yan T, Li J, Cheng C, Song B, Niu X, Liu X, Xu E, Hu X, Qian Y, Wang F, Li H, Ma Y, Yang J, Liu Y, Zhai Y, Wang Y, Zhang Y, Liu H, Liu J, Wang J, Cui Y, Cheng X. EP300 as an oncogene correlates with poor prognosis in esophageal squamous carcinoma. J Cancer. 2019;10:5413-5426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Alsamri H, Hasasna HE, Baby B, Alneyadi A, Dhaheri YA, Ayoub MA, Eid AH, Vijayan R, Iratni R. Carnosol Is a Novel Inhibitor of p300 Acetyltransferase in Breast Cancer. Front Oncol. 2021;11:664403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 19. | Li J, Huang C, Xiong T, Zhuang C, Li Y, Ye J, Gui Y. A CRISPR Interference of CBP and p300 Selectively Induced Synthetic Lethality in Bladder Cancer Cells In Vitro. Int J Biol Sci. 2019;15:1276-1286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 20. | Du C, Huang D, Peng Y, Yao Y, Zhao Y, Yang Y, Wang H, Cao L, Zhu WG, Gu J. 5-Fluorouracil targets histone acetyltransferases p300/CBP in the treatment of colorectal cancer. Cancer Lett. 2017;400:183-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 21. | Okuda Y, Shimura T, Iwasaki H, Fukusada S, Nishigaki R, Kitagawa M, Katano T, Okamoto Y, Yamada T, Horike SI, Kataoka H. Urinary microRNA biomarkers for detecting the presence of esophageal cancer. Sci Rep. 2021;11:8508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Liu CC, Chou KT, Hsu JW, Lin JH, Hsu TW, Yen DH, Hung SC, Hsu HS. High metabolic rate and stem cell characteristics of esophageal cancer stem-like cells depend on the Hsp27-AKT-HK2 pathway. Int J Cancer. 2019;145:2144-2156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 23. | Wang X, Kutschat AP, Yamada M, Prokakis E, Böttcher P, Tanaka K, Doki Y, Hamdan FH, Johnsen SA. Bromodomain protein BRDT directs ΔNp63 function and super-enhancer activity in a subset of esophageal squamous cell carcinomas. Cell Death Differ. 2021;28:2207-2220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 24. | Liu H, Zhang Z, Huang Y, Wei W, Ning S, Li J, Liang X, Liu K, Zhang L. Plasma HSP90AA1 Predicts the Risk of Breast Cancer Onset and Distant Metastasis. Front Cell Dev Biol. 2021;9:639596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 25. | Lindhiem O, Petersen IT, Mentch LK, Youngstrom EA. The Importance of Calibration in Clinical Psychology. Assessment. 2020;27:840-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |