Published online Jul 26, 2022. doi: 10.12998/wjcc.v10.i21.7517
Peer-review started: December 21, 2021
First decision: February 8, 2022
Revised: February 21, 2022
Accepted: June 3, 2022
Article in press: June 3, 2022
Published online: July 26, 2022
Processing time: 201 Days and 19 Hours
Xia–Gibbs syndrome (XGS, OMIM: 615829), caused by mutations within the AT-Hook DNA-binding motif-containing protein 1 (AHDC1) gene (OMIM: 615790), located on the short arm of chromosome 1 within the cytogenetic band 1p36.11, contains five noncoding 5 exons, a single 4.9-kb coding exon, and a noncoding 3 exon.
In this case report, we diagnosed and treated a 6-mo-old girl with XGS. The primary clinical symptoms included global developmental delay, hypotonia, and mild dysmorphic features. Using high-throughput whole-exosome sequencing to sequence the patient and her parents, and the results showed a novel frameshift mutation of c.1155dupG (p.Arg386Alafs*3) in the AHDC1 gene. The paternal gene was wild type.
This report extends the mutation spectrum of the AHDC1 gene to provide the diagnostic basis for genetic counseling in families with XGS.
Core Tip: We report a 6-mo-old girl with Xia–Gibbs syndrome (XGS). The main clinical manifestations were global developmental delay, hypotonia, and other mild dysmorphic features. DNA sequencing showed that there was a novel frameshift mutation of c.1155dupG (p.Arg386Alafs*3) in the AT-Hook DNA-binding motif-containing protein 1 (AHDC1) gene. This study extends the mutation spectrum of the AHDC1 gene, and provides a molecular basis for the etiological diagnosis of XGS and genetic consultation for the family.
- Citation: Lin SZ, Xie HY, Qu YL, Gao W, Wang WQ, Li JY, Feng XC, Jin CQ. Novel frameshift mutation in the AHDC1 gene in a Chinese global developmental delay patient: A case report. World J Clin Cases 2022; 10(21): 7517-7522
- URL: https://www.wjgnet.com/2307-8960/full/v10/i21/7517.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i21.7517
Xia–Gibbs syndrome (XGS) is an autosomal dominant genetic disease caused by mutation of the AT-Hook DNA-binding motif-containing protein 1 (AHDC1) gene. Typical features include global developmental delay, hypotonia, obstructive sleep apnea, seizures, delayed myelination, micrognathia, and other mild dysmorphic features[1,2]. The AHDC1 gene is located on the short arm of chromosome 1 within the cytogenetic band 1p36.11, and consists of seven exons with only one coding exon (exon 6), five noncoding 5 exons, and a noncoding 3 exon[3].
In this case, a novel frameshift mutation of c.1155dupG (p.Arg386Alafs*3) in the AHDC1 gene was found by high-throughput whole-exosome sequencing (WES) in a patient with global developmental delay, hypotonia, micrognathia, and other mild dysmorphic features.
A 6-mo-old female patient presented to our hospital due to global developmental delay.
In June 2021, the patient presented with a 6-mo history of global developmental delay. Since birth, the child had lagged behind in developmental milestones. She could not raise her head until she was age 4 mo, and she still shook her head sometimes. She could not sit alone without the help of others, and turning over was not flexible. She could not crawl, and the stability of her lower limbs was too poor to support her weight. She had no difficulty in pronunciation.
The patient weighed 3.7 kg and was 52 cm long after full-term normal delivery without a history of asphyxiation and resuscitation. The patient did not have a history of feeding difficulties. There was no history of encephalitis and brain trauma since disease onset.
Her parents were clinically normal. Genetic history or family history of infectious diseases were denied.
At her visit at 6 mo of age, we performed a physical examination. Her weight was 9.0 kg and length 70.0 cm. Mild dysmorphic features were observed, such as micrognathia. Grasping reflex was present, and limb muscle tension showed hypotonia. Ankle clonus was positive. Muscle strength was level 4. Physiological reflexes were present.
We performed a series of examinations of the patient, including liver function, kidney function, electrolytes, and organic acids in blood and urine, which showed no abnormalities.
In June 2021, brain magnetic resonance imaging showed bilateral frontal subdural effusion, hydrocephalus, and hydrops in the mastoid area. Electroencephalography (EEG) was abnormal, showing sharp waves and spike waves in the Rolandic area or right forehead region.
The patient underwent a Peabody Developmental Motor Scales (PDMS-2) test on May 14, 2021[4]. The Fine Motor Quotient (FMQ) was 9 points, with a motor quotient of 67, and the Gross Motor Quotient (GMQ) was 21 points with a motor quotient of 81; both measures were worse than those of her peers. Griffiths Mental Development Scales (GMDS) showed that motor ability was equivalent to that of a 3-mo-old child, with a development quotient (DQ) of 46 points; human-social ability was equivalent to that of a 4-mo-old child, with a DQ of 62 points; hearing and language ability was equivalent to that of a 4.5-mo-old child with a DQ of 69 points, and hand and eye coordination was equivalent to that of a 3.5-mo-old child, with a DQ of 54 points. The patients’ overall performance was equivalent to that of a 3.5-mo-old child, with a DQ of 54 points.
Informed parental consent was obtained for WES, mitochondrial sequencing, and publication of photographs on behalf of the proband. DNA samples were extracted from the peripheral blood of the child and her parents to detect whole-exome sequences and whole-genome copy number variants. The results revealed a novel frameshift mutation of c.1155dupG (p.Arg386Alafs*3) in the AHDC1 gene. Polymorphic sites were detected in each sample data using GATK software and statistical analysis was performed on 1000 human genomes and ExAC databases. The reported pathogenic locus has been confirmed using the Human Gene Mutation Database (HGMD) and the Human Online Mendelian Genetic Database (OMIM). The pathogenicity of the mutation locus was comprehensively assessed using the American College of Medical Genetics and Genomics (ACMG) standards and guidelines for the interpretation of sequence variation.
The WES results showed the presence of a novel frameshift variant in the AHDC1 gene, which was an unreported frameshift mutation, c.1155dupG (p.Arg386Alafs*3), and may result in altered gene function. The frequency at which variation occurs in the normal population database is unknown, and is a low-frequency variation. The results of protein function prediction are unknown, and are not reported in the HGMD database.
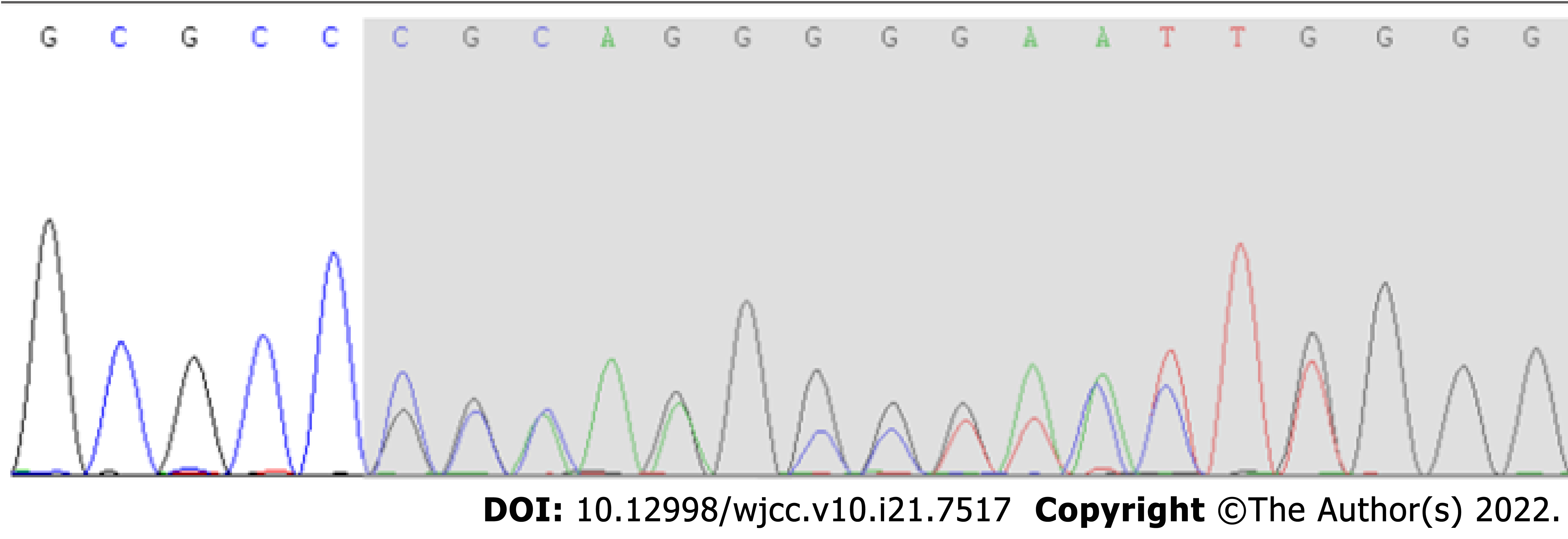
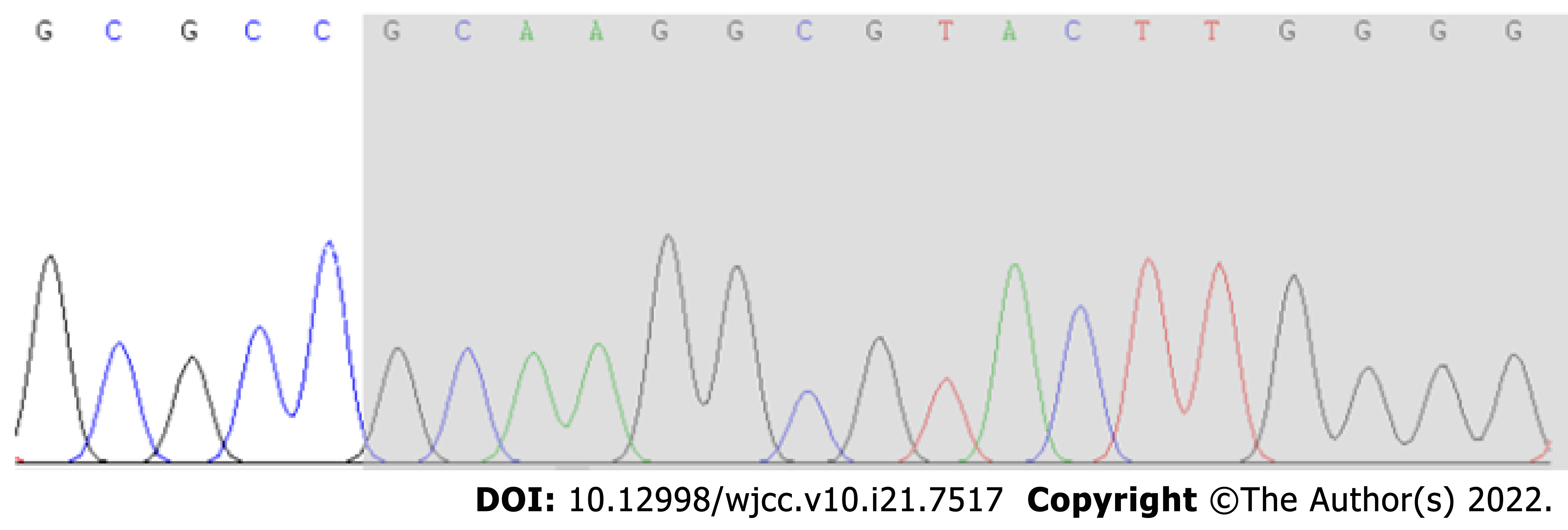
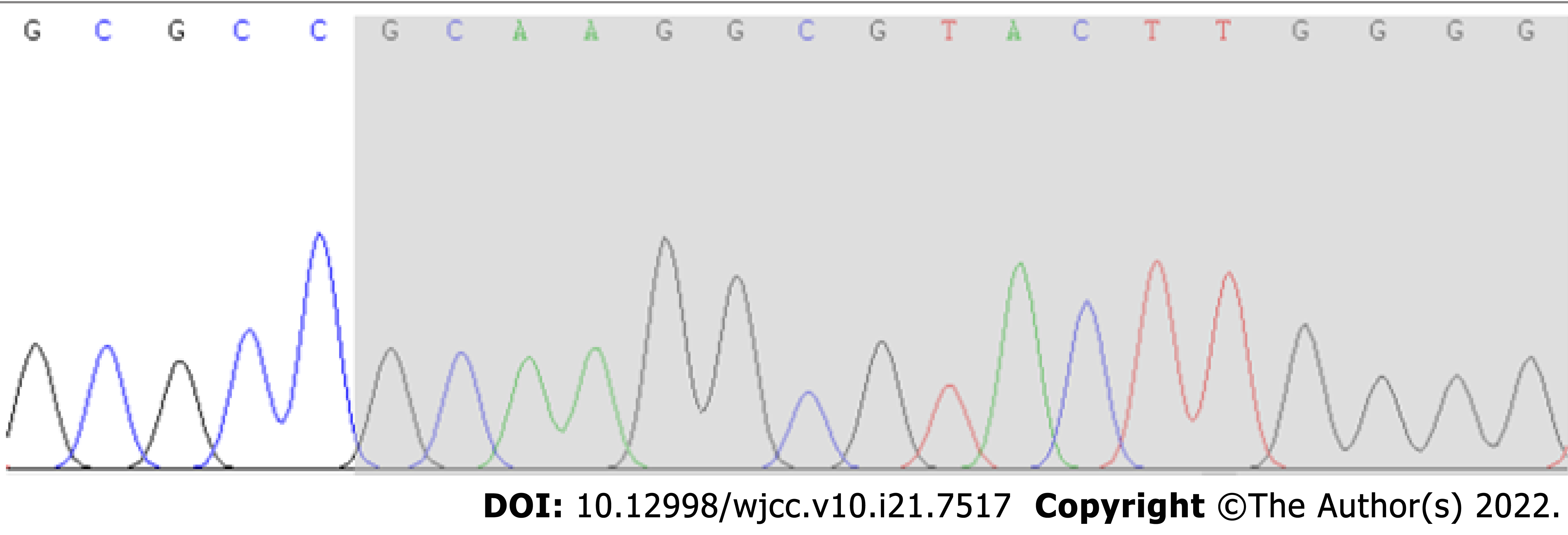
According to Sanger sequencing, the variation occurred in the child, while the parental genes were wild type (Figures 1-3). The mutation was suspected to be a pathogenic according to the ACMG guidelines[5].
Sanger sequencing showed that there was a novel frameshift variation of c.1155dupG (p.Arg386Alafs*3) in the AHDC1 gene. Based on clinical presentation, laboratory tests and gene sequencing results, the clinical diagnosis was XGS.
To improve quality of life of the patient, she has been provided with rehabilitation training and behavioral guidance therapy since June 2021.
The patient had been receiving rehabilitation treatment for nearly 6 mo. The patient is now age 11 mo, her weight is 11.0 kg and length is 74.0 cm. She can sit alone, and her hands can engage in more elaborate movements, and have increased responses to external stimuli, and she is able to speak some simple vowels. The patient underwent the PDMS-2 test in November 2021. The FMQ was 13 points with a motor quotient of 79, and GMQ was 17 points with a motor quotient of 72. GMDS showed that motor ability was equivalent to that of a 5.3-mo-old child, with a DQ of 46 points. Performance was equivalent to that of a 6.5-mo-old child, with a DQ of 56 points.
The AHDC1 gene is located on chromosome 1p36.11, and likely functions in DNA binding. The AHDC1 gene is part of the CBX family of proteins associated with human chromodomain-containing Polycomb proteins. It encodes a protein of 1603 amino acids, consisting of five noncoding 5 exons, a noncoding 3 exon and a single 4.9-kb coding exon (exon 6) containing 2 AT hooks[1,3]. Previous studies have shown that AHDC1 interacts with nuclear proteins involved in epigenetic regulation during development, mainly at neural loci and neuronal protein transport. Mutation of the AHDC1 gene can lead to XGS.
XGS (OMIM: 615829) is an autosomal dominant genetic disease caused by mutation of the AHDC1 gene. Typical features include global developmental delay, intellectual disability, structural abnor
The present patient first attended hospital due to delayed motor milestones over 2 mo when she was age 6 mo. In order to clarify the cause of the disease, we used high-throughput WES and identified an unreported mutation of the AHDC1 gene.
According to previous studies, more than 90% of patients had motor and speech delay, hypotonia occurred in approximately 85% of patients, and less than 40% of patients had short stature. About 30% of patients had symptoms of autism. Some patients showed sleep apnea (34.33%), laryngomalacia (14.93%), and other manifestations. Approximately 35% of patients developed epileptic seizures[8,12,13].
In this case report, the child conformed to the typical clinical manifestations of XGS. The child developed significant motor delay and hypotonia, but speech ability was not delayed. She was able to respond positively to external stimuli. There was no sufficient evidence of sleep apnea and laryngomalacia. Although the child’s EEG showed the distribution of spike waves and sharp waves, she did not appear to have the related actions, and she did not have other related manifestations of epilepsy[14,15].
Our patient has not yet developed growth hormone deficiency or short stature. Height and weight were within the mean range of normal, in contrast to two previously reported Chinese children who exhibited partial growth hormone deficiency and attended hospital due to short stature[16].
In this report, we describe a Chinese patient with XGS, with a new mutation c.1155dupG (p.Arg386Alafs*3) in the AHDC1 gene identified by WES, and compared the results with those from two Chinese cases reported in the literature, to better understand the clinical phenotype and the association with the AHDC1 gene.
Previous studies have shown that global developmental delay occurs in the AHDC1-related phenotype of XGS. Our patient was found to have a novel frameshift variation of c.1155dupG (p.Arg386Alafs*3) in the AHDC1 gene, which was an unreported frameshift mutation. Typical features of XGS include global developmental delay, hypotonia, obstructive sleep apnea, seizures, delayed myelination, micrognathia, and other mild dysmorphic features. This report result showed clinicians could consider XGS in patients with similar clinical characteristics. Genetic testing can help physicians confirm the diagnosis and help with further genetic counseling.
We would like to thank the family members of the child for agreeing to participate in this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Pediatrics
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mesquita J, Portugal; Sahin Y, Turkey A-Editor: Bedane DA, Ethiopia S-Editor: Chang KL L-Editor: Filipodia P-Editor: Chang KL
| 1. | Xia F, Bainbridge MN, Tan TY, Wangler MF, Scheuerle AE, Zackai EH, Harr MH, Sutton VR, Nalam RL, Zhu W, Nash M, Ryan MM, Yaplito-Lee J, Hunter JV, Deardorff MA, Penney SJ, Beaudet AL, Plon SE, Boerwinkle EA, Lupski JR, Eng CM, Muzny DM, Yang Y, Gibbs RA. De novo truncating mutations in AHDC1 in individuals with syndromic expressive language delay, hypotonia, and sleep apnea. Am J Hum Genet. 2014;94:784-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 2. | Ellis C, Pai GS, Wine Lee L. Atypical aplasia cutis in association with Xia Gibbs syndrome. Pediatr Dermatol. 2021;38:533-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Yang H, Douglas G, Monaghan KG, Retterer K, Cho MT, Escobar LF, Tucker ME, Stoler J, Rodan LH, Stein D, Marks W, Enns GM, Platt J, Cox R, Wheeler PG, Crain C, Calhoun A, Tryon R, Richard G, Vitazka P, Chung WK. De novo truncating variants in the AHDC1 gene encoding the AT-hook DNA-binding motif-containing protein 1 are associated with intellectual disability and developmental delay. Cold Spring Harb Mol Case Stud. 2015;1:a000562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Griffiths A, Toovey R, Morgan PE, Spittle AJ. Psychometric properties of gross motor assessment tools for children: a systematic review. BMJ Open. 2018;8:e021734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 5. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19696] [Cited by in RCA: 22532] [Article Influence: 2253.2] [Reference Citation Analysis (0)] |
| 6. | Yang S, Li K, Zhu MM, Yuan XD, Jiao XL, Yang YY, Li J, Li L, Zhang HN, Du YH, Wei YX, Qin YW. Rare Mutations in AHDC1 in Patients with Obstructive Sleep Apnea. Biomed Res Int. 2019;2019:5907361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | Gumus E. Extending the phenotype of Xia-Gibbs syndrome in a two-year-old patient with craniosynostosis with a novel de novo AHDC1 missense mutation. Eur J Med Genet. 2020;63:103637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 8. | Della Vecchia S, Milone R, Cagiano R, Calderoni S, Santocchi E, Pasquariello R, Battini R, Muratori F. Focusing on Autism Spectrum Disorder in Xia-Gibbs Syndrome: Description of a Female with High Functioning Autism and Literature Review. Children (Basel). 2021;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Wang Q, Huang X, Liu Y, Peng Q, Zhang Y, Liu J, Yuan H. Microdeletion and microduplication of 1p36.11p35.3 involving AHDC1 contribute to neurodevelopmental disorder. Eur J Med Genet. 2020;63:103611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Cardoso-Dos-Santos AC, Oliveira Silva T, Silveira Faccini A, Woycinck Kowalski T, Bertoli-Avella A, Morales Saute JA, Schuler-Faccini L, de Oliveira Poswar F. Novel AHDC1 Gene Mutation in a Brazilian Individual: Implications of Xia-Gibbs Syndrome. Mol Syndromol. 2020;11:24-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Goyal C, Naqvi W, Sahu A. Xia-Gibbs Syndrome: A Rare Case Report of a Male Child and Insight into Physiotherapy Management. Cureus. 2020;12:e9622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Ritter AL, McDougall C, Skraban C, Medne L, Bedoukian EC, Asher SB, Balciuniene J, Campbell CD, Baker SW, Denenberg EH, Mazzola S, Fiordaliso SK, Krantz ID, Kaplan P, Ierardi-Curto L, Santani AB, Zackai EH, Izumi K. Variable Clinical Manifestations of Xia-Gibbs syndrome: Findings of Consecutively Identified Cases at a Single Children's Hospital. Am J Med Genet A. 2018;176:1890-1896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Jiang Y, Wangler MF, McGuire AL, Lupski JR, Posey JE, Khayat MM, Murdock DR, Sanchez-Pulido L, Ponting CP, Xia F, Hunter JV, Meng Q, Murugan M, Gibbs RA. The phenotypic spectrum of Xia-Gibbs syndrome. Am J Med Genet A. 2018;176:1315-1326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Park HY, Kim M, Jang W, Jang DH. Phenotype of a Patient With a 1p36.11-p35.3 Interstitial Deletion Encompassing the AHDC1. Ann Lab Med. 2017;37:563-565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Khayat MM, Li H, Chander V, Hu J, Hansen AW, Li S, Traynelis J, Shen H, Weissenberger G, Stossi F, Johnson HL, Lupski JR, Posey JE, Sabo A, Meng Q, Murdock DR, Wangler M, Gibbs RA. Phenotypic and protein localization heterogeneity associated with AHDC1 pathogenic protein-truncating alleles in Xia-Gibbs syndrome. Hum Mutat. 2021;42:577-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Cheng X, Tang F, Hu X, Li H, Li M, Fu Y, Yan L, Li Z, Gou P, Su N, Gong C, He W, Xiang R, Bu D, Shen Y. Two Chinese Xia-Gibbs syndrome patients with partial growth hormone deficiency. Mol Genet Genomic Med. 2019;7:e00596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |