Published online Sep 16, 2022. doi: 10.12998/wjcc.v10.i26.9285
Peer-review started: April 2, 2022
First decision: June 16, 2022
Revised: June 30, 2022
Accepted: July 20, 2022
Article in press: July 20, 2022
Published online: September 16, 2022
Processing time: 152 Days and 14.5 Hours
Currently, there are many therapeutic methods for lung adenocarcinoma (LUAD), but the 5-year survival rate is still only 15% at later stages. Epithelial– mesen
To further elucidate the value of EMT-related genes in LUAD prognosis.
Univariate, least absolute shrinkage and selection operator, and multivariate Cox regression analyses were applied to establish and validate a new EMT-related gene signature for predicting LUAD prognosis. The risk model was evaluated by Kaplan–Meier survival analysis, principal component analysis, and functional enrichment analysis and was used for nomogram construction. The potential structures of drugs to which LUAD is sensitive were discussed with respect to EMT-related genes in this model.
Thirty-three differentially expressed genes related to EMT were found to be highly associated with overall survival (OS) by using univariate Cox regression analysis (log2FC ≥ 1, false discovery rate < 0.001). A prognostic signature of 7 EMT-associated genes was developed to divide patients into two risk groups by high or low risk scores. Kaplan–Meier survival analysis showed that the OS of patients in the high-risk group was significantly poorer than that of patients in the low-risk group (P < 0.05). Multivariate Cox regression analysis showed that the risk score was an independent risk factor for OS (HR > 1, P < 0.05). The results of receiver operator characteristic curve analysis suggested that the 7-gene signature had a perfect ability to predict prognosis (all area under the curves > 0.5).
The EMT-associated gene signature classifier could be used as a feasible indicator for predicting OS.
Core Tip: Lung cancer is one of the major causes of death associated with malignancy, and lung cancer is associated with approximately 2 million new cases and 1.76 million deaths every year. Although some reports have shown that suppression or knockdown of some genes in lung adenocarcinoma (LUAD) could reverse the epithelial–mesenchymal transition (EMT) process that inhibits tumor occurrence and metastasis, the correlations of these genes with overall survival in patients with LUAD remain largely unclear. A cohort of 884 LUAD patients was used to construct and validate a novel predictive model comprising a 7-EMT-related gene signature, which can be used to predict the prognosis of LUAD.
- Citation: Zhou DH, Du QC, Fu Z, Wang XY, Zhou L, Wang J, Hu CK, Liu S, Li JM, Ma ML, Yu H. Development and validation of an epithelial–mesenchymal transition-related gene signature for predicting prognosis. World J Clin Cases 2022; 10(26): 9285-9302
- URL: https://www.wjgnet.com/2307-8960/full/v10/i26/9285.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i26.9285
Worldwide, lung cancer is one of the major causes of death associated with malignancy, and lung cancer is associated with approximately 2 million new cases and 1.76 million deaths every year[1]. Approximately 80%-85% of lung carcinomas are non-small cell lung cancer, and the two major histopathological types are lung adenocarcinoma (LUAD) and lung squamous cell carcinoma[2]. Among patients with lung cancer, 40% have LUAD, which has a 5-year survival rate of only 15% at later stages[3]. Currently, there are many therapeutic methods for LUAD, including curative resection, radiotherapy, chemo-therapy, targeted therapy, and immunotherapy. Although the therapeutic prognosis of lung cancer has improved significantly because of advances in technology and selection strategies, the prognosis for lung cancer patients remains poor, as 70%-80% of patients are diagnosed in advanced stages[4,5]. In modern clinical oncology, histopathology is commonly effective in predicting the prognosis of lung cancer patients; however, its value is limited, as individual distinctions in patients with the identical pathological characteristics result in different conclusions. Therefore, more effective biomarkers for the early diagnosis of LUAD and more accurate prognostic predictions are necessary.
Epithelial–mesenchymal transition (EMT) is a multistep process in which morphological changes occur and is involved in neoplastic invasion, metastasis, embryogenesis and wound healing; during EMT, epithelial cells acquire mesenchymal characteristics and gradually lose their epithelial features[6]. EMT has been shown to be closely associated with local dissemination and subsequent metastasis of solid tumors[7]. Exosomes also frequently facilitate communication between cells and the extracellular matrix, complete the EMT process, and increase the transformation of nontumor stem cells into tumor stem cells, which accelerates the development of cancer[8]. There are many EMT states that can occur in cancer, and these states contribute differently to carcinogenesis, invasion, and metastasis. Mixed EMT states have a higher propensity for metastasis[9]. Pinin was shown by Qiao et al[10] to induce EMT by controlling m6A modification, and it might also be a potential anticancer target for hepatocellular carcinoma treatment. Previous studies suggested that highly proliferative non-EMT cells are sensitive to chemotherapy, and recurrent EMT-derived metastases were observed after treatment[11]. However, the underlying mechanism of EMT in the occurrence and development of non-small cell lung cancer, especially LUAD, remains largely unclear. Therefore, understanding the roles of EMT-related genes in LUAD diagnosis and prognostic prediction might be useful for identifying prognostic targets.
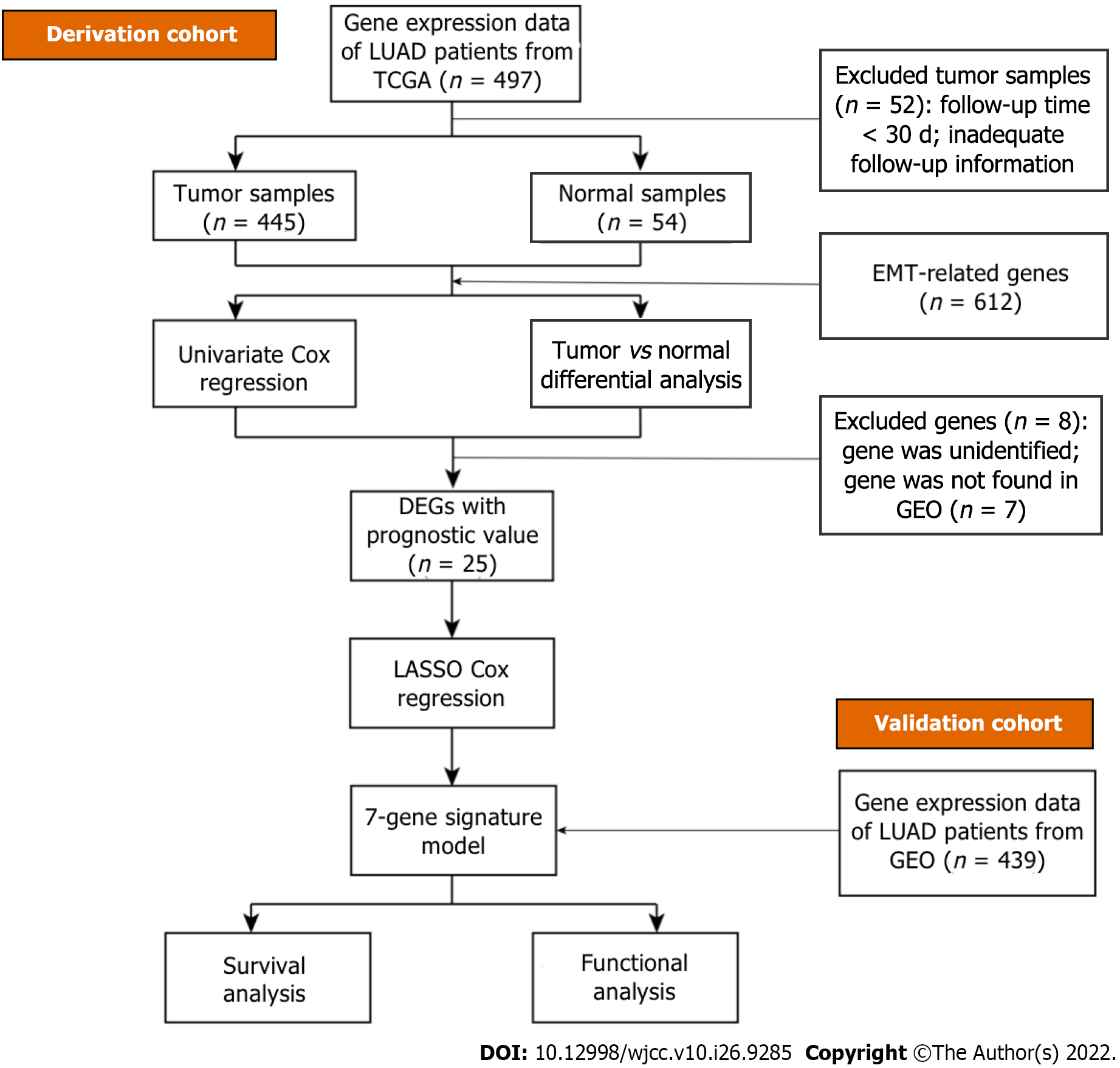
However, studies based on the transcriptome that systematically assess the EMT-related gene signature and its correlation with overall survival (OS) in LUAD patients remains rare. In this study, we explored available gene expression data from LUAD patient samples (n = 497) and extracted a total of 54774 candidate genes. In addition, GSEA identified 612 EMT-related functional gene sets that presented differential expression between tumor samples and normal samples. Then, we became the first to develop a novel prognostic model composed of differentially expressed EMT-related genes based on LUAD samples. This model based on external data (n = 443) was further validated to be an independent prognostic factor related to OS in LUAD patients.
The LUAD dataset, including whole-transcriptome RNA sequencing data and the corresponding clinical data of LUAD patients up to July 15, 2021, was downloaded from The Cancer Genome Atlas (https://cancergenome.nih.gov/). The clinical data of the LUAD patients included sex, grade, age, stage, and tumor, node and metastasis (TNM) classification. Data normalization was carried out utilizing the limma (Linear Models for Microarray Data) package in the R environment (version 4.1.0,2021–05-18, R Foundation for statistical computing, Vienna, Austria). Gene expression profiles were obtained using raw data, and log2 normalization was performed for each candidate gene for further statistical analysis. If one gene occupied more than one row, we took the mean of its expression value. All genes with an expression value of 0 were deleted from the sample. RNA-seq data and the corresponding clinical baseline characteristics for 443 LUAD patient samples were collected from a Gene Expression Omnibus dataset (GSE68465, http://www.ncbi.nlm.nih.gov/geo/). All 443 candidate patients underwent surgery. Normalized count values were utilized. All the data in this article obtained from The Cancer Genome Atlas and the Gene Expression Omnibus dataset were open access and freely available. Therefore, the study did not need approval from the local ethics committee. All methods were carried out following the related policies, regulations and guidelines provided by The Cancer Genome Atlas, Gene Expression Omnibus and other databases for analysis of their data and presentation of the related findings.
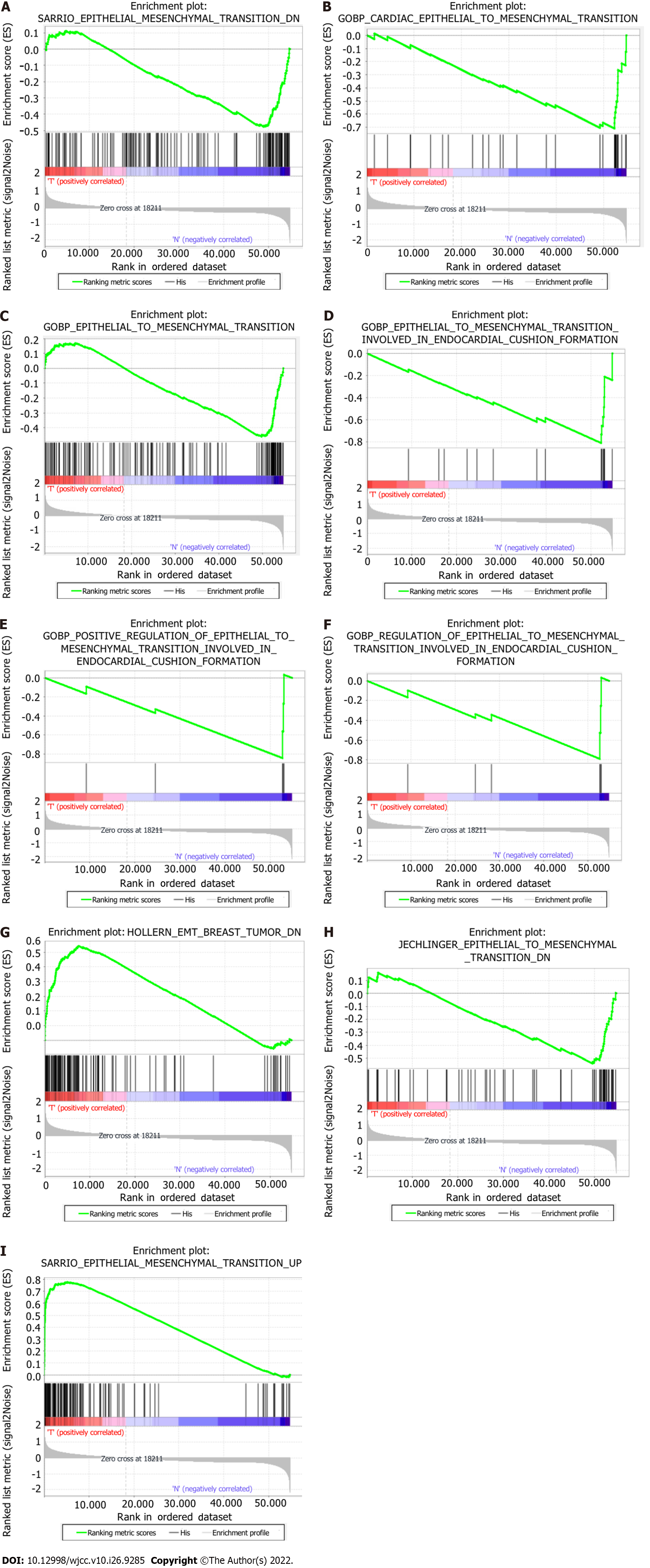
The hallmark gene sets, including 612 specific gene sets, comprised significantly expressed signature genes obtained from Molecular Signatures Database (version 7.4) gene sets to show perfectly defined biological states or processes. Gene set enrichment analysis (GSEA) was carried out using GSEA software (v4.1.0) to establish whether a fixed set of genes exhibited a statistically significant difference between certain phenotypes[12]. We acquired nine EMT-related gene sets: SARRIO_ EPITHELIAL_MES
| GS: follow link to MSigDB | Size | NES | Nom P value | FDR Q value |
| SARRIO_EPITHELIAL_MESENCHYMAL_TRANSITION_DN | 144 | -1.66 | 0.040 | 0.040 |
| GOBP_CARDIAC_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | 30 | -1.98 | 0.006 | 0.006 |
| GOBP_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | 150 | -1.71 | 0.045 | 0.045 |
| GOBP_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_INVOLVED_ | 16 | -2.05 | < 0.001 | < 0.001 |
| GOBP_POSITIVE_REGULATION_OF_EPITHELIAL_TO_MESENCHYMAL_ | 5 | -1.65 | 0.006 | 0.006 |
| GOBP_REGULATION_OF_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_ | 6 | -1.61 | 0.029 | 0.029 |
| HOLLERN_EMT_BREAST_TUMOR_DN | 121 | 1.90 | 0.020 | 0.020 |
| JECHLINGER_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_DN | 62 | -1.80 | 0.042 | 0.042 |
| SARRIO_EPITHELIAL_MESENCHYMAL_TRANSITION_UP | 166 | 2.03 | 0.008 | 0.008 |
To identify differentially expressed genes (DEGs) between cancer tissues and paracancerous tissues, we utilized the “limma” package in the R environment. The criteria used to identify DEGs in The Cancer Genome Atlas-LUAD cohort were log2FC ≥ 1 and false discovery rate (FDR) < 0.001. Univariate Cox regression analysis was carried out to evaluate OS-related genes (P values < 0.05). To predict correlations among DEGs, we constructed a PPI network using the online STRING database (version 11.0, https://string-db.org/). To reduce the risk of overfitting, we utilized the least absolute shrinkage and selection operator (LASSO) Cox regression model (“glmnet” R package), and the minimum criteria were selected[13,14]. The independent variable in the LASSO regression analysis was the standardized expression matrix of candidate prognostic DEGs associated with EMT, and the dependent variable was the OS (or other survival) status of LUAD patients in The Cancer Genome Atlas cohort. The optimal penalty parameter lambda (λ) and the matching coefficients were confirmed by 10-fold cross-validation according to the minimum criteria in the LASSO Cox regression algorithm. Lambda (λ) was the minimum partial likelihood deviance that was used for feature selection. The linear combination of gene expression values weighted by the corresponding regression coefficients was used to calculate the risk scores of LUAD patients according to the following formula: Risk score = esum (expression value of each gene × corresponding regression coefficient). LUAD patients were classified into a low-risk group and a high-risk group according to the median risk score. Principal component analysis (PCA) was carried out with the “prcomp” function in the R package “stats”. The t-distributed stochastic neighbor embedding (t-SNE) method in the R package “Rtsne” was performed for cluster visualization. For survival analysis based on each gene, the “surv_cutpoint” function in the “survminer” package was used to identify the best cutoff value of the risk score. The R package “survivalROC” was used to assess the predictive prognostic performance of the EMT-associated gene signature via time-dependent receiver operator characteristic curve (ROC) curve estimation.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses based on the prognostic DEGs (P value < 0.05, FDR < 0.05) between the high-risk group and the low-risk group were carried out using the R package “clusterProfiler”. Our GO functional enrichment analysis covered three ontologies: biological process, cellular component and molecular function. KEGG pathway enrichment analysis based on a hypergeometric algorithm was carried out to determine the significant KEGG pathways, and the results were visualized with the “ggplot2” package in the R environment. Adjusted P values were calculated by the Benjamini–Hochberg method.
R software was used to develop a novel nomogram model containing predictors (age, risk score, and TNM stage) related to OS at 1, 2 and 3 years. Calibration curves were generated to estimate the agreement of the model between the actual outcomes and predicted outcomes for one-year OS, two-year OS and three-year OS.
We uploaded the prognostic DEGs with a P value of < 0.05 to CMap (https://portals.broadinstitute.org/cmap), which is a biological application database of small molecule drugs, gene expression profiles, and diseases. The CMap database was built based on DEGs in human cells treated with small molecule drugs. The enrichment score based on similarity was ultimately calculated. A positive correlation score showed that a small molecule drug increased the risk of death in OS patients. In contrast, a negative correlation score showed that a drug reduced the risk of death. Drugs with negative connectivity scores had a potential therapeutic application. We obtained the three-dimensional structures of the candidate small molecule therapeutic drugs from PubChem (https://pubchem.ncbi.nlm.nih.gov/).
Student’s t test was performed to identify differences in gene expression between cancer tissues and adjacent paracancerous tissues. Differences between proportions were evaluated by the χ2 test. Overall survival was evaluated by Kaplan–Meier survival analysis, and the survival of the groups was compared using the log-rank test. Univariate and multivariate Cox regression analyses were carried out to explore independent prognostic factors of OS in LUAD patients. All statistical analyses were performed using SPSS software (version: 23.0, IBM Corporation, 2015, United States) and R software. All P values were two-tailed, and statistical significance was defined as P < 0.05.
The overall workflow of our research in light of model development and subsequent analyses is visualized in Figure 2. All 445 patients with LUAD from The Cancer Genome Atlas cohort and 439 patients from the gene set enrichment analysis cohort (GSE68465) were finally combined. The specific clinical baseline data of these participants are presented in Table 2.
| Variable | TCGA cohort | GEO cohort |
| Patients (n) | 445 | 439 |
| Sex | ||
| Female | 245 (55.1%) | 218 (49.7%) |
| Male | 200 (44.9%) | 221 (50.3%) |
| Age, yr | ||
| < 65 | 199 (44.7%) | 213 (48.5%) |
| ≥ 65 | 246 (55.3%) | 226 (51.5%) |
| TNM stage | ||
| Ι | 241 (54.2%) | 274 (62.4%) |
| Ⅱ | 107 (24.0%) | 95 (21.6%) |
| Ⅲ | 74 (16.4%) | 67 (15.3%) |
| Ⅳ | 24 (5.4%) | 0 |
| Unknown | 0 | 3 (0.7%) |
| T stage | ||
| T1 | 154 (34.6%) | 149 (33.9%) |
| T2 | 234 (52.6%) | 248 (56.5%) |
| T3 | 37 (8.3%) | 28 (6.4%) |
| T4 | 17 (3.8%) | 11 (2.5%) |
| Tx | 3 (0.7%) | 3 (0.7%) |
| Survival status | ||
| OS time, median days | 653 | 1418 |
| Censored (%) | 170 (38.2%) | 233 (53.1%) |
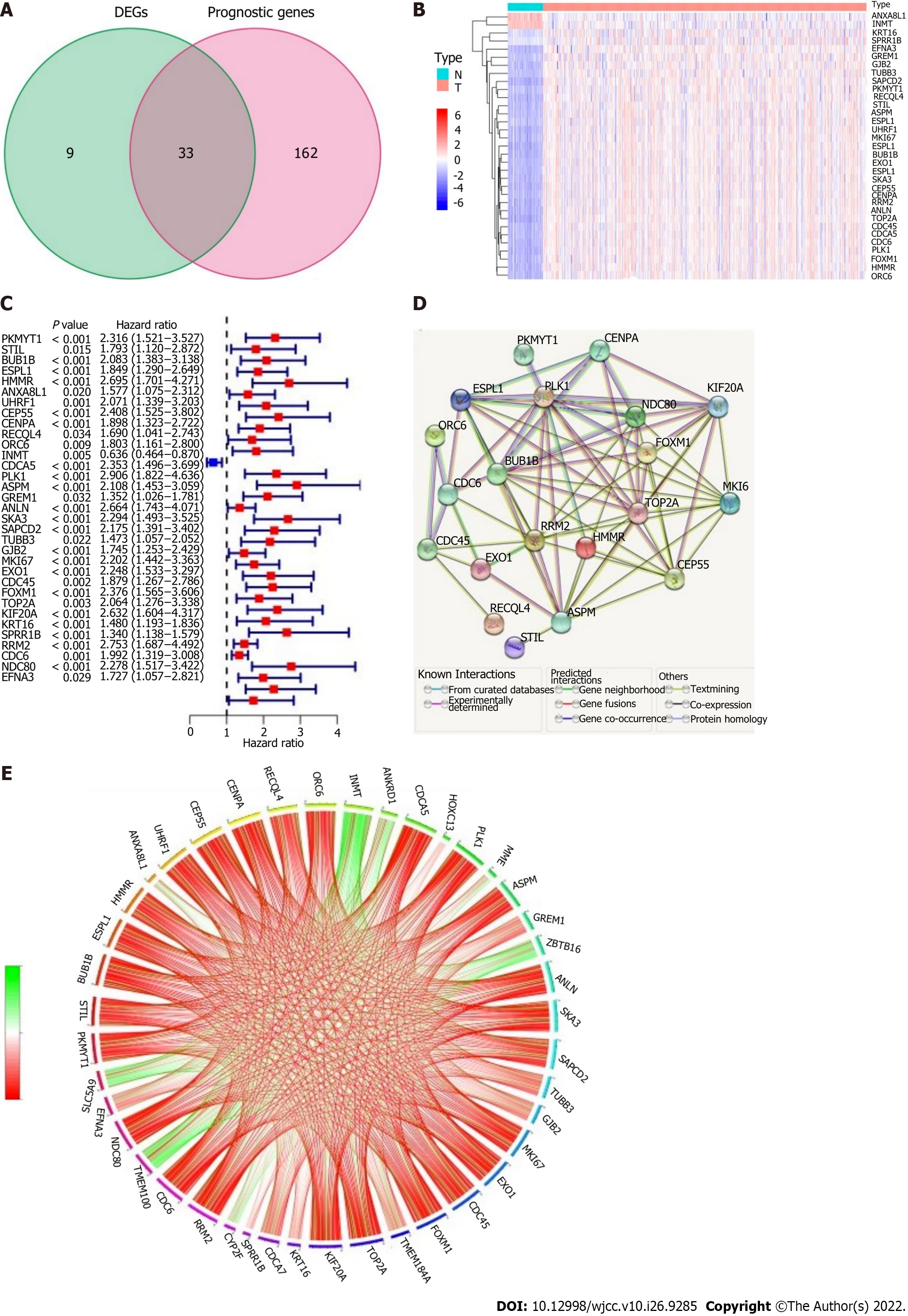
EMT-associated genes (42/612, 6.86%) were differentially expressed between cancer tissues and adjacent paracancerous tissues, and univariate Cox regression analysis showed that there were 33 genes associated with OS in LUAD (Figure 3A). The heatmap showed that ANXA8L1 and INMT were highly expressed in adjacent paracancerous tissues (Figure 3B). However, the forest plots showed that the expression of ANXA8L1 was upregulated in cancer tissues, and the results of univariate Cox regression analysis showed that there was a statistically significant association between ANXA8L1 gene expression and OS (Figure 3C). Therefore, ANXA8L1 was excluded from further study. In addition, 7 genes (UHRF1, INMT, ANLN, SKA3, SAPCD2, GJB2 and CDCA5) were not found in the gene expression omnibus cohort. Finally, a total of 25 EMT-related DEGs associated with prognosis were used in subsequent analyses (all P values < 0.05, Figure 3B-E). The protein–protein interaction network of these 25 genes showed that PLK1, NDC80 and BUB1B were considered the hub genes (Figure 3D). The mutual connections among the 25 genes are shown in Figure 3E.
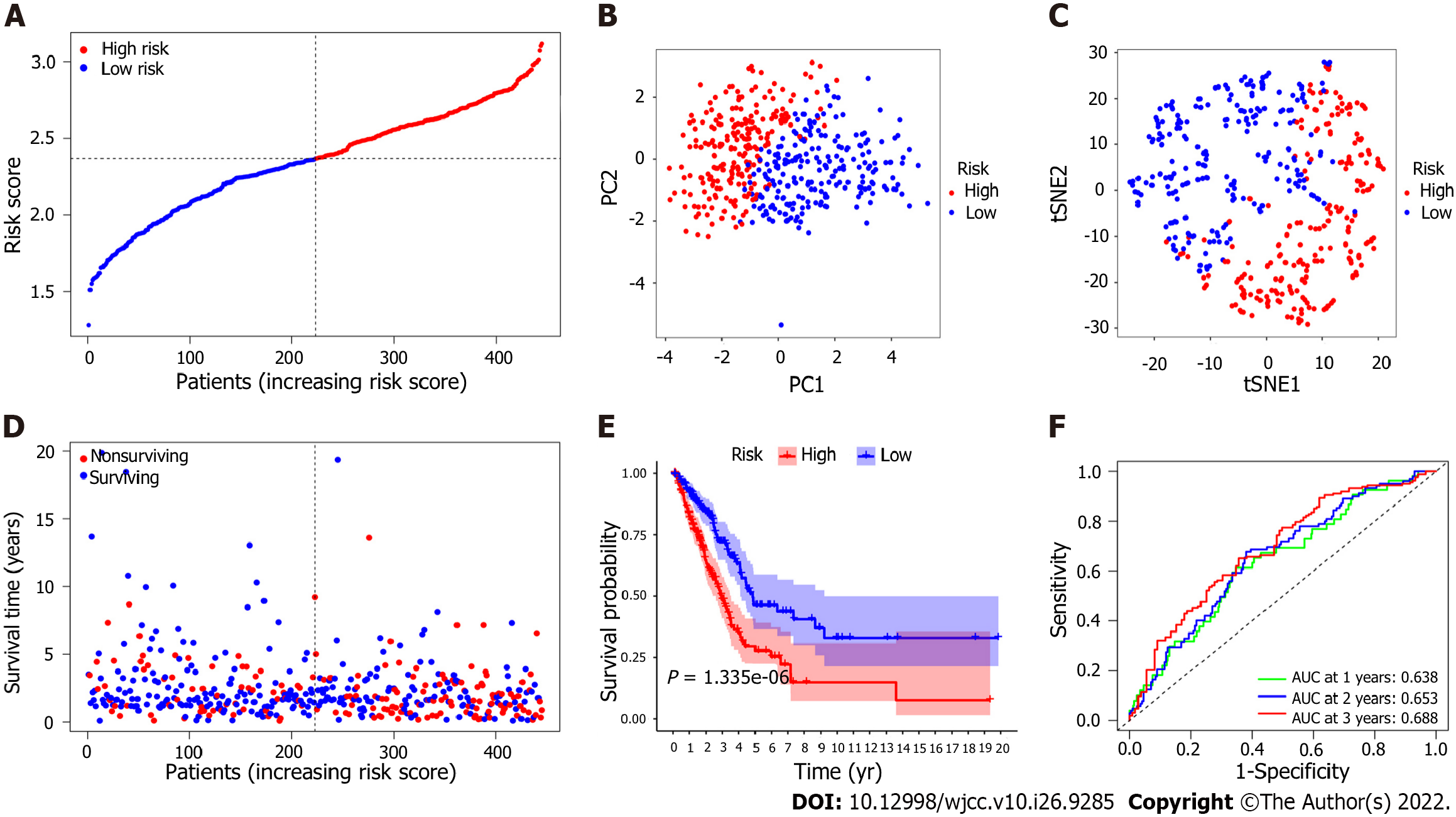
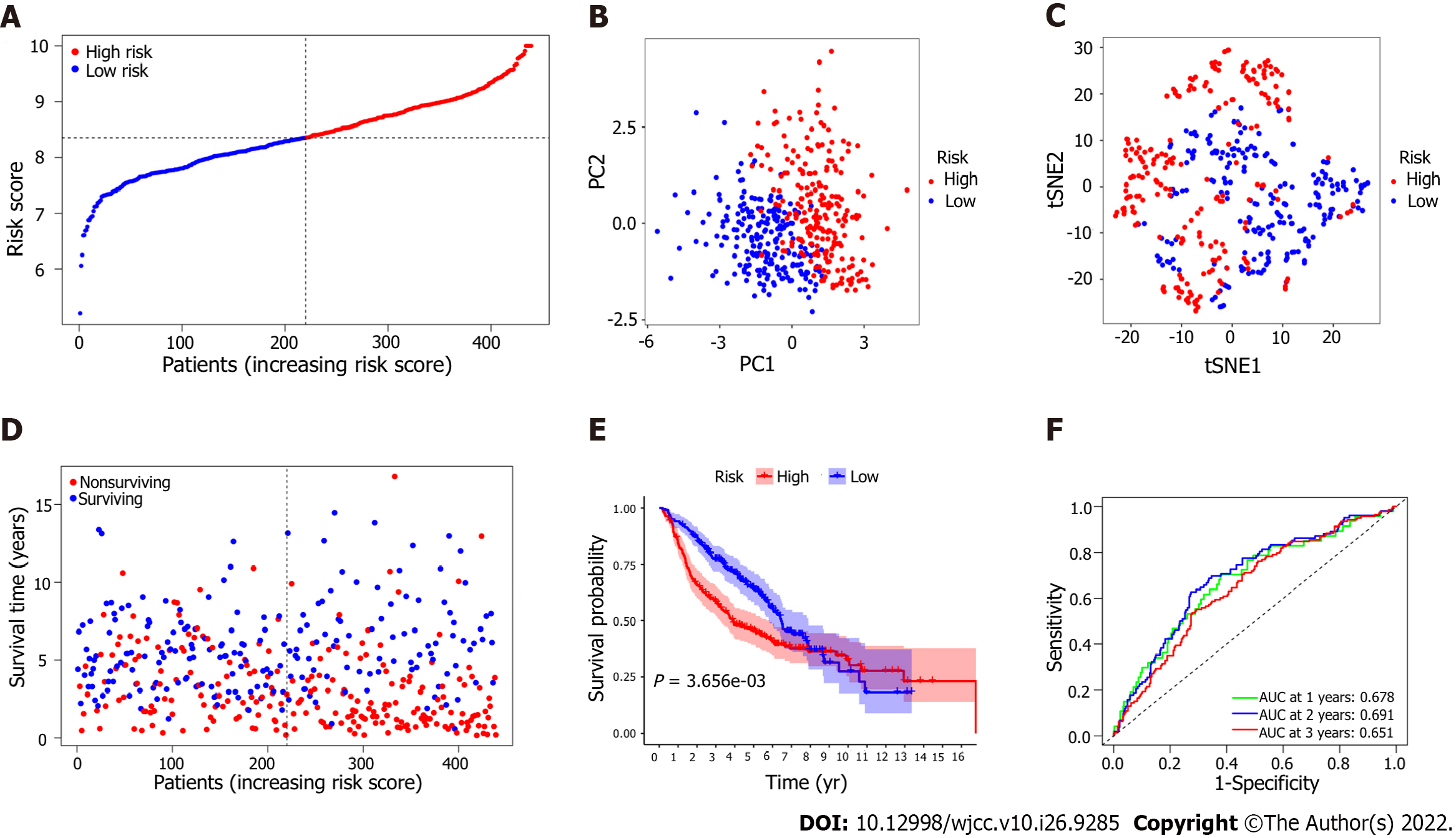
To assess the risk probability for each patient, LASSO Cox regression analysis was performed to develop a predictive model based on prognostic genes utilizing the expression profiles of the 25 EMT-related genes indicated above. According to the optimum value of lambda (λ), 7 possible factors (HMMR, PLK1, ASPM, EXO1, KRT16, SPRR1B and EFNA3) were finally confirmed. Survival analysis according to the optimum threshold expression value of each gene showed that there was a significant correlation between high expression of these genes and poor prognosis (P value < 0.05). The risk score formula was as follows: Risk score = e(0.229 × expression level of HMMR + 0.484 × expression level of PLK1 + 0.018 × expression level of ASPM + 0.085 × expression level of EXO1 + 0.075 × expression level of KRT16 + 0.178 × expression level of SPRR1B + 0.086 × expression level of EFNA3). The LUAD patients were classified into the high-risk group (n = 222) or the low-risk group (n = 223) on the basis of the median cutoff value (Figure 4A). Patients in the high-risk group were significantly more likely to have a more advanced TNM stage (Table 3). PCA and t-SNE analyses were carried out to ascertain the difference in the distribution between the two risk groups (Figure 4B and C). The results showed that patients with higher risk scores had shorter survival times than those with lower risk scores (Figure 4D). Kaplan–Meier analysis showed that the OS of patients in the high-risk group was significantly poorer than that of patients in the low-risk group (P = 1.335 × 10-6, Figure 4E). Time-dependent ROC curve analysis showed that the model for predicting prognosis had favorable performance in predicting OS [area under the curve (AUC) for one-year OS = 0.638, AUC for two-year OS = 0.653, AUC for three-year OS = 0.688; Figure 4F].
| Variable | Derivation cohort | Validation cohort | ||||
| High risk | Low risk | P value | High risk | Low risk | P value | |
| Sex | 0.235 | 0.015 | ||||
| Female | 116 | 129 | 96 | 122 | ||
| Male | 106 | 94 | 123 | 98 | ||
| Age, yr | 0. 742 | 0.416 | ||||
| < 65 | 101 | 98 | 102 | 111 | ||
| ≥ 65 | 121 | 125 | 117 | 109 | ||
| TNM stage | 0.001 | 0.017 | ||||
| Ι + Ⅱ | 159 | 189 | 174 | 195 | ||
| Ⅲ + Ⅳ | 63 | 34 | 44 | 23 | ||
| Unknown | 1 | 2 | ||||
| T stage | 0.002 | < 0.001 | ||||
| T1 | 57 | 97 | 49 | 100 | ||
| T2 | 132 | 102 | 142 | 106 | ||
| T3 | 23 | 14 | 21 | 7 | ||
| T4 | 9 | 8 | 6 | 5 | ||
| Tx | 1 | 2 | 1 | 2 | ||
| Status | < 0.001 | 0.003 | ||||
| Surviving | 116 | 159 | 87 | 119 | ||
| Non-surviving | 106 | 64 | 132 | 101 | ||
Survival analysis based on the 7 EMT-associated genes in the signature proved that these genes were associated with poorer OS. To confirm the robustness of the predictive model, the LUAD patients in the external cohort were also classified into the high-risk group and the low-risk group according to the median risk score calculated by the same formula used in the derivation cohort (Figure 5A). Patients in the high-risk group in this validation cohort were also more likely to have a more advanced TNM stage, and the evidence for this association was entirely consistent (Table 3). Moreover, the results of PCA and t-SNE analysis showed that the LUAD patients in the different risk groups were distributed in two directions (Figures 5B and C). Similarly, we found the number of nonsurviving patients was much higher in the high-risk group, revealing that patients with high risk scores might have poor survival outcomes (P = 3.656 × 10-3, Figure 5D and E). Additionally, the AUCs for survival in the validation model were 0.678 at one year, 0.691 at two years, and 0.651 at three years (Figure 5F).
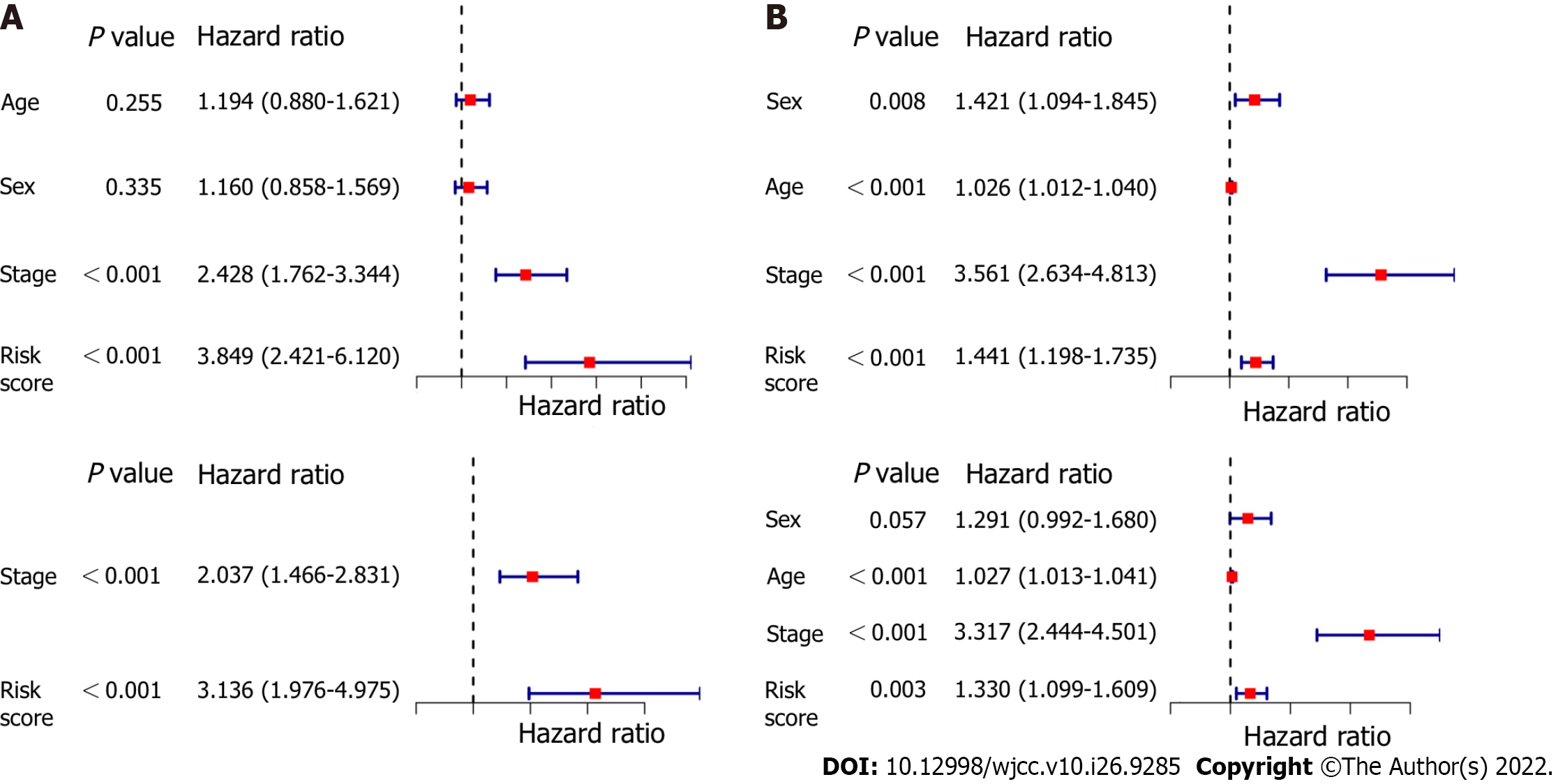
The OS-associated indicators confirmed by univariate Cox regression analysis were subsequently determined utilizing multivariate Cox regression analysis. The results of univariate Cox regression analysis showed that the risk score was considerably associated with OS in both the modeling cohort and the validation cohort (modeling cohort: HR = 3.849, 95%CI: 2.421-6.120, P < 0.001; validation cohort: HR = 1.441, 95%CI: 1.198-1.735, P < 0.001; Figure 6A and C). After controlling for confounding effects, multivariate Cox regression analysis showed that the risk score was an independent predictor for OS in LUAD patients (derivation cohort: HR = 3.136, 95%CI: 1.976-4.975, P < 0.001; validation cohort: HR = 1.330, 95%CI: 1.099-1.609, P = 0.003, respectively) (Figure 6B and D).
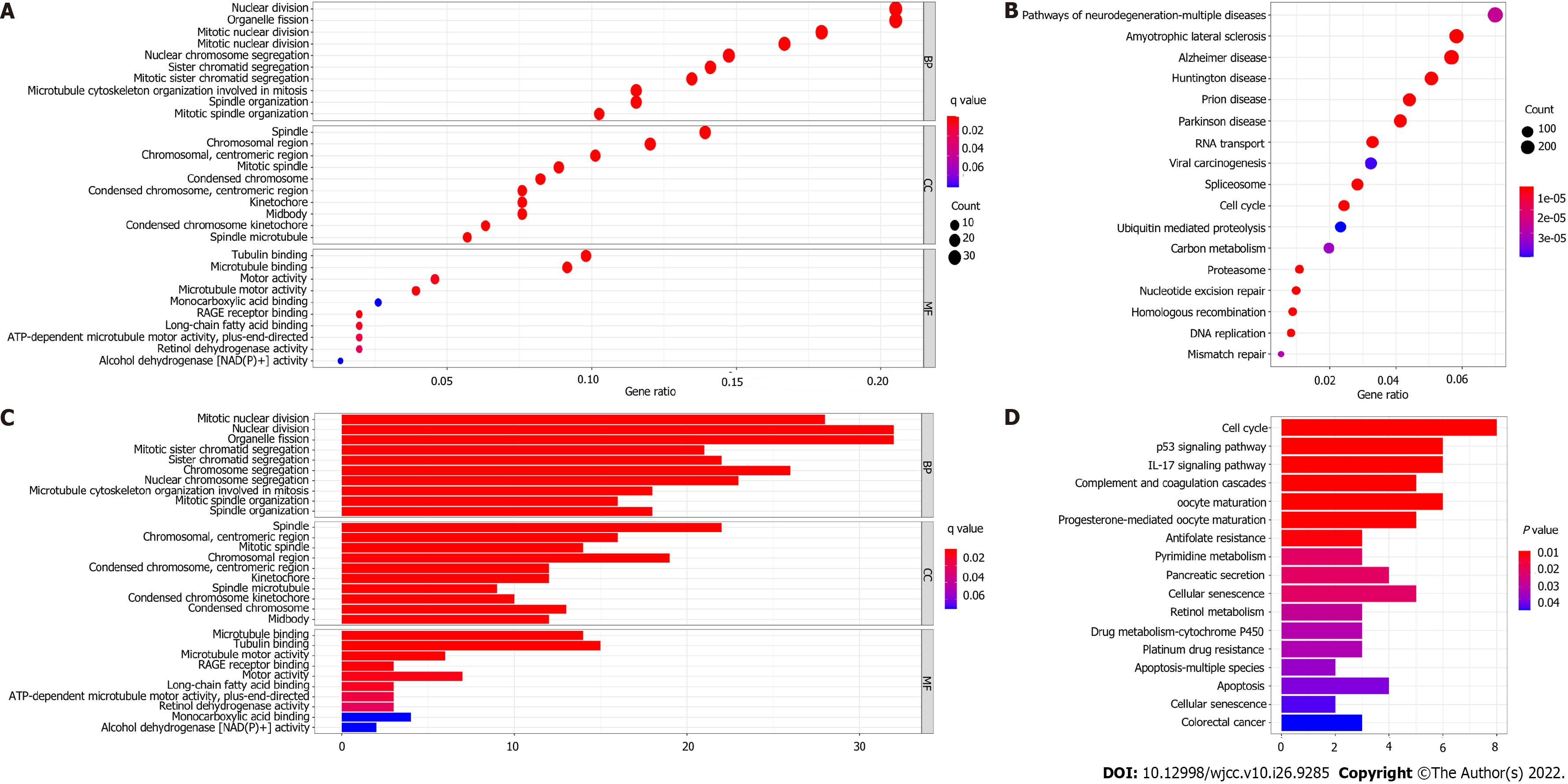
To further elucidate the biological functions and relevant pathways associated with the risk score, GO functional enrichment analysis and KEGG pathway enrichment analysis were carried out using the DEGs between the high-risk group and the low-risk group. The GO molecular function terms enriched with the DEGs in the modeling cohort and validation cohort were mainly associated with the cell cycle and cell division and included nuclear division, organelle fission, and mitotic nuclear division (P < 0.05, Figure 7A and C). Surprisingly, the results of GO functional enrichment analysis were completely consistent between the modeling cohort and validation cohort. The results of KEGG pathway enrichment analysis showed that the cell cycle pathway was enriched in the two cohorts (P < 0.05, Figure 7B and D).
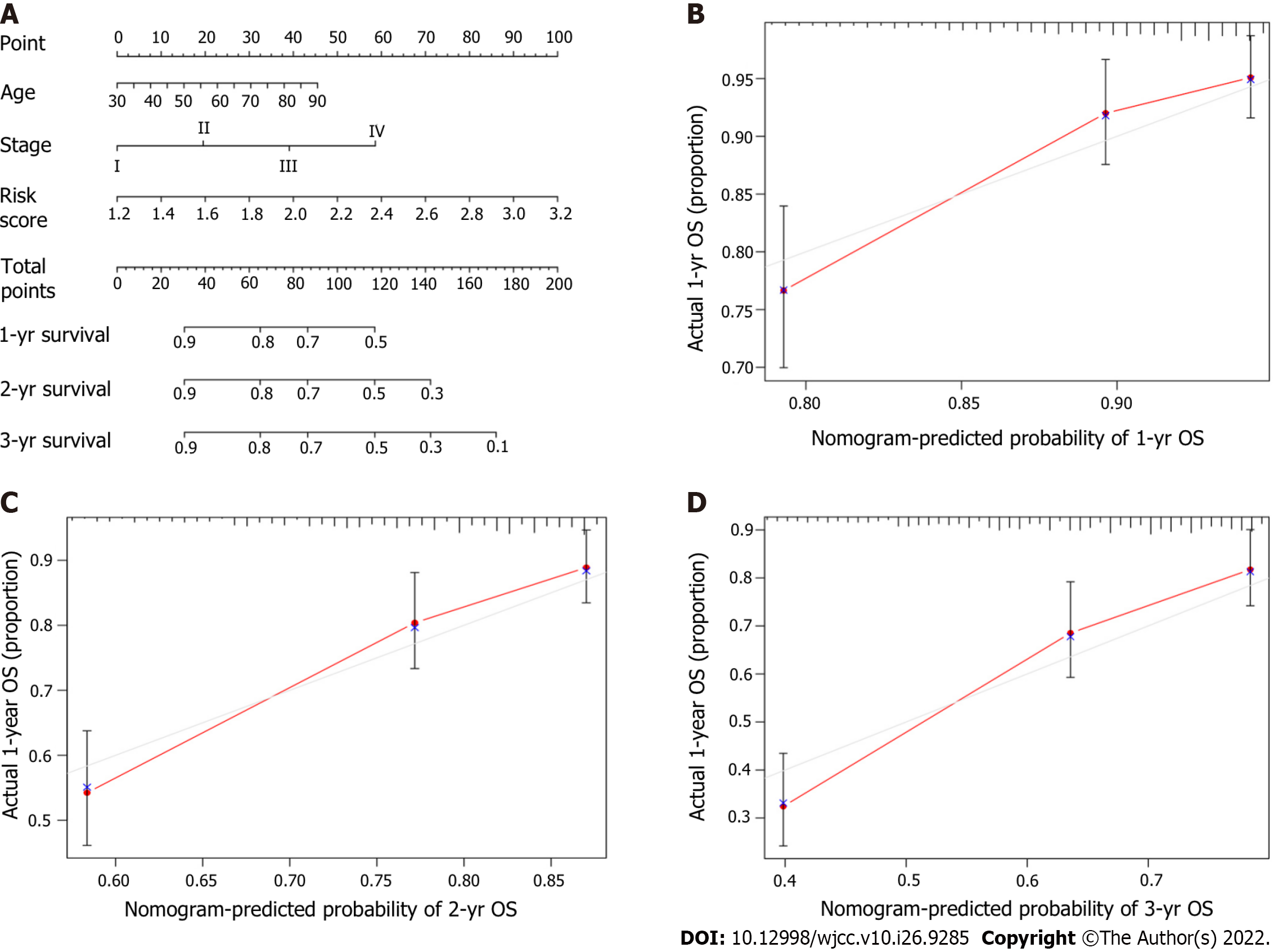
A novel prognostic nomogram including the risk score and clinical risk factors was developed to evaluate one-year, two-year, and three-year OS. Combined with clinical risk factors, the risk level in the model for predicting prognosis had significant predictive ability in the nomogram (Figure 8A). The calibration plots for the novel nomogram for predicting one-year, two-year and three-year OS are shown, and all exhibited satisfactory accuracy (Figure 8B-D).
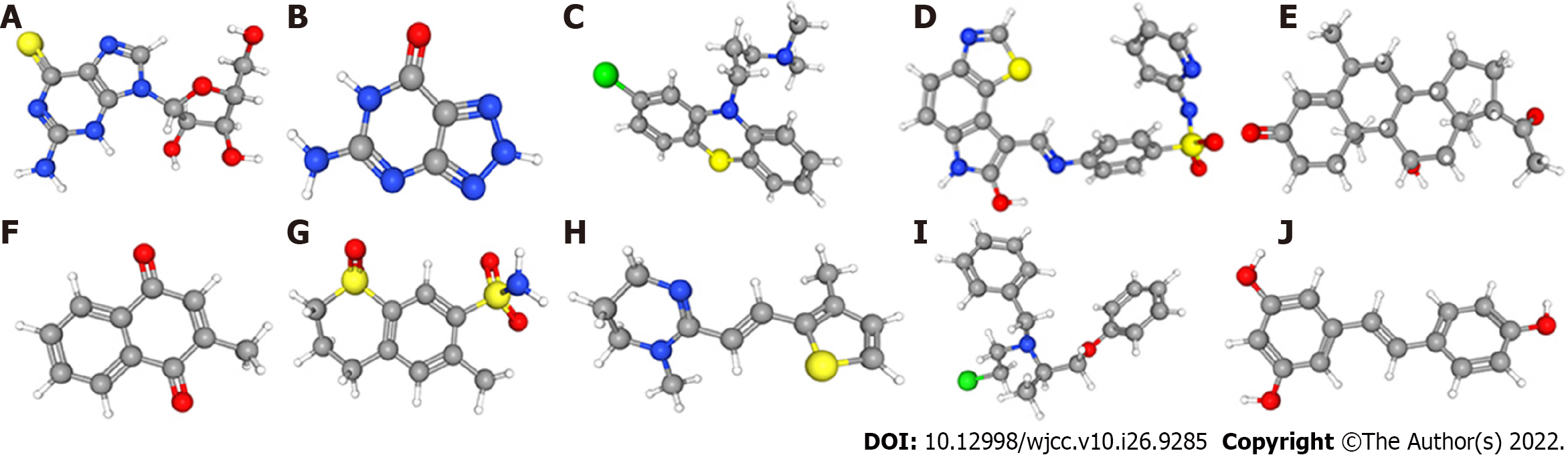
We uploaded 12012 DEGs with P value < 0.05, namely, 1186 downregulated genes and 4982 upregulated genes, into the CMap network tool. According to the screening criteria of an enrichment score < 0 and a P value < 0.05, 66 small molecule drugs were found to have significant antitumor effects. Among these highly significantly correlated molecules, GW-8510, menadione, 6-thioguanosine, phenoxybenzamine, medrysone, chlorpromazine, 8-azaguanine, meticrane, morantel and resveratrol were among the 10 drugs with the highest negative correlations with high-risk patients (Figure 9). The three-dimensional structure of 0175029-0000 was not found in the CMap network tool; thus, this drug was not included.
LUAD is recognized as the most common and aggressive malignant tumor with variable prognosis and is a threat to health and life[15]. Epithelial cells can lose their epithelial phenotypes, including cell polarity and connection to the basement membrane, and acquire mesenchymal phenotypes with potent migration, invasion, anti-apoptotic and extracellular matrix degradation abilities, thus acquiring important biological behaviors of migration and invasion[16]. To the best of our knowledge, no prior research has explored the correlation between EMT and LUAD growth. Surprisingly, EMT-related genes were differentially expressed in tumor and normal tissues, and these genes were substantially correlated with OS (33/42, 78.57%). This suggests that EMT might have a role in LUAD prognosis and that this risk score is useful for predicting LUAD survival. Previous studies have also reported that the malignant progression of non-small cell lung cancer is determined by EMT and PD-L1 and is related to poor survival in patients with cancer[17].
GSEA is a gene set enrichment analysis approach that combines information from several sources and levels. Using expression data for 54774 genes from 551 individuals with lung cancer, we attempted to conduct GSEA in this study and discovered significant differences in 9 functions. EMT, with a statistically significant P value, was chosen for further study based on the normalized enrichment score, size, and P value. Via GSEA, we concentrated on a particular function to select genes for predicting patient survival and thoroughly investigated these genes. A combination of 7 genes with prognostic significance for patients with LUAD, rather than a single gene, was found through univariate and multivariate Cox regression analyses. This selected risk signature might be slightly more focused and have greater predictive power than certain other known prognostic biomarkers, supporting clinical outcomes and serving as a useful categorization tool for LUAD patients. Using the TCGA LUAD dataset, genes associated with EMT were identified, and their expression was compared to that in adjacent noncancerous tissues. High risk scores were linked to worse survival according to Kaplan–Meier analysis. These findings implied that identifying and determining the risk scores for LUAD patients has significant prognostic relevance and might improve the methods currently used to predict patient survival and prognosis. The risk score could also aid physicians in selecting the most appropriate course of action.
Stevens et al[18] reported that overexpression of the hyaluronic acid receptor HMMR in LUAD was related to molecular characteristics of inflammation and poor prognosis. In LUAD cells, decreasing HMMR expression decreased their ability to induce the development of lung carcinomas and distant metastases. In addition, HMMR is important for the orientation of the mitotic spindle in human mitotic cells and could regulate spindle assembly in mitotic cells and meiotic extracts[19]. Cell cycle analysis showed that in PLK1-silenced cells, the cell cycle was arrested at the G2/M phase transition. There was a positive connection between the expression levels of PLK1 and KPNB1 in LUAD cell lines. A decrease in KPNB1 expression was induced by inhibition of PLK1 and resulted in apoptosis in LUAD cells[20]. ASPM has been reported to be widely expressed in a variety of cancer tissues and is involved in the occurrence and progression of many malignant tumors, including lung tumors[21]. The results of Yuan et al[21] showed that ASPM depletion significantly suppressed the proliferation of lung squamous cell carcinoma cells, consistent with significant decreases in cyclin D1 and cyclin-dependent kinase 4 expression. Liu et al[22] reported that ASPM was positively related to progression and poor prognosis in prostate and hepatocellular carcinoma, but the relationship between ASPM and prognosis in LUAD has not been reported. Previous reports suggested that functional polymorphisms of EXO1 might be related to the occurrence of lung cancer and that EXO1 might be a novel biomarker for the diagnosis and treatment of lung carcinoma[23]. The KRT family is closely associated with tumor development and metastasis in various cancers, including lung, breast, and colon cancers. Han et al[24] suggested that KRT16 was overexpressed in primary melanoma and metastatic melanoma and indicated significant differences in KRT16 expression in diverse T stages and postoperative pathological stages[25]. Downregulation of SPRR1B suppressed the proliferation, metastasis and invasion of LUAD cells and induced G2/M arrest in vitro[26]. Studies have shown that hypoxia has a significant inhibitory effect on the increase in EFNA3 expression, but there are few studies on EFNA3 in tumor cells[27]. However, hypoxia is clearly associated with prognosis and metastasis in many cancers, including LUAD[28]. All 7 genes were upregulated in LUAD tissues and were related to poor prognosis in this study.
We performed GO functional enrichment analysis based on the differentially expressed genes between the different risk groups and found that these genes were unexpectedly enriched in many processes and cytoskeletal features involved in the cell division cycle. It was rational to hypothesize that EMT might be closely related to the cell division cycle. Interestingly, there were significant differences in the enrichment of mitotic nuclear division processes between the low-risk group and the high-risk group in our research. Wang et al[29] showed that Axl inhibition was particularly synergistic with antimitotic drugs in eliminating tumor cells that had undergone EMT. In particular, tumor cells stopped dividing when they underwent EMT; thus, EMT acted as a suppressor of cancer cell division and then ended proliferation[30]. Therefore, how EMT influences tumor metastasis by affecting cell cycle processes, including mitotic nuclear division, requires further study.
Although many attempts have been made to improve the prognosis of OS, the results of these changes have not been obvious. Here, we identified 10 small molecules (GW-8510, menadione, 6-thioguanosine, phenoxybenzamine, medrysone, chlorpromazine, 8-azaguanine, meticrane, morantel and resveratrol) that have potential therapeutic benefits in OS. GW-8510 is a CDK2 inhibitor that usually inhibits the activity of cyclin/CDK complexes and negatively regulates cell cycle progression[31]. Menadione, also named VK3, is a redox cycling agent that has an antitumor effect against prostate, hepatic, lung, and breast cancer[32,33]. A previous study indicated that 6-thioguanosine, which inhibits PTHrP transcription, was used in the treatment of leukemia[34]. Phenoxybenzamine, a nonselective first-generation α1- and α2-AR antagonist, has been administered to treat lower urinary tract symptoms[35]. Medrysone is a corticosteroid usually administered to cure eye inflammation[36]. Chlorpromazine is a traditional antipsychotic or neuroleptic that is used for the treatment of schizophrenia and other psychiatric disorders[37]. Liberante et al[38] showed that the concurrent targeting of RNA metabolism and splicing by 8-azaguanine offered curative prospects for SF3B1 mutant myelodysplastic syndromes. A study found that meticrane can target the PPAR signaling pathway and inhibit the reabsorption of sodium and chloride ions in distal convoluted tubules[39]. Morantel is the classic competitive antagonist of dihydro-β-erythroidine, which noncompetitively inhibits morantel-evoked currents[40]. Resveratrol has been reported to treat a variety of cancers, including papillary thyroid cancer and liver cancer[41,42]. Overall, GW-8510, menadione, 6-thioguanosine, phenoxybenzamine, medrysone, chlorpromazine, 8-azaguanine, meticrane, morantel and resveratrol exhibit high clinical potential and warrant further study, particularly for the improvement of OS through mechanisms affecting the cell cycle.
Undoubtedly, the current research has some limitations that should be addressed. First, this was a retrospective study for the establishment of gene signatures based on public databases. Next, we must use our own esophageal cancer specimens to confirm the expression of the seven EMT-related genes in tumor tissues and adjacent tissues, as well as their correlations with clinicopathological parameters and prognosis. Second, a well-defined EMT mechanism is lacking in lung adenocarcinoma research. The biological functions of the predicted genes were estimated by computational methods, and further study is needed to reveal their mechanisms of action in tumorigenesis. In the future, EMT-related genes could be overexpressed or silenced in animal models to study tumor growth and metastasis. Additionally, we investigated how different EMT states influence tissue canceration, invasion, and metastasis in lung adenocarcinoma. Exosomes need to be studied for their role in lung adenocarcinoma initiation, development, and EMTs. Therefore, other large external multicenter experimental studies are needed to further confirm our results.
In conclusion, we constructed and validated a 7-gene prognostic risk prediction signature based on EMT-related genes in LUAD. The risk score determined by the prognostic model of EMT-related genes was an independent risk factor for OS in LUAD. We developed a nomogram for clinicians to help predict OS in LUAD patients. According to the results of functional enrichment analysis, the pathways involved in the cell cycle and cell division were enriched in the high-risk group. In addition, we analyzed the molecular structure of potential therapeutic agents and obtained sufficient evidence indicating that EMT-related genes could be used to predict biomarkers and targeted therapies for LUAD.
The transformation between epithelial and stromal cells during cancer progression is important in cancer progression. Understanding the relationship between epithelial-mesenchymal transition (EMT)-related genes and lung adenocarcinoma is valuable for improving its prognosis.
To develop an EMT-related gene signature to predict the prognosis of lung adenocarcinoma.
To construct and validate prognostic polygenic signatures of differentially expressed genes associated with EMT.
We constructed a prognostic model based on differentially expressed EMT-related genes from 445 lung adenocarcinoma samples and validated its feasibility with another 439 Lung adenocarcinoma samples.
Seven EMT-related prognostic gene signatures were developed and validated. Kaplan–Meier survival analysis showed that the overall survival of patients in the high-risk group was statistically significantly poorer than that of patients in the low-risk group. The risk score based on EMT-associated genes was an independent risk factor for overall survival.
The EMT-related gene signature could be used as a feasible indicator to predict the overall survival of lung adenocarcinoma patients. The molecular structures of potential therapeutic agents associated with EMT genes that could be used to treat lung adenocarcinoma were identified.
More accurate genetic markers and models are needed to predict prognosis because of the low survival rate of lung adenocarcinoma.
We thank The Cancer Genome Atlas database (https://cancergenome.nih.gov/) and the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) for providing us with analytical data.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Respiratory system
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Qin Y, China; Wang J, China S-Editor: Zhang H L-Editor: A P-Editor: ChenYX
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64360] [Article Influence: 16090.0] [Reference Citation Analysis (176)] |
| 2. | Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 1415] [Article Influence: 353.8] [Reference Citation Analysis (0)] |
| 3. | Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, Bray F. Cancer statistics for the year 2020: An overview. Int J Cancer. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2411] [Cited by in RCA: 2937] [Article Influence: 734.3] [Reference Citation Analysis (7)] |
| 4. | Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, Zhou C, Reungwetwattana T, Cheng Y, Chewaskulyong B, Shah R, Cobo M, Lee KH, Cheema P, Tiseo M, John T, Lin MC, Imamura F, Kurata T, Todd A, Hodge R, Saggese M, Rukazenkov Y, Soria JC; FLAURA Investigators. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med. 2020;382:41-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1120] [Cited by in RCA: 1901] [Article Influence: 380.2] [Reference Citation Analysis (0)] |
| 5. | Shen J, Zhuang W, Xu C, Jin K, Chen B, Tian D, Hiley C, Onishi H, Zhu C, Qiao G. Surgery or Non-surgical Treatment of ≤8 mm Non-small Cell Lung Cancer: A Population-Based Study. Front Surg. 2021;8:632561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Giannopoulou AF, Velentzas AD, Anagnostopoulos AK, Agalou A, Papandreou NC, Katarachia SA, Koumoundourou DG, Konstantakou EG, Pantazopoulou VI, Delis A, Michailidi MT, Valakos D, Chatzopoulos D, Syntichaki P, Iconomidou VA, Tsitsilonis OE, Papassideri IS, Voutsinas GE, Hatzopoulos P, Thanos D, Beis D, Anastasiadou E, Tsangaris GT, Stravopodis DJ. From Proteomic Mapping to Invasion-Metastasis-Cascade Systemic Biomarkering and Targeted Drugging of Mutant BRAF-Dependent Human Cutaneous Melanomagenesis. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 7. | Kryczka J, Papiewska-Pajak I, Kowalska MA, Boncela J. Cathepsin B Is Upregulated and Mediates ECM Degradation in Colon Adenocarcinoma HT29 Cells Overexpressing Snail. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 8. | Jiang J, Li J, Zhou X, Zhao X, Huang B, Qin Y. Exosomes Regulate the Epithelial-Mesenchymal Transition in Cancer. Front Oncol. 2022;12:864980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 9. | Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019;29:212-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 920] [Cited by in RCA: 1887] [Article Influence: 269.6] [Reference Citation Analysis (0)] |
| 10. | Qiao K, Chen C, Liu H, Qin Y. Pinin Induces Epithelial-to-Mesenchymal Transition in Hepatocellular Carcinoma by Regulating m6A Modification. J Oncol. 2021;2021:7529164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Chaudhary KR, Kinslow CJ, Cheng H, Silva JM, Yu J, Wang TJ, Hei TK, Halmos B, Cheng SK. Smurf2 inhibition enhances chemotherapy and radiation sensitivity in non-small-cell lung cancer. Sci Rep. 2022;12:10140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | Tan Z, Chen K, Wu W, Zhou Y, Zhu J, Wu G, Cao L, Zhang X, Guan H, Yang Y, Zhang W, Li J. Overexpression of HOXC10 promotes angiogenesis in human glioma via interaction with PRMT5 and upregulation of VEGFA expression. Theranostics. 2018;8:5143-5158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 13. | Zhou ZR, Wang WW, Li Y, Jin KR, Wang XY, Wang ZW, Chen YS, Wang SJ, Hu J, Zhang HN, Huang P, Zhao GZ, Chen XX, Li B, Zhang TS. In-depth mining of clinical data: the construction of clinical prediction model with R. Ann Transl Med. 2019;7:796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 198] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 14. | Mathew B, Hauptmann A, Léon J, Sillanpää MJ. NeuralLasso: Neural Networks Meet Lasso in Genomic Prediction. Front Plant Sci. 2022;13:800161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | Rong H, Chen B, Wei X, Peng J, Ma K, Duan S, He J. Long non-coding RNA XIST expedites lung adenocarcinoma progression through upregulating MDM2 expression via binding to miR-363-3p. Thorac Cancer. 2020;11:659-671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Jasemi S, Emaneini M, Fazeli MS, Ahmadinejad Z, Nomanpour B, Sadeghpour Heravi F, Sechi LA, Feizabadi MM. Toxigenic and non-toxigenic patterns I, II and III and biofilm-forming ability in Bacteroides fragilis strains isolated from patients diagnosed with colorectal cancer. Gut Pathog. 2020;12:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 17. | Manjunath Y, Upparahalli SV, Avella DM, Deroche CB, Kimchi ET, Staveley-O'Carroll KF, Smith CJ, Li G, Kaifi JT. PD-L1 Expression with Epithelial Mesenchymal Transition of Circulating Tumor Cells Is Associated with Poor Survival in Curatively Resected Non-Small Cell Lung Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 18. | Stevens LE, Cheung WKC, Adua SJ, Arnal-Estapé A, Zhao M, Liu Z, Brewer K, Herbst RS, Nguyen DX. Extracellular Matrix Receptor Expression in Subtypes of Lung Adenocarcinoma Potentiates Outgrowth of Micrometastases. Cancer Res. 2017;77:1905-1917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 19. | He Z, Mei L, Connell M, Maxwell CA. Hyaluronan Mediated Motility Receptor (HMMR) Encodes an Evolutionarily Conserved Homeostasis, Mitosis, and Meiosis Regulator Rather than a Hyaluronan Receptor. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 20. | Sekimoto N, Suzuki Y, Sugano S. Decreased KPNB1 Expression is Induced by PLK1 Inhibition and Leads to Apoptosis in Lung Adenocarcinoma. J Cancer. 2017;8:4125-4140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Yuan YJ, Sun Y, Gao R, Yin ZZ, Yuan ZY, Xu LM. Abnormal spindle-like microcephaly-associated protein (ASPM) contributes to the progression of Lung Squamous Cell Carcinoma (LSCC) by regulating CDK4. J Cancer. 2020;11:5413-5423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 22. | Liu J, Zhou S, Li S, Jiang Y, Wan Y, Ma X, Cheng W. Eleven genes associated with progression and prognosis of endometrial cancer (EC) identified by comprehensive bioinformatics analysis. Cancer Cell Int. 2019;19:136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 23. | Zhang M, Zhao D, Yan C, Zhang L, Liang C. Associations between Nine Polymorphisms in EXO1 and Cancer Susceptibility: A Systematic Review and Meta-Analysis of 39 Case-control Studies. Sci Rep. 2016;6:29270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Han W, Hu C, Fan ZJ, Shen GL. Transcript levels of keratin 1/5/6/14/15/16/17 as potential prognostic indicators in melanoma patients. Sci Rep. 2021;11:1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 25. | Zhou CS, Feng MT, Chen X, Gao Y, Chen L, Li LD, Li DH, Cao YQ. Exonuclease 1 (EXO1) is a Potential Prognostic Biomarker and Correlates with Immune Infiltrates in Lung Adenocarcinoma. Onco Targets Ther. 2021;14:1033-1048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 26. | Zhang Z, Shi R, Xu S, Li Y, Zhang H, Liu M, Zhu G, Chen C, Pan Z, Liu H, Chen J. Identification of small proline-rich protein 1B (SPRR1B) as a prognostically predictive biomarker for lung adenocarcinoma by integrative bioinformatic analysis. Thorac Cancer. 2021;12:796-806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 27. | Zajkowska M, Mroczko B. Angiopoietin-like Proteins in Colorectal Cancer-A Literature Review. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 28. | Koren A, Rijavec M, Krumpestar T, Kern I, Sadikov A, Čufer T, Korošec P. Gene Expression Levels of the Prolyl Hydroxylase Domain Proteins PHD1 and PHD2 but Not PHD3 Are Decreased in Primary Tumours and Correlate with Poor Prognosis of Patients with Surgically Resected Non-Small-Cell Lung Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 29. | Wang C, Jin H, Wang N, Fan S, Wang Y, Zhang Y, Wei L, Tao X, Gu D, Zhao F, Fang J, Yao M, Qin W. Gas6/Axl Axis Contributes to Chemoresistance and Metastasis in Breast Cancer through Akt/GSK-3β/β-catenin Signaling. Theranostics. 2016;6:1205-1219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 30. | Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525-530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1357] [Cited by in RCA: 1636] [Article Influence: 163.6] [Reference Citation Analysis (0)] |
| 31. | Kuang Y, Kang J, Li H, Liu B, Zhao X, Li L, Jin X, Li Q. Multiple functions of p21 in cancer radiotherapy. J Cancer Res Clin Oncol. 2021;147:987-1006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Ren X, Santhosh SM, Coppo L, Ogata FT, Lu J, Holmgren A. The combination of ascorbate and menadione causes cancer cell death by oxidative stress and replicative stress. Free Radic Biol Med. 2019;134:350-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 33. | Ivanova D, Zhelev Z, Getsov P, Nikolova B, Aoki I, Higashi T, Bakalova R. Vitamin K: Redox-modulation, prevention of mitochondrial dysfunction and anticancer effect. Redox Biol. 2018;16:352-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 34. | Gładysz M, Andrałojć W, Czapik T, Gdaniec Z, Kierzek R. Thermodynamic and structural contributions of the 6-thioguanosine residue to helical properties of RNA. Sci Rep. 2019;9:4385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 35. | Shah PA, Shrivastav PS. Novel LC-MS/MS method for the quantification of silodosin and its metabolites in human plasma. Bioanalysis. 2018;10:7-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 36. | Lo YC, Senese S, France B, Gholkar AA, Damoiseaux R, Torres JZ. Computational Cell Cycle Profiling of Cancer Cells for Prioritizing FDA-Approved Drugs with Repurposing Potential. Sci Rep. 2017;7:11261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 37. | Nikvarz N, Vahedian M, Khalili N. Chlorpromazine vs penfluridol for schizophrenia. Cochrane Database Syst Rev. 2017;9:CD011831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 38. | Liberante FG, Lappin K, Barros EM, Vohhodina J, Grebien F, Savage KI, Mills KI. Altered splicing and cytoplasmic levels of tRNA synthetases in SF3B1-mutant myelodysplastic syndromes as a therapeutic vulnerability. Sci Rep. 2019;9:2678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 39. | Tragni V, Cotugno P, De Grassi A, Cavalluzzi MM, Mincuzzi A, Lentini G, Sanzani SM, Ippolito A, Pierri CL. Targeting Penicillium expansum GMC Oxidoreductase with High Affinity Small Molecules for Reducing Patulin Production. Biology (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 40. | Keiser J, Häberli C. Assessment of FDA-approved drugs against Strongyloides ratti in vitro and in vivo to identify potentially active drugs against strongyloidiasis. Parasit Vectors. 2021;14:615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 41. | Bian P, Hu W, Liu C, Li L. Resveratrol potentiates the anti-tumor effects of rapamycin in papillary thyroid cancer: PI3K/AKT/mTOR pathway involved. Arch Biochem Biophys. 2020;689:108461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 42. | Rahman M, Almalki WH, Afzal O, Alfawaz Altamimi AS, Kazmi I, Al-Abbasi FA, Choudhry H, Alenezi SK, Barkat MA, Beg S, Kumar V, Alhalmi A. Cationic Solid Lipid Nanoparticles of Resveratrol for Hepatocellular Carcinoma Treatment: Systematic Optimization, in vitro Characterization and Preclinical Investigation. Int J Nanomedicine. 2020;15:9283-9299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |