Published online Oct 6, 2022. doi: 10.12998/wjcc.v10.i28.10017
Peer-review started: December 13, 2021
First decision: April 16, 2022
Revised: April 28, 2022
Accepted: August 25, 2022
Article in press: August 25, 2022
Published online: October 6, 2022
Processing time: 288 Days and 10.5 Hours
Insulin-like growth factor-1 receptor (IGF-1R) is over-expressed in hepatocellular carcinoma (HCC). However, the relationship between IGF-1R activation and HCC progression remains unidentified.
To investigate the effects of editing IGF-1R on the biological features of HCC cells.
Immunohistochemistry analyzed the expressions of IGF-1R and P-glyco protein (P-gp) in HCC tissues and their distal non-cancerous tissues (non-Ca). IGF-1R was edited with Crispr/Cas9 system, screened specific sgRNAs, and then transfected into HepG2 cells. CCK-8, scratch wound test detected cell proliferation, migration, invasion and transwell assays, respectively. Alterations of IGF-1R and P-gp were confirmed by Western blotting. Alterations of anti-cancer drug IC50 values were analyzed at the cell level.
The positive rates of IGF-1R (93.6%, χ2 = 63.947) or P-gp (88.2%, χ2 = 58.448) were significantly higher (P < 0.001) in the HCC group than those (36.6% in IGF-1R or 26.9% in P-gp) in the non-Ca group. They were positively correlated between high IGF-1R and P-gp expression, and they were associated with hepatitis B virus infection and vascular invasion of HCC. Abnormal expressions of circulating IGF-1R and P-gp were confirmed and associated with HCC progression. Biological feature alterations of HCC cells transfected with specific sgRNA showed IGF-1R expression down-regulation, cell proliferation inhibition, cell invasion or migration potential decreasing, and enhancing susceptibility of HepG2 cells to anti-cancer drugs.
Edited oncogenic IGF-1R was useful to inhibit biological behaviors of HepG2 cells.
Core Tip: Abnormal expression of insulin-like growth factor-1 receptor (IGF-1R) was associated with hepatocellular carcinoma (HCC). IGF-1R level was significantly higher in HCC more than that in their noncancerous tissues. Circulating IGF-1R continued to increase from benign liver disease to HCC. Down-regulating expression of IGF-1R with a specific sgRNA was markedly affected on biological behaviors of HCC cells, including inhibiting cell proliferation, decreasing cell migration or invasion potential, increasing cell apoptosis and enhancing cell susceptibility to anti-tumor drugs and indicated that oncogenic IGF-1R should be a promising targeted-molecule for HCC therapy.
- Citation: Yao M, Cai Y, Wu ZJ, Zhou P, Sai WL, Wang DF, Wang L, Yao DF. Effects of targeted-edited oncogenic insulin-like growth factor-1 receptor with specific-sgRNA on biological behaviors of HepG2 cells. World J Clin Cases 2022; 10(28): 10017-10030
- URL: https://www.wjgnet.com/2307-8960/full/v10/i28/10017.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i28.10017
Hepatocellular carcinoma (HCC) is still one of the most common malignant tumors worldwide and its increasing incidence might be associated with nonalcoholic fatty liver disease because of the effective control of hepatitis B virus (HBV) or hepatitis C virus infection[1-3]. The limited effective therapies available for advanced HCC patients are considered with aberrant gene transcriptions that govern crucial signal pathways, high migration or invasion potential, or multi-drug resistance (MDR) formation[4]. Previous studies have indicated that the related-signal molecules of the insulin-like growth factor (IGF) axis play a crucial role in malignant transformation of hepatocytes or HCC progression. Recently, the genomic sequencing studies of IGFs, type 1 IGF receptor (IGF-1R) and IGF binding proteins (IGFBPs) have confirmed that IGFs regulate multiple physiological processes, including mammalian development, metabolism and growth[5,6]. IGF-1R attributes to high-energy intake, cell proliferation, or apoptosis, and promotes mitogenic pathway activation via tyrosine kinase, conformational changes, tyrosine autophosphorylation and insulin-receptor substrate proteins[7,8] suggesting that a high IGF-1R level might affect ineffective treatment of HCC[9,10].
High IGF-1R expression and MDR with increasing P-glyco protein (P-gp) are major obstacles to the successful treatment of HCC with chemotherapy[11,12]. High IGF-1R mediated mitogenic, differentiating and anti-apoptotic features were found in HCC cells but not in mature hepatocytes[13]. The P-gp, as a member of the superfamily of ATP-binding cassette transporters is encoded in MDR 1. MDR emergence is still a complex problem for effective treatment and a high P-gp level is closely related to MDR in HCC. Therefore, it is essential to establish a new technology of MDR reversal, selectively inhibit P-gp or interfere with the activation of related genes to improve the curative effect or prognosis of HCC treatment[14,15]. Although high IGF-1R was associated with malignant transformation of hepatocytes[16], IGF-1R activation could be inhibited by small hairpin RNA (shRNA)[9]. However, the editing of IGF-1R on biological behaviors or MDR of HCC remain to be explored. This study aimed to investigate the clinicopathological features of IGF-1R or P-gp expression in HCC, further analyze IGF-1R on the effects of biological behaviors of HepG2 cells and the synergistic role with anti-cancer drugs on the reversal MDR of HCC.
According to the self-controlled method, a total of 93 pairs of HCC- and their distal non-cancerous (3 cm to cancer, non-Ca) tissues were collected after the HCC patient operation from Feb 2015 to Aug 2016. The prior written informed consent was obtained from all patients according to the Helsinki Declaration of World Medical Association. This study was approved by the Ethics Committee permission (TDFY2013008) at the Affiliated Hospital Nantong University, China. Based on medical records, the cases were 78 males and 15 females within 35-80-years-old (average 55.8 years ± 16.1 years). There were 74 cases with a single tumor and 19 cases with multiples; 22 cases with tumor size > 5.0 cm and 71 cases ≤ 5.0 cm; 36 cases with vascular invasion and 57 cases without vascular invasion. Differentiation degree were well (n = 21), middle (n = 49), and poor (n = 23) on the Edmondson grading system. Clinical staging was 58 cases at I-II and 35 at III-IV on the tumor-node-metastasis (TNM) classification of the International Union against Cancer. There were 64.5% (60 of 93) cases with positive HBV surface antigen and 71.0% (66 of 93) with liver cirrhosis. Serum α-fetoprotein (AFP) levels were 34 cases with ≥ 400 μg/L and 59 with < 400 μg/L. All cases had regular follow-up from operation to death until Aug 2021. Criteria of HCC diagnosis were set by the Chinese Collaborative Liver Cancer Research Group[17].
Liver sections from HCC or non-Ca tissues were made for constructing a tissue microarray (TMA). They were boiled for antigen retrieval in pH 6.0 citrate buffer and incubated for 2 h with 1st mouse anti-IGF-1R (Abcam, United Kingdom) or anti-P-gp antibodies (Santa Cruz, United States), washing with phosphate-buffered saline (PBS), and then incubated with horseradish peroxidase (HRP) conjugated goat anti-mouse IgG (Dako, Carpentaria, United States) and diaminobenzidine solution, counterstained with Hematoxylin & Xylene. Negative control used PBS instead of 1st antibodies. Two independent pathologists examined TMA staining. Positive cells (%) were scored into: 0 (negative, 0%), 1 (weak, 1%-33%), 2 (moderate, 34%-66%), and 3 (strong, 67%-100%). Liver IGF-1R or P-gp levels were divided into low (0-1) and high (2-3) scores according to immunohistochemistry (IHC) staining[18].
Purified proteins from 50 mg of liver tissue or five × 104 cells were quantitatively detected using BCA assay (Beyotime, China) for the specific concentration/per mg wet liver or cell protein analysis. Phenyl-methanesulfonyl fluoride (Byotime, China) was added in case of protein degradation. Proteins (50 μg/case) from transfected cells were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, United States) and blocked in 5% skimmed milk at room temperature, incubated with antibodies of mouse anti-human IGF-1R or anti-β- actin (Abcam, United States), and anti-human P-gp (Santa Cruz, United States) at 4 ℃ overnight and goat anti-mouse HRP-conjugated IgG (Dako, United States) at room temperature for 2 h. Protein bands were captured by enhanced chemiluminescence kit (Millipore, United States) and analyzed by Quantity-one software (Bio-Rad, United States).
Human LO2 cells, HCC Bel-7404, Bel-7402, HepG2, and HeH-7 cell lines were obtained from the Chinese Academy of Sciences (Shanghai, China). HepG2 cell lines were grown in DMEM (Hyclone, United States) containing 10% FBS (Gibco, United States) at 37 ℃ in a humidified atmosphere of 5% CO2. Three candidate sgRNAs targeting human IGF-1R (NM000875) with Crispr/Cas9 system along with vectors (Lenti-CAS9-puro and Lenti-sgRNA-EGFP) were designed and constructed by GeneChem (Shanghai, China). Their sequences were listed as follows: sgRNA1: 5’-TCAGTACGCCGTTTACGTCA-3’ (PCA00469); sgRNA2:5’-TGTTTCCGAAATTTACCGCA-3’(PCA00470); and sgRNA3: 5’-GGCTCTCTCCCCGTTGTTCC-3’ (PCA00471). HepG2 cells were divided into blank control (Con), negative sgRNA-CON244 (sgRNA-Neg), and IGF-1R-sgRNA (sgRNA) groups. Cell transfection, briefly, five × 104 HepG2 cells at 50% confluency in 6-well plates were transfected with Lenti-CAS9-puro vector for 48 h and then screened by puromycin. Following harvest, Lenti-sgRNA-EGFP with different sgRNAs against IGF-1R was transfected into Cas9-transfected cells. Then the transfection efficacy was analyzed under a fluorescence microscope (Olympus, Japan).
DNA of samples with sgRNAs was harvested for PCR and nuclease digestion for analysis using SURVEYOR Mutation Detection Kits (IDT, United States)[19]. DNA fragments were electrophoresed on 2% agarose gel.
Cell proliferation were analyzed by CCK-8 assay using the Cell Proliferation Kit I (Dojindo, Japan). According to the manufacturer’s instruction (Beyotime, China), the dark blue formazan crystal was formed. Then the medium was carefully removed, dimethyl sulfoxide added, and the detected A values read with Multi-Detection Microplate Reader (Bio-Tek, United States) at 570 nm. Cells (2 × 105) or transfected cells after 48 h were placed on the bottom with glass coverslips for 24 h, fixed in 4% paraformaldehyde in PBS (pH 7.5). Cover slips were first immersed for one h in a blocking solution containing 5% bovine serum albumin in PBS and incubated overnight at 4 ℃ with mouse antibodies against IGF-1R (CST, United States). DNA was counterstained with 4',6- diamidino-2-phenylindole and observed under inverted fluorescence micro-scope. Levels of IGF-1R expression in culture supernatant were quantitatively detected with an ELISA kit at 450 nm.
Effective apoptosis of transfected cells was evaluated by the PE-labeled Annexin- V/7-AAD assay according to the manufacturer’s protocol. In brief, HepG2 cells (2 × 105) were collected, rinsed twice in cold PBS, and added with binding buffer; then, apoptosis rate was analyzed. After 24 h, they were collected (including dead cells in culture medium) and processed according to the instructions of Annexin V-FITC/PI Double stain apoptosis detection kit (#4101-2, Bestbio, Shanghai, China). The fluorescence intensity on the flow cytometer was collected to immediately check apoptotic rate (Cytoflex Beckman, China). Three independent experiments were performed.
Transwell chambers (8.0 μm pore size, Corning, United States) were adopted for testing cell migration and invasion assays. Cells were seeded in the upper chamber after transfection for 24 h. Lower chambers had 600 μL medium added. After 24 h, cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The upper chamber cells were wiped with cotton. During the invasion assay, the upper chambers needed to be pretreated with a 0.3% Matrigel matrix (#356234, Corning, United States) and incubated at 37 ℃, 4 h. Three independent experiments were performed.
Quantitative and qualitative analysis of HepG2 cells migration were assessed by in vitro Transwell assay with modified Boyden Chambers and Transwell-coated Matrigel membrane filter (BD Biosciences, Bedford, MA, United States). Cells (5 × 103) from Con, Neg, and MiR groups (n = 3/per group) were plated onto the upper compartment without FBS or 10% FBS in the lower chamber as a chemoattractant. Fluorescent images of nuclear Hoechst staining (10 μg/mL) were captured at 24 h of incubation in a 5% CO2 humidified at 37 ℃. Percentages of migrated cells in each group were counted from 10 random microscope fields for each sample in 3 independent experiments. The modified Boyden Chambers without the Transwell-precoated Matrigel membrane filter in the above method was performed for cell migration analysis.
HCC cells were seeded to 96-well plates (5000 cells/well) and placed in the incubator for 24 h under cell culture conditions. The culture medium is then replaced by serum-free medium containing varying dosages of Sorafenib (Bayer Corporation, Germany) from 0.001 to 1000 μmol/L, and oxaliplatin (Jiangsu AoSaikang pharmaceutical Co. China) from 0.001 to 1000 nmol/L, where appropriate. Dose-response curve and IC50 calculation were analyzed by evaluating the cell viability using the Cell Counting Kit, CCK-8 (Tojindo), 48 h after drug treatment with the nonlinear regression method by GraphPad Prism 5.0 software (San Diego, CA, United States).
Continuous variables are expressed as means ± SD. Data were analyzed by using SPSS software (version 20.0). Favorable rates and clinicopathological features of IGF-IR or P-gp expression were evaluated by χ2 test, Wilcoxon signed-rank test in paired tissue samples; Between-group differences were assessed by t-test or one-way analysis of variance. A P value less than 0.05 value was considered statistically significant.
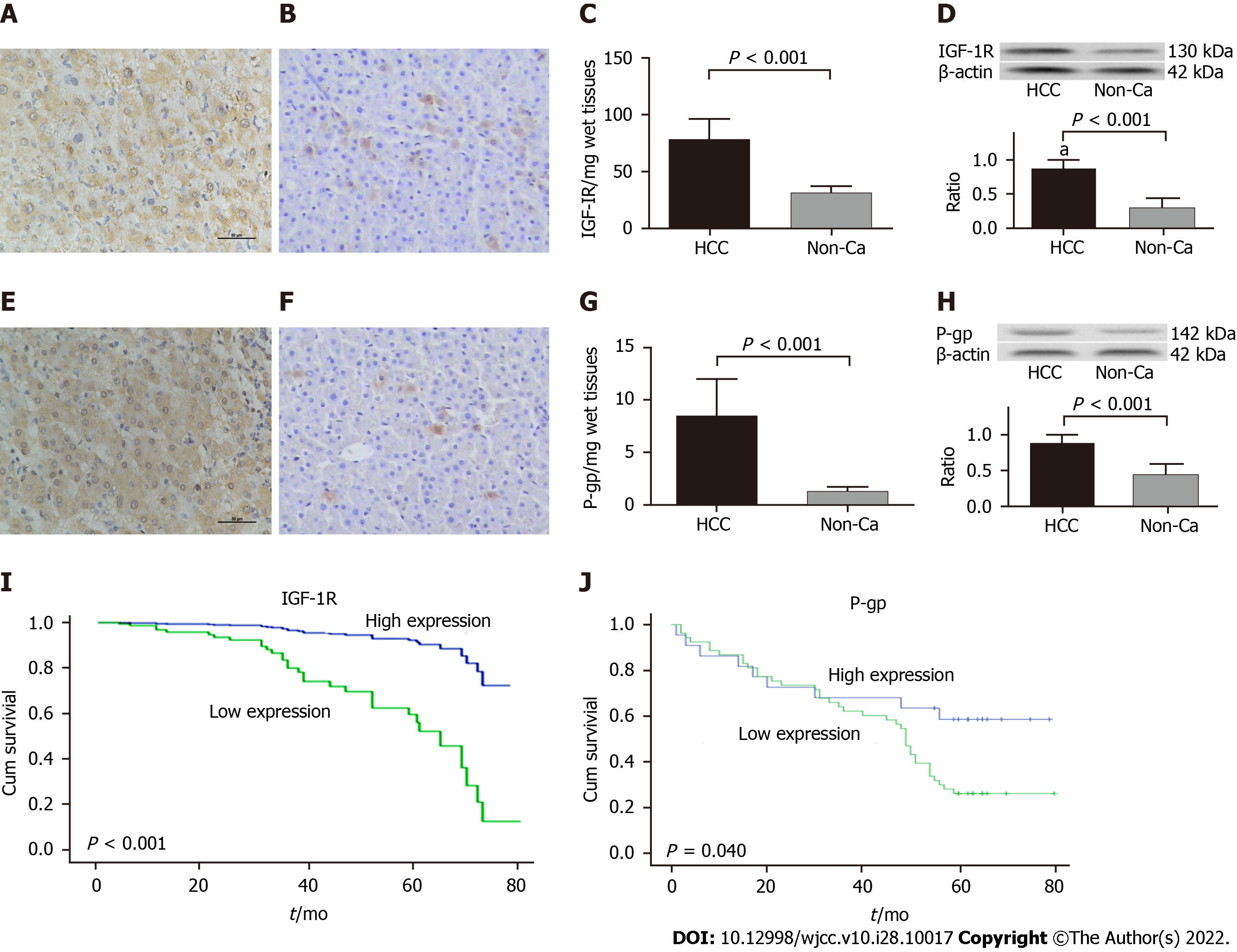
Comparative analysis of the representational IHC staining of IGF-IR or P-gp from the 93 pairs of HCC or their distal non-cancerous tissues (non-Ca) is shown in Figure 1. According to the hepatic IGF-IR or P-gp staining, there were the stronger in the HCC group (Figure 1A and E), the lighter or non-expression in the non-Ca group (Figure 1B and F). Both brown positive staining particles were mainly located in the cytosol with the clearly heterogeneous distribution. Specific concentrations of IGF-1R/per mg wet tissues (t = 23.451, P < 0.001, Figure 1C) or P-gp/per mg wet tissues (t = 19.832, P < 0.001, Figure 1G) were quantitatively detected, with significantly higher expression in the HCC group more than those in the non-Ca group. Abnormal expressions of IGF-1R (Figure 1D, 130 kDa) or P-gp (Figure 1H, 142 kDa) were confirmed by Western blotting, with high ratios (P < 0.001) from IGF-1R to β-actin (Figure 1D) or P-gp to β-actin (Figure 1H) expression in the HCC group more than those in the non-Ca group. According to the cumulative survival curves, there were shorter survival times in HCC patients with high IGF-1R (Figure 1I) or P-gp (Figure 1J) expression.
The summary of hepatic IGF-IR or P-gp expression and its scores between HCC and D-can tissues are shown in Table 1. The positive rates of IGF-IR expression were 93.6% in the HCC group, with significantly higher (χ2 = 63.947, P < 0.001) than that in the D-can group (36.6%). Furthermore, the higher IGF-IR with 2-3 scores in the HCC group (82.8%) was significantly higher (Z = 9.682, P < 0.001) than that in the D-can group (9.7%). Similar to IGF-1R expression, the positive rates of P-gp expression in the HCC group (88.2%) was significantly higher (χ2 = 58.448, P < 0.001) more than that in the D-can group (26.9%), and the higher P-gp with 2-3 scores in the HCC group was 74.2% was significantly higher (Z = 8.941, P < 0.001) more than that in the D-can group (0%). The clinicopathological features of IGF-IR or P-gp expression in HCC are shown in Table 2. The higher expression of IGF-1R in the HCC group was closely related to vascular invasion, HBV infection and middle or poor differentiation degree but, not to age or sex of patients, AFP level, TNM stage, liver cirrhosis and tumor size. Also, the abnormal P-gp levels in the HCC group were closely related to vascular invasion, HBV infection, poor differentiation degree and TNM stage but not to age or sex of patients, AFP level, liver cirrhosis and tumor size or number.
| Group | n | IHC | χ2 value | P value | IHC score | Z value | P value | ||||
| Neg. | Pos. | 0 | 1 | 2 | 3 | ||||||
| IGF-IR | 63.947 | < 0.001 | 9.682 | < 0.001 | |||||||
| HCC | 93 | 6 | 87 | 6 | 10 | 67 | 10 | ||||
| Non-Ca | 93 | 59 | 34 | 59 | 25 | 9 | 0 | ||||
| P-gp | 58.448 | < 0.001 | 8.941 | < 0.001 | |||||||
| HCC | 93 | 11 | 82 | 11 | 14 | 57 | 12 | ||||
| Non-Ca | 93 | 68 | 25 | 68 | 22 | 2 | 0 | ||||
| Group | n | IGF-IR | P-gp | ||||
| Pos. n (%) | χ2 value | P value | Pos. n (%) | χ2 value | P value | ||
| Sex | |||||||
| Male | 78 | 73 (93.6) | 0.288 | 0.591 | 69 (88.5) | 0.811 | 0.057 |
| Female | 15 | 14 (93.3) | 13 (86.7) | ||||
| Age | |||||||
| ≤ 50 yr | 68 | 63(92.6) | 0.012 | 0.914 | 59 (86.8) | 1.611 | 0.204 |
| > 50 yr | 25 | 24(96.0) | 23 (92.0) | ||||
| HBsAg | |||||||
| Positive | 60 | 60 (100) | 8.844 | 0.003 | 58 (96.6) | 9.517 | 0.002 |
| Negative | 33 | 27 (81.8) | 24 (72.7) | ||||
| AFP | |||||||
| ≤ 400 μg/L | 59 | 55 (93.2) | 0.104 | 0.747 | 51 (86.4) | 0.154 | 0.695 |
| > 400 μg/L | 34 | 32 (94.1) | 31 (91.2) | ||||
| Tumor diameter | |||||||
| ≤ 5.0 cm | 71 | 67 (94.4) | 3.550 | 0.060 | 63 (88.7) | 3.208 | 0.073 |
| > 5.0 cm | 22 | 20 (90.9) | 19 (86.4) | ||||
| Differentiation | |||||||
| Well | 21 | 17 (81.0) | 4.201 | 0.040 | 16 (76.2) | 1.484 | 0.223 |
| Middle | 49 | 47 (95.9) | 43 (87.8) | ||||
| Poor | 23 | 23 (100) | 4.819 | 0.028 | 23 (100) | 6.178 | 0.022 |
| Cirrhosis | |||||||
| With | 66 | 62 (93.9) | 1.014 | 0.314 | 58 (87.9) | 0.782 | 0.377 |
| Without | 27 | 25 (92.6) | 24 (88.9) | ||||
| TNM staging | |||||||
| I-II | 58 | 52 (89.7) | 2.3461 | 0.1256 | 47 (81.0) | 6.161 | 0.013 |
| III-IV | 35 | 35 (100) | 35 (100) | ||||
| Vascular invasion | |||||||
| With | 36 | 35 (97.2) | 25.363 | < 0.001 | 36 (100) | 24.158 | < 0.001 |
| Without | 57 | 42 (73.7) | 46 (80.7) | ||||
| Tumor number | |||||||
| One | 74 | 69 (93.2) | 0.082 | 0.774 | 65 (87.8) | 0.041 | 0.841 |
| More | 19 | 18 (94.7) | 17 (89.5) | ||||
Comparative analysis of IGF-1R and P-gp levels in sera of patients with liver diseases are shown in Table 3. Significant differences were found in circulating IGF-1R (F = 154.501, P < 0.001) or P-gp (F = 66.182, P < 0.001) levels among different liver disease groups, with high IGF-1R or P-gp levels in the HCC group more than those in the liver cirrhosis or chronic hepatitis or healthy control group. The incidences of IGF-1R over 600 pg/mL or P-gp over nine ng/mL were 86% or 75% in HCC patients, and less than 20% in patients with other diseases, with an increasing tendency from chronic hepatitis, liver cirrhosis to HCC. IGF-1R or P-gp expressions were 93.0% or 87.2% in sera of HCC patients, 25.5% or 26% in sera of cases with liver cirrhosis, and 5% or 8% in sera of cases with chronic hepatitis, respectively. A closely positive correlation (r = 0.682, P < 0.001) was found between IGF-1R or P-gp expression and liver disease progression.
| Group | n | IGF-1R (pg/mL) | P-gp (ng/mL) |
| HCC | 93 | 758.6 ± 126.4 | 11.6 ± 5.1 |
| Liver cirrhosis | 40 | 521.4 ± 78.3 | 7.3 ± 6.3 |
| Chronic hepatitis | 40 | 456.8 ± 82.1 | 3.7 ± 1.4 |
| Health control | 40 | 421.8 ± 58.6 | 1.0 ± 0.5 |
| F value | 154.501 | 66.182 | |
| q value1 | 0.689 | 0.487 | |
| P value | < 0.001 | < 0.001 | |
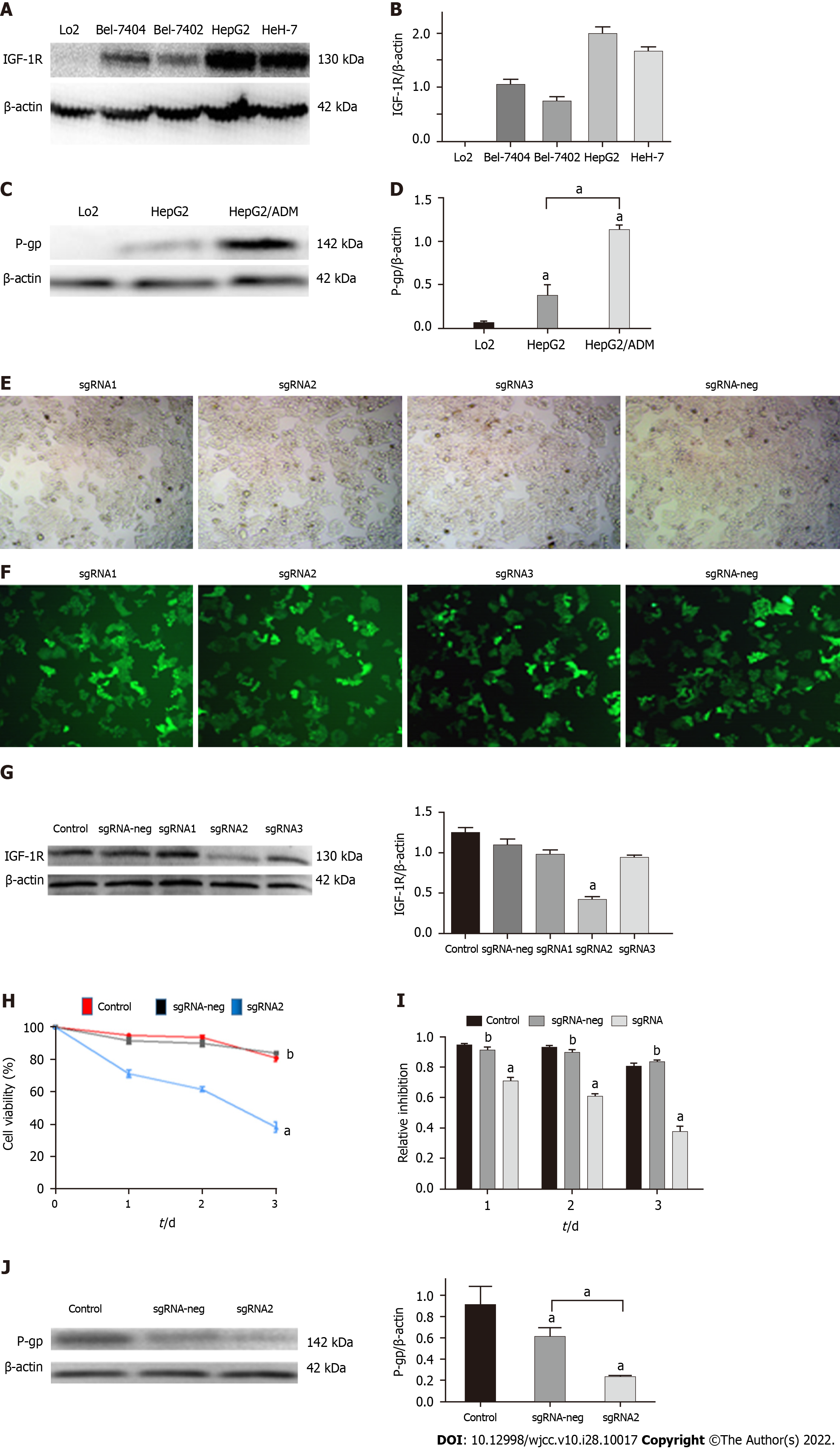
Human HepG2 cells were divided into control, sgRNA-neg, and sgRNA2 groups. Using the CRISPR/Cas9 system, human IGF-IR in HepG2 cells genetically modified and the editing IGF-1R on effects of HepG2 cell proliferation and P-gp expression are shown in Figure 2. The IGF-1R in HepG2 cells was the strongest expression among LO2 cells (Figure 2A and B), increasing P-gp levels after HepG2 cells plus drug treatment (HepG2/ADM cells, Figure 2C and D). Comparative analysis of the constructed-sgRNA1-3 and sgRNA-neg plasmids into HepG2 cell lines, the transfected efficiencies were 91% for sgRNA1, 90% for sgRNA2, 88% for sgRNA3, and 86% for sgRNA-neg plasmids, respectively (Figure 2E and F). The IGF-IR expression in the transfected HepG2 cells were confirmed by Western bolting (Figure 2G, Upper), and the relative ratios of IGF-1R to β-actin were 1.32 ± 0.13 in the control group, 1.14 ± 1.23 in the sgRNA-neg group, 1.01 ± 0.94 in the sgRNA1 group, 0.43 ± 0.79 in the sgRNA2 group, and 0.99 ± 0.82 in the sgRNA3 group (Figure 2G, Down), respectively. Specific sgRNA were chosen to study further, the curves of the transfected HepG2 cell proliferation were significantly inhibited (P < 0.01) in a time dependence manner, and the inhibiting rate were 0.31 ± 0.08 on the 1st day, 0.42 ± 0.14 on the 2nd day, and 0.62 ± 0.21 in the 3rd day (Figure 2H and I). Also, the decreasing P-gp level in the HepG2/ADM cells is transfected with sgRNA2 (Figure 2J).
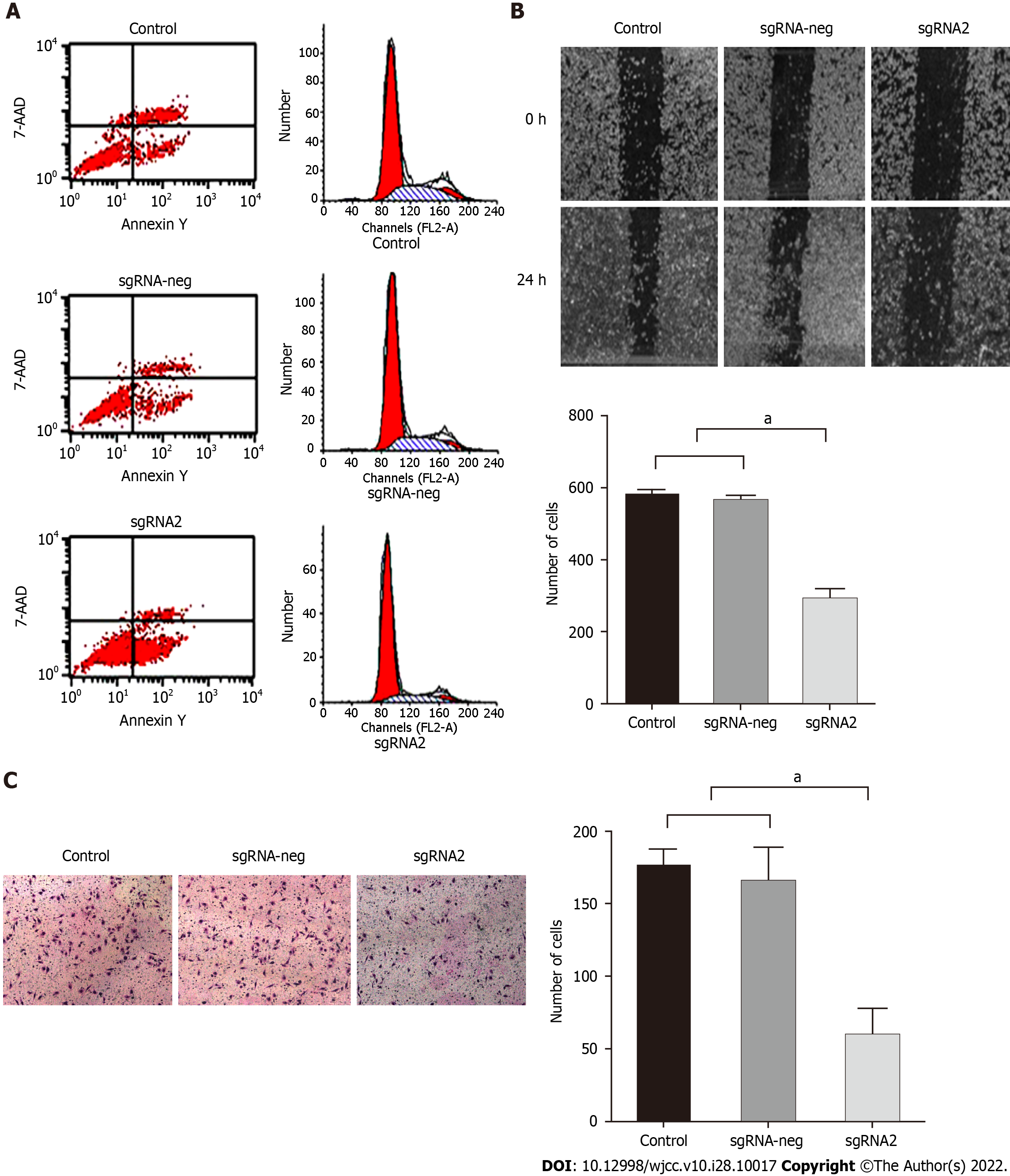
Editing IGF-IR on effects of HepG2 cell apoptosis, migration and invasion are shown in Figure 3. HepG2 cells transfected with specific sgRNA2 besides inhibiting proliferation, targeting IGF-IR also directly affected the biological function of HepG2 cells by the analysis of flow cytometry. Apoptotic rates of HepG2 cells in the sgRNA2 group (56.25%) were significantly higher (P < 0.01, Figure 3A Left) than those in the sgRNA-neg group (5.98%) or control group (5.66%). Edited IGF-1R led to cell cycle arrest in G1 phase of HepG2 cells (P < 0.01, Figure 3A Right). The numbers of cell migration were 292.3 ± 28.6 in the sgRNA2 group and significantly lower (F = 391.322, P < 0.001) than those in the sgRNA-neg (564.5 ± 15.8, q = 35.241, P < 0.001) or the control (580.5 ± 15.2, q = 35.214, P < 0.001) group (Figure 3B). The numbers of cell invasion in the sgRNA2 group (59.3 ± 19.1) was significantly lower (F = 69.510, P < 0.001) than those in the sgRNA-neg (165.5 ± 24.8, q =13.684, P < 0.001) or control (176.1 ± 12.2, q =15.102, P < 0.001) group (Figure 3C). There was no significant differences of cell migration or invasion between the control group and the sgIGF-1R-neg group. These data indicated that edited IGF-IR might be obviously decreasing invasion or migration potential of HepG2 cells.
The synergistic effect of specific sgRNA with anti-cancer drugs on inhibition of HepG2 cell growth is shown in Table 4. HepG2 cells with or without transfected sgRNA were exposed to sorafenib from 0.0 nmol/L to 80.0 nmol/L or oxaliplatin from 0.0 μmol/L to 40 μmol/L for 24 h, and cell survival rates were assessed using the CCK-8 assay. IGF-1R expression was down-regulated by sgRNA in a dose-dependent manner, especially at 20 nmol/L with high inhibiting effects on HCC cells. IC50 values were 16.9 nmol/L in the control group, 16.5 nmol/L in the sgRNA-neg group, and 4.4 nmol/L in the sgRNA group. Similar to sorafenib, the specific sgRNA plus oxaliplatin had higher inhibitory effects on the cells. IC50 values were 14.6 nmol/L in the control group, 14.2 nmol/L in the sgRNA-neg group, and 8.2 nmol/L in the sgRNA group. These data indicated that HCC cells transfected with specific sgRNA could be more sensitive to anti-cancer drugs.
| Group | Control | Neg-sgRNA | Sg-IGF1R2 | F value | q value1 | P value |
| Sorafenib | ||||||
| 0.0 nmol/L | 0.297 ± 0.06 | 0.310 ± 0.07 | 0.199 ± 0.07 | 1.361 | 2.604 | 0.026 |
| 2.5 nmol/L | 0.326 ± 0.10 | 0.335 ± 0.11 | 0.188 ± 0.05 | 100.00 | 3.364 | 0.007 |
| 5.0 nmol/L | 0.337 ± 0.11 | 0.318 ± 0.05 | 0.191 ± 0.06 | 3.316 | 2.854 | 0.017 |
| 10.0 nmol/L | 0.331 ± 0.05 | 0.284 ± 0.17 | 0.152 ± 0.09 | 1.494 | 3.085 | 0.012 |
| 20.0 nmol/L | 0.093 ± 0.03 | 0.084 ± 0.02 | 0.033 ± 0.04 | 6.499 | 0.464 | 0.009 |
| Oxaliplation | ||||||
| 0.0 μmol/L | 2.391 ± 0.30 | 2.351 ± 0.17 | 1.429 ± 0.27 | 51.91 | 0.874 | 0.001 |
| 5.0 μmol/L | 1.064 ± 0.21 | 1.014 ± 0.05 | 0.677 ± 0.08 | 6.891 | 4.218 | 0.002 |
| 10.0 μmol/L | 0.659 ± 0.16 | 0.575 ± 0.13 | 0.417 ± 0.14 | 1.515 | 2.768 | 0.020 |
| 20.0 μmol/L | 0.288 ± 0.12 | 0.280 ± 0.08 | 0.148 ± 0.08 | 36.000 | 2.819 | 0.018 |
| 40.0 μmol/L | 0.164 ± 0.05 | 0.162 ± 0.07 | 0.099 ± 0.04 | 1.000 | 2.815 | 0.018 |
Accumulating data of basic and clinical studies have indicated that many kinds of signaling are associated with HCC hypoxic microenvironment and oncogenesis[20]. IGF-IR activation was related to malignant transformation of hepatocytes[11], specific shRNA for inhibiting IGF-IR[10], IGF-1R tyrosine kinase inhibitors and vitamin K1 enhancing the antitumor effects of regorafenib[8], and miRNA-187 inhibits HCC growth and metastasis via targeting IGF-1R[9] have been explored. This study confirmed high IGF-1R expression in cancerous tissues that were related to poorly differentiated degree, shorter survival, TNM stage and lymph node metastasis of HCC; also, serum IGF-1R level in the HCC group was significantly higher than those in chronic hepatitis, liver cirrhosis or control group, indicating that IGF-1R should be an essential signaling for the formation, progression and poor prognosis of HCC[21].
IGF-1R signaling regulates cell differentiation, organ development and tissue regeneration during embryonic development. Also, IGF-1R as a critical molecule of the IGF axis was associated with the progression and drug resistance of HCC and it could promote cell proliferation and activate HCC reprogramming in the liver, especially in patients with chronic liver diseases[22]. Dysregulation of IGF-1R signaling might activate expression of HCC stemness that leads to hepatocytes malignant trans
Hepatic IGF-1R as a multifunctional regulatory factor plays an important and vital role in HCC occurrence and progression. Abnormal activation of IGF-1R signaling promotes the proliferation, dissemination and aggressive behaviors of HCC cells[24,25]. In this study, higher IGF-1R expression is consistent with the level in HCC tissues, circulating blood and HepG2 cells. Down-regulating IGF-1R has an anti-cancer effect through increasing apoptosis, inhibiting growth of HCC cell proliferation, decreasing migration or invasion of HepG2 cells and cell cycle arrest in the G1 phase, because of IGF-1R as an essential transmembrane protein for the related-pathway activation via its tyrosine kinase[26], suggesting that oncogenic IGF-IR could be a molecule-targeted for HCC therapy.
Acquiring resistance to chemotherapy remains a significant hurdle to effective HCC treatment[27,28]. Biomarker P-gp as an MDR-associated protein 1 (MRP1/ABCC1) is considered a prime factor for MDR induction[29]. The critical role of nuclear factor-kappa B (NF-κB) is to induce MDR in tumor models characterized precisely by innate or acquired MDR, particularly HCC[4,30]. Different pharmacological approaches have been employed to reduce the expression/activation of this transcriptional factor and thus to restore chemosensitivity. Scientific evidence was found by the most significant clinical trials regarding NF-κB and new perspectives on the possibility to consider this transcriptional factor a valid drug target in neoplastic diseases[31]. Both high P-gp and IGF-1R levels are common in HCC. In this study, the specific sgRNA plus anti-tumor drugs had higher inhibitory effects on HCC cells and the drug IC50 values were significantly decreased with inhibiting cell proliferation of HCC in the sgRNA group indicating that HCC cells transfected with sgRNA could be more sensitive to anti-cancer drugs via inhibiting the NF-κB pathway[4,32].
In conclusion, the high IGF-1R or P-gp expression has been confirmed as related to the progression or therapeutic effect of HCC. Although the accurate mechanism for IGF-1R reactivation in HCC remains be explored, the specific edited oncogenic IGF-1R gene is promising for inhibiting proliferation, altering biological behaviors, or as potential modulators for reversal MDR of HCC cells. Further studies should clarify the exact relationship between up-regulating IGF-1R and MDR formation. However, IGF-1R as a critical signaling molecule of the IGF axis might be a novel effective target for inhibiting HCC growth or reversal MDR of HCC.
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related death with a high incidence and mortality rate in China. Insulin-like growth factor 1 receptor (IGF-1R) signaling triggers cell proliferation, liver growth and tissue regeneration during embryonic development. Unbalanced IGF-1R signaling can promote HCC cell proliferation and regulating IGF-1R gene transcription should be useful as a potential therapy targeting HCC.
IGF-1R as key signaling of the IGF axis in HCC progression was investigated in sera or tissues from HBV-related HCC patients. We analyzed the relationship between IGF-1R and multi-drug resistance (MDR) and edited the IGF-1R gene for downing-regulating expression to confirm effects on proliferation and a potential therapeutic role for HCC cells.
The expressing statues and clinicopathological characteristics of IGF-1R or P-glyco protein (P-gp) were investigated in the circulating blood and tissues of HCC patients and editing IGF-1R gene at the mRNA transcription level to observe effects on biological behaviors HepG2 cells and their synergistic role with anti-cancer drugs on reversal MDR of HCC.
Comparative analysis of IGF-1R and P-gp expression in tissues or sera of HCC patients were analyzed by immunohistochemistry and confirmed by Western blotting. Specific sgRNA was screened among editing IGF-1R gene with Crispr/Cas9 system and then transfected into HepG2 cells. CCK-8, scratch wound test detected HCC cell proliferation, migration, invasion and transwell assay, respectively.
Abnormal over-expression of IGF-1R and P-gp were confirmed in tissues or sera of HCC patients with a positive close correlation between IGF-1R and P-gp and related to HBV infection or vascular invasion during HCC progression. HepG2 cell biological features were altered by specific IGF-1R-sgRNA with down-regulation, cell proliferation inhibition, cell invasion or migration potential decreasing and enhancing cell susceptibility to anti-cancer drugs.
Based on this these studies, high IGF-1R or P-gp expression has been confirmed related to the progression or therapeutic effect of HCC. Although the accurate mechanism for IGF-1R reactivation in HCC remains to be explored, specific edited oncogenic IGF-1R gene is promising for inhibiting proliferation, altering biological features or as potential modulators for reversal MDR of HCC cells.
Abnormal expression of hepatic IGF-1R level was associated with HCC progression. Inhibiting IGF-1R expression could markedly affect the biological behaviors of HCC cell proliferation, migration or invasion, cell apoptosis and drug susceptibility suggesting that the IGF-1R gene could be a promising targeted molecule for HCC therapy.
All authors thank Professor Zhizhen Dong for comments on earlier manuscript drafts.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D, D
Grade E (Poor): 0
P-Reviewer: Limaiem F, Tunisia; Suda T, Japan; Virarkar M, United States S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Gong ZM
| 1. | McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology. 2021;73 Suppl 1:4-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 1346] [Article Influence: 336.5] [Reference Citation Analysis (1)] |
| 2. | Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 3. | Huang DQ, El-Serag HB, Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2021;18:223-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 840] [Cited by in RCA: 1217] [Article Influence: 304.3] [Reference Citation Analysis (0)] |
| 4. | Shi Y, Wang SY, Yao M, Sai WL, Wu W, Yang JL, Cai Y, Zheng WJ, Yao DF. Chemosensitization of HepG2 cells by suppression of NF-κB/p65 gene transcription with specific-siRNA. World J Gastroenterol. 2015;21:12814-12821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 5. | Forbes BE, Blyth AJ, Wit JM. Disorders of IGFs and IGF-1R signaling pathways. Mol Cell Endocrinol. 2020;518:111035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 6. | Blyth AJ, Kirk NS, Forbes BE. Understanding IGF-II Action through Insights into Receptor Binding and Activation. Cells. 2020;9. [PubMed] [DOI] [Full Text] |
| 7. | Chen RS, Zhang XB, Zhu XT, Wang CS. The crosstalk between IGF-1R and ER-α in the proliferation and anti-inflammation of nucleus pulposus cells. Eur Rev Med Pharmacol Sci. 2020;24:5886-5894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 8. | Refolo MG, D'Alessandro R, Lippolis C, Carella N, Cavallini A, Messa C, Carr BI. IGF-1R tyrosine kinase inhibitors and Vitamin K1 enhance the antitumor effects of Regorafenib in HCC cell lines. Oncotarget. 2017;8:103465-103476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 9. | Han X, Wang X, Zhao B, Chen G, Sheng Y, Wang W, Teng M. MicroRNA-187 inhibits tumor growth and metastasis via targeting of IGF-1R in hepatocellular carcinoma. Mol Med Rep. 2017;16:2241-2246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Wang L, Yao M, Zheng W, Fang M, Wu M, Sun J, Dong Z, Yao D. Insulin-like Growth Factor I Receptor: A Novel Target for Hepatocellular Carcinoma Gene Therapy. Mini Rev Med Chem. 2019;19:272-280. [PubMed] [DOI] [Full Text] |
| 11. | Yan XD, Yao M, Wang L, Zhang HJ, Yan MJ, Gu X, Shi Y, Chen J, Dong ZZ, Yao DF. Overexpression of insulin-like growth factor-I receptor as a pertinent biomarker for hepatocytes malignant transformation. World J Gastroenterol. 2013;19:6084-6092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (1)] |
| 12. | Kumar A, Jaitak V. Natural products as multidrug resistance modulators in cancer. Eur J Med Chem. 2019;176:268-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 228] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 13. | Aleem E, Nehrbass D, Klimek F, Mayer D, Bannasch P. Upregulation of the insulin receptor and type I insulin-like growth factor receptor are early events in hepatocarcinogenesis. Toxicol Pathol. 2011;39:524-543. [PubMed] [DOI] [Full Text] |
| 14. | Dong Z, Yao M, Wang L, Yan X, Gu X, Shi Y, Yao N, Qiu L, Wu W, Yao D. Abnormal expression of insulin-like growth factor-I receptor in hepatoma tissue and its inhibition to promote apoptosis of tumor cells. Tumour Biol. 2013;34:3397-3405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 15. | Ceballos MP, Rigalli JP, Ceré LI, Semeniuk M, Catania VA, Ruiz ML. ABC Transporters: Regulation and Association with Multidrug Resistance in Hepatocellular Carcinoma and Colorectal Carcinoma. Curr Med Chem. 2019;26:1224-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 16. | Bie CQ, Liu XY, Cao MR, Huang QY, Tang HJ, Wang M, Cao GL, Yi TZ, Wu SL, Xu WJ, Tang SH. Lentivirus-mediated RNAi knockdown of insulin-like growth factor-1 receptor inhibits the growth and invasion of hepatocellular carcinoma via down-regulating midkine expression. Oncotarget. 2016;7:79305-79318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 17. | Xie DY, Ren ZG, Zhou J, Fan J, Gao Q. Critical appraisal of Chinese 2017 guideline on the management of hepatocellular carcinoma. Hepatobiliary Surg Nutr. 2017;6:387-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 18. | Zhang HJ, Yao DF, Yao M, Huang H, Wu W, Yan MJ, Yan XD, Chen J. Expression characteristics and diagnostic value of annexin A2 in hepatocellular carcinoma. World J Gastroenterol. 2012;18:5897-5904. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 35] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Hua Y, Wang C, Huang J, Wang K. A simple and efficient method for CRISPR/Cas9-induced mutant screening. J Genet Genomics. 2017;44:207-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 20. | Qin Y, Liu HJ, Li M, Zhai DH, Tang YH, Yang L, Qiao KL, Yang JH, Zhong WL, Zhang Q, Liu YR, Yang G, Sun T, Yang C. Salidroside improves the hypoxic tumor microenvironment and reverses the drug resistance of platinum drugs via HIF-1α signaling pathway. EBioMedicine. 2018;38:25-36. [PubMed] [DOI] [Full Text] |
| 21. | Xu J, Bie F, Wang Y, Chen X, Yan T, Du J. Prognostic value of IGF-1R in lung cancer: A PRISMA-compliant meta-analysis. Medicine (Baltimore). 2019;98:e15467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Ngo MT, Jeng HY, Kuo YC, Diony Nanda J, Brahmadhi A, Ling TY, Chang TS, Huang YH. The Role of IGF/IGF-1R Signaling in Hepatocellular Carcinomas: Stemness-Related Properties and Drug Resistance. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 23. | Fang M, Yao M, Yang J, Zheng WJ, Wang L, Yao DF. Abnormal CD44 activation of hepatocytes with nonalcoholic fatty accumulation in rat hepatocarcinogenesis. World J Gastrointest Oncol. 2020;12:66-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 24. | Ji Y, Wang Z, Chen H, Zhang L, Zhuo F, Yang Q. Serum from Chronic Hepatitis B Patients Promotes Growth and Proliferation via the IGF-II/IGF-IR/MEK/ERK Signaling Pathway in Hepatocellular Carcinoma Cells. Cell Physiol Biochem. 2018;47:39-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 25. | Chung W, Kim M, de la Monte S, Longato L, Carlson R, Slagle BL, Dong X, Wands JR. Activation of signal transduction pathways during hepatic oncogenesis. Cancer Lett. 2016;370:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Cheng W, Huang PC, Chao HM, Jeng YM, Hsu HC, Pan HW, Hwu WL, Lee YM. Glypican-3 induces oncogenicity by preventing IGF-1R degradation, a process that can be blocked by Grb10. Oncotarget. 2017;8:80429-80442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Yao WF, Liu JW, Sheng GL, Huang DS. Blockade of IGF-IR exerts anticancer effects in hepatocellular carcinoma. Mol Med Rep. 2011;4:719-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Chen S, Cao Q, Wen W, Wang H. Targeted therapy for hepatocellular carcinoma: Challenges and opportunities. Cancer Lett. 2019;460:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 178] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 29. | Wang Y, Jiang F, Jiao K, Ju L, Liu Q, Li Y, Miao L, Li Z. De-methylation of miR-148a by arsenic trioxide enhances sensitivity to chemotherapy via inhibiting the NF-κB pathway and CSC like properties. Exp Cell Res. 2020;386:111739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 30. | Qiu Y, Dai Y, Zhang C, Yang Y, Jin M, Shan W, Shen J, Lu M, Tang Z, Ju L, Wang Y, Jiao R, Xia Y, Huang G, Yang L, Li Y, Zhang J, Wong VKW, Jiang Z. Arsenic trioxide reverses the chemoresistance in hepatocellular carcinoma: a targeted intervention of 14-3-3η/NF-κB feedback loop. J Exp Clin Cancer Res. 2018;37:321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 31. | Hwang JW, Cho H, Lee JY, Jeon Y, Kim SN, Lee SJ, Bae GU, Yoon S, Jeon R, Kim YK. The synthetic ajoene analog SPA3015 induces apoptotic cell death through crosstalk between NF-κB and PPARγ in multidrug-resistant cancer cells. Food Chem Toxicol. 2016;96:35-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 32. | Tovar V, Cornella H, Moeini A, Vidal S, Hoshida Y, Sia D, Peix J, Cabellos L, Alsinet C, Torrecilla S, Martinez-Quetglas I, Lozano JJ, Desbois-Mouthon C, Solé M, Domingo-Domenech J, Villanueva A, Llovet JM. Tumour initiating cells and IGF/FGF signalling contribute to sorafenib resistance in hepatocellular carcinoma. Gut. 2017;66:530-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 157] [Article Influence: 19.6] [Reference Citation Analysis (0)] |