Published online Jul 26, 2022. doi: 10.12998/wjcc.v10.i21.7224
Peer-review started: January 17, 2022
First decision: March 16, 2022
Revised: March 24, 2022
Accepted: June 4, 2022
Article in press: June 4, 2022
Published online: July 26, 2022
Processing time: 174 Days and 19.3 Hours
The therapeutic effects of a combination of Chinese medicines called Biyu decoction have been clinically verified, although its molecular targets in psoriasis remain unknown.
To explore the molecular mechanisms of Biyu decoction for psoriasis treatment.
In this network pharmacology and molecular docking study, the Traditional Chinese Medicine Systems Pharmacology database was searched for Biyu decoction active ingredients. GeneCards, Online Mendelian Inheritance in Man, PharmGkb, Therapeutic Target Database, and DrugBank databases were searched for psoriasis-related genes. The genes targeted by the decoction’s active ingredient and disease genes were intersected to obtain predictive targets of the drug during psoriasis treatment. Cytoscape 3.8.0 was used to construct a drug component/ target disease network. The The functional protein association networks database and Cytoscape were used to construct a protein-protein interaction network and streamline the core network. The Gene Ontology and Kyoto Encyclopedia of Genes and Genomes were used for pathway enrichment analysis. Molecular docking technology was used to verify the drug component/target disease network.
We screened 117 major active ingredients, including quercetin, kaempferol, naringenin, and acetyl-shikonin, and identified 213 gene targets, such as MAPK3, JUN, FOS, MYC, MAPK8, STAT3, and NFKBIA. Using a molecular docking analysis, the main active ingredients demonstrated good binding to the core targets. The Gene Ontology analysis showed that these ingredients were significantly associated with biological activities, such as transcription factor DNA binding, RNA polymerase II-specific DNA binding of transcription factors, and cytokine receptor binding; responses to lipopolysaccharides, molecules of bacterial origin, and oxidative stress; and were mainly distributed in membrane rafts, microdomains, and regions. The Kyoto Encyclopedia of Genes and Genomes analysis showed that decoction ingredients act on Th17 cell differentiation, tumor necrosis factor and mitogen-activated protein signaling pathways, the interleukin-17 signaling pathway, and the PI3K-Akt signaling pathway.
Biyu decoction may be effective against psoriasis through multi-component, multi-target, and multi-channel synergy.
Core Tip: Biyu decoction has significant effects on psoriasis; however, its molecular targets in psoriasis remain unknown. We conducted a network pharmacology and molecular docking study to determine whether Biyu decoction ingredients target molecules and signaling pathways related to psoriasis pathogenesis. The main active ingredients identified include quercetin, kaempferol, beta-sitosterol, naringenin, and acetyl-shikonin. Target genes included MAPK3, JUN, FOS, MYC, MAPK8, STAT3, and NFKBIA, which can regulate the inflammatory state mediated by psoriasis immune cells and mediate the expression of factors that adjust local skin inflammation. Our results confirm that Biyu decoction can treat psoriasis through multi-component, multi-target, and multi-channel synergy.
- Citation: Wang Z, Zhang HM, Guo YR, Li LL. Molecular mechanisms of Biyu decoction as treatment for psoriasis: A network pharmacology and molecular docking study. World J Clin Cases 2022; 10(21): 7224-7241
- URL: https://www.wjgnet.com/2307-8960/full/v10/i21/7224.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i21.7224
Psoriasis (PSO) is an immune-mediated chronic and recurrent inflammatory disease that affects the skin or joints. The global incidence of PSO is approximately 2%[1]. It also affects approximately 0.47% of the Chinese population[2]. Although gender does not have a clear effect on the incidence of PSO, genetic susceptibility and increasing age are known to increase its incidence[3].Because of the disfigurement and teratogenicity associated with this disease that seriously affect the quality of life, PSO confers a marked psychological burden[4]. The physiological burden is related to recurrent episodes of itching and several accompanying diseases[5]. Plaque PSO (PSO vulgaris) accounts for more than 90% of cases. The clinical manifestations are well-defined erythema and silvery scaly skin, which can occur anywhere on the body[6].
Conventional treatment for PSO includes topical glucocorticoids, vitamin D derivatives, calcineurin inhibitors, corticosteroids, phototherapy, and systemic therapy[7]. Practicability (the time required to apply treatment), convenience, and adverse reactions (e.g., skin irritation) limit the use of topical drugs, whereas drug interactions and accumulated organ toxicity limit systemic treatment. PSO is mediated by dendritic and T cells in a complex feedback loop involving antigen-presenting cells, neutrophils, keratinocytes, vascular endothelial cells, and the skin’s nervous system. Tumor necrosis factor (TNF)-α, interferon (IFN)-γ, interleukin (IL)-1, and other cytokines mediate innate and adaptive immune responses, and resident immune cell interaction disorder in the skin is the main PSO pathogenesis[8]. Therefore, targeted therapy blocking the PSO-related immune pathway is effective; however, the cost of biological agents is high and thus difficult to popularize.
In the current medical environment in China, traditional Chinese medicine (TCM) has a high penetration rate among psoriasis patients due to various advantages, such as less adverse reactions, long-term use, discontinuation at any time, and low long-term recurrence rate[9]. According to Chinese medicine, "blood-heat" runs through psoriasis from beginning to end[10]. "Blood heat" is mainly characterized by bleeding and fiery heat. "Heat" stagnates blood collaterals, and erythema is seen on the skin. "Heat" forces blood to rush out of the veins, and causes spot-like bleeding. These correspond to the bright red patches, bleeding spots of a psoriatic rash. Biyu decoction (BYT) is a combination of Chinese medicines with significant effects on PSO; its selection is based on Xiaoyinjiedu granules[10] by Professor Jin Qifeng (National TCM Doctor of Dongzhimen Hospital, Beijing University of Chinese Medicine) and our preliminary research results. BYT comprises Zicao[11,12] (Lithospermum Erythrorhizon), Diyu[13] (Radix Sanguisorbae), Cebaiye[14] (Platycladi Cacumen), and Gancao[15] (licorice), which are Chinese herbal medicines with cooling blood effect, and in vitro and in vivo studies have revealed their therapeutic effects on immunity and inflammation. The therapeutic effects of BYT on PSO have been verified clinically. However, the BYT molecular targets of PSO remain unknown. To improve PSO treatment, we investigated whether BYT ingredients target molecules and signaling pathways related to PSO pathogenesis.
Drug data were obtained from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database (http: //tcmspw.com/tcmsp.php). Gene data were obtained from the Human Gene Database (GeneCards; https://www.genecards.org), Online Mendelian Inheritance in Man (OMIM; https://omim.org), Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB; https://www.pharmgkb.org), Therapeutic Target Database (TTD; http://db.idrblab.net/ttd), and Drugbank database (Drugbank; https://www.drugbank.ca). The functional protein association networks database (STRING; https://string-db.org) was used to construct the protein-protein interaction (PPI) network. The molecular structure data of the receptors and ligands required for molecular docking were obtained from PubChem (PubChem; https://pubchem.ncbi.nlm.nih.gov) and Protein Data Bank (PDB; http://www.rcsb.org). The above data were obtained in May 2021. In addition, we conducted a relevant search by Reference Citation Analysis (https://www.referencecitationanalysis.com).
We searched the TCMSP database for active components of BYT, namely, the chemical components of Zicao, Diyu, Cebaiye, and Gancao. After performing screen filtering by oral bioavailability (OB; > 30%) and drug-like properties (DL; > 0.18) and removing duplicate components, we obtained effective drug components. The TCMSP database was searched to obtain the molecular targets of each active ingredient and identify molecular targets corresponding to all the active ingredients of BYT.
Genes related to PSO were identified by searching the following five databases: GeneCards, OMIM, PharmGKB, TTD, and Drugbank. After removing the duplicate and false-positive targets, genes related to PSO were identified. The intersection of the retrieved molecular targets of the BYT active ingredients and these PSO-related genes were identified to retrieve PSO-related target genes of BYT.
We used Cytoscape (version 3.8.0) software to establish the drug-disease network based on the BYT components targeting PSO-related genes obtained from TCMSP and the PSO-related drug targets of BYT obtained from the gene database. The network comprised drug nodes, target nodes, and related events. The larger the number of associated events, the larger the corresponding target node. The attributes of different traditional Chinese medicine sources in the drug nodes were represented by different colors; however, the drug nodes could show multiple colors because they were derived from multiple drugs.
A PPI network was established based on the selected BYT target genes in PSO. The lines between gene nodes defined the interaction between proteins. After filtering out free gene nodes, a large PPI network was obtained. Using the CytoNCA plug-in of the Cytoscape software, a topology analysis was performed to filter out the central network. We analyzed the following six indicators: Betweenness centrality (BC), closeness centrality (CC), degree centrality (DC), eigenvector centrality (EC), local average connectivity (LAC), and network centrality (NC); these provide a standard for the in-depth analysis of the attributes of each node. A higher quantitative value for each index indicated that the node was more important in the network and allowed identification of the core network.
The PDB database was searched for core genes to perform molecular docking experiments. The structures of the corresponding proteins were downloaded. PyMOL (version 2.4.0) software was used to remove water molecules and small ligand molecules from these receptor macromolecular structures. AutoDockTools (version 1.5.6) software was used to add polar hydrogen ions and set the grid box to determine the search range of the active pockets of the proteins, which were unknown. To include as many receptor structures as possible, the size of the active pocket had to be increased appropriately to improve the docking results. According to the core genes identified, the drug components that interacted with the core genes were sought in the constructed drug-disease network. The two-dimensional structures of these small ligand molecules were downloaded from the PubChem database, and the MM2 calculation tool in ChemBio3D (version 14.0) software was used to optimize the three-dimensional structure of the small ligand molecule with the smallest free energy. Vina (version 1.1.2) software was used to perform the docking work, and the optimal model of molecular docking was calculated.
We used R (version 4.0.3) software to perform a Gene Ontology (GO) biological functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathway enrichment analysis of the intersection genes. These were filtered using P < 0.05 (with the q-value for correction if they were not in compliance). GO term enrichment was performed for biological processes (BP), CC, and molecular function (MF), and the GO term most likely to be related to the target gene was identified. We explored which functions in PSO were regulated by BYT and determined the connections between functional units and their potential significance in biological system networks. We also selected PSO-related pathways during the KEGG enrichment analysis and drew pathway diagrams.
According to the TCMSP database, BYT contained a total of 458 ingredients (Zicao, 51; Diyu, 41; Cebaiye, 86; and Gancao, 280). After screening, based on OB and DL values, 124 active ingredients were obtained (Zicao, 12; Diyu, 13; Cebaiye, 7; and Gancao, 92). After deleting the compounds with no action targets and duplicated components, 117 active ingredients were obtained (Table 1). Based on these ingredients, 2410 pairs of ingredient-predicted target combinations were identified (Zicao, 83 pairs; Diyu, 269 pairs; Cebaiye, 289 pairs; and Gancao, 1769 pairs). After removing duplicate targets, 213 targets of BYT Chinese medicine were obtained.
| Drug | Mol ID | Mol name | OB | DL |
| Cebaiye | MOL002005 | Hinokinin | 56.5 | 0.64 |
| Cebaiye | MOL002034 | (5aR, 8aS, 9R)-9-(3, 4, 5-trimethoxyphenyl)-5a, 6, 8a, 9-tetrahydro-5H-isobenzofurano[5, 6-f][1, 3]benzodioxol-8-one | 52.7 | 0.83 |
| Cebaiye | MOL000358 | Beta-sitosterol (Diyu) | 36.91 | 0.75 |
| Cebaiye | MOL002032 | Di-n-octylphthalate | 40.59 | 0.4 |
| Cebaiye | MOL002039 | Isopimaric acid | 36.2 | 0.28 |
| Cebaiye | MOL000422 | Kaempferol (Diyu, Gancao) | 41.88 | 0.24 |
| Cebaiye | MOL000098 | Quercetin (Diyu, Gancao) | 46.43 | 0.28 |
| Diyu | MOL005858 | 3, 7, 8-tri-O-methylellagic acid | 37.54 | 0.57 |
| Diyu | MOL005860 | 3-O-galloylprocyanidin B-3 | 30.06 | 0.33 |
| Diyu | MOL005399 | Alexandrin_qt | 36.91 | 0.75 |
| Diyu | MOL005869 | Daucostero_qt | 36.91 | 0.75 |
| Diyu | MOL005883 | Gambiriin B-3 | 34.99 | 0.75 |
| Diyu | MOL000211 | Mairin (Gancao) | 55.38 | 0.78 |
| Diyu | MOL005862 | Methyl 4, 6-di-O-galloyl-beta-D-glucopyranoside | 48.07 | 0.68 |
| Diyu | MOL005853 | Methyl-2, 3, 6-tri-O-galloyl-β-D-glucopyranoside | 44.95 | 0.67 |
| Diyu | MOL005864 | Methyl-6-O-galloyl-β-D-glucopyranoside | 44.85 | 0.29 |
| Diyu | MOL005880 | Sauvissimoside R1 | 37.39 | 0.31 |
| Gancao | MOL004924 | (-)-medicocarpin | 40.99 | 0.95 |
| Gancao | MOL004941 | (2R)-7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one | 71.12 | 0.18 |
| Gancao | MOL004805 | (2S)-2-[4-hydroxy-3-(3-methylbut-2-enyl)phenyl]-8, 8-dimethyl-2, 3-dihydropyrano[2, 3-f]chromen-4-one | 31.79 | 0.72 |
| Gancao | MOL004824 | (2S)-6-(2, 4-dihydroxyphenyl)-2-(2-hydroxypropan-2-yl)-4-methoxy-2, 3-dihydrofuro[3, 2-g]chromen-7-one | 60.25 | 0.63 |
| Gancao | MOL004945 | (2S)-7-hydroxy-2-(4-hydroxyphenyl)-8-(3-methylbut-2-enyl)chroman-4-one | 36.57 | 0.32 |
| Gancao | MOL004815 | (E)-1-(2, 4-dihydroxyphenyl)-3-(2, 2-dimethylchromen-6-yl)prop-2-en-1-one | 39.62 | 0.35 |
| Gancao | MOL004898 | (E)-3-[3, 4-dihydroxy-5-(3-methylbut-2-enyl)phenyl]-1-(2, 4-dihydroxyphenyl)prop-2-en-1-one | 46.27 | 0.31 |
| Gancao | MOL004914 | 1, 3-dihydroxy-8, 9-dimethoxy-6-benzofurano[3, 2-c]chromenone | 62.9 | 0.53 |
| Gancao | MOL004913 | 1, 3-dihydroxy-9-methoxy-6-benzofurano[3, 2-c]chromenone | 48.14 | 0.43 |
| Gancao | MOL005013 | 18α-hydroxyglycyrrhetic acid | 41.16 | 0.71 |
| Gancao | MOL004959 | 1-methoxyphaseollidin | 69.98 | 0.64 |
| Gancao | MOL004866 | 2-(3, 4-dihydroxyphenyl)-5, 7-dihydroxy-6-(3-methylbut-2-enyl)chromone | 44.15 | 0.41 |
| Gancao | MOL004978 | 2-[(3R)-8, 8-dimethyl-3, 4-dihydro-2H-pyrano[6, 5-f]chromen-3-yl]-5-methoxyphenol | 36.21 | 0.52 |
| Gancao | MOL004849 | 3-(2, 4-dihydroxyphenyl)-8-(1, 1-dimethylprop-2-enyl)-7-hydroxy-5-methoxy-coumarin | 59.62 | 0.43 |
| Gancao | MOL004863 | 3-(3, 4-dihydroxyphenyl)-5, 7-dihydroxy-8-(3-methylbut-2-enyl)chromone | 66.37 | 0.41 |
| Gancao | MOL004905 | 3, 22-dihydroxy-11-oxo-delta(12)-oleanene-27-alpha-methoxycarbonyl-29-oic acid | 34.32 | 0.55 |
| Gancao | MOL004966 | 3'-hydroxy-4'-O-methylglabridin | 43.71 | 0.57 |
| Gancao | MOL004974 | 3'-methoxyglabridin | 46.16 | 0.57 |
| Gancao | MOL004864 | 5, 7-dihydroxy-3-(4-methoxyphenyl)-8-(3-methylbut-2-enyl)chromone | 30.49 | 0.41 |
| Gancao | MOL004989 | 6-prenylated eriodictyol | 39.22 | 0.41 |
| Gancao | MOL004990 | 7, 2', 4'-trihydroxy-5-methoxy-3-arylcoumarin | 83.71 | 0.27 |
| Gancao | MOL004991 | 7-acetoxy-2-methylisoflavone | 38.92 | 0.26 |
| Gancao | MOL003896 | 7-methoxy-2-methyl isoflavone | 42.56 | 0.2 |
| Gancao | MOL004838 | 8-(6-hydroxy-2-benzofuranyl)-2, 2-dimethyl-5-chromenol | 58.44 | 0.38 |
| Gancao | MOL004993 | 8-prenylated eriodictyol | 53.79 | 0.4 |
| Gancao | MOL000417 | Calycosin | 47.75 | 0.24 |
| Gancao | MOL005020 | Dehydroglyasperins C | 53.82 | 0.37 |
| Gancao | MOL001792 | Dihydroxyflavanone | 32.76 | 0.18 |
| Gancao | MOL004806 | Euchrenone | 30.29 | 0.57 |
| Gancao | MOL004915 | Eurycarpin A | 43.28 | 0.37 |
| Gancao | MOL000392 | Formononetin | 69.67 | 0.21 |
| Gancao | MOL004996 | Gadelaidic acid | 30.7 | 0.2 |
| Gancao | MOL004856 | Gancaonin A | 51.08 | 0.4 |
| Gancao | MOL004857 | Gancaonin B | 48.79 | 0.45 |
| Gancao | MOL005000 | Gancaonin G | 60.44 | 0.39 |
| Gancao | MOL005001 | Gancaonin H | 50.1 | 0.78 |
| Gancao | MOL004910 | Glabranin | 52.9 | 0.31 |
| Gancao | MOL004911 | Glabrene | 46.27 | 0.44 |
| Gancao | MOL004908 | Glabridin | 53.25 | 0.47 |
| Gancao | MOL004912 | Glabrone | 52.51 | 0.5 |
| Gancao | MOL004828 | Glepidotin A | 44.72 | 0.35 |
| Gancao | MOL004829 | Glepidotin B | 64.46 | 0.34 |
| Gancao | MOL004808 | Glyasperin B | 65.22 | 0.44 |
| Gancao | MOL004811 | Glyasperin C | 45.56 | 0.4 |
| Gancao | MOL004810 | Glyasperin F | 75.84 | 0.54 |
| Gancao | MOL005007 | Glyasperins M | 72.67 | 0.59 |
| Gancao | MOL004879 | Glycyrin | 52.61 | 0.47 |
| Gancao | MOL002311 | Glycyrol | 90.78 | 0.67 |
| Gancao | MOL004917 | Glycyroside | 37.25 | 0.79 |
| Gancao | MOL005008 | Glycyrrhiza flavonol A | 41.28 | 0.6 |
| Gancao | MOL004835 | Glypallichalcone | 61.6 | 0.19 |
| Gancao | MOL004907 | Glyzaglabrin | 61.07 | 0.35 |
| Gancao | MOL004957 | 3-(4-hydroxyphenyl)-7-methoxychromen-4-one | 38.37 | 0.21 |
| Gancao | MOL004985 | Icos-5-enoic acid | 30.7 | 0.2 |
| Gancao | MOL001484 | Inermine | 75.18 | 0.54 |
| Gancao | MOL004980 | Inflacoumarin A | 39.71 | 0.33 |
| Gancao | MOL004948 | Isoglycyrol | 44.7 | 0.84 |
| Gancao | MOL004949 | Isolicoflavonol | 45.17 | 0.42 |
| Gancao | MOL000354 | Isorhamnetin | 49.6 | 0.31 |
| Gancao | MOL004814 | Isotrifoliol | 31.94 | 0.42 |
| Gancao | MOL000239 | Jaranol | 50.83 | 0.29 |
| Gancao | MOL004988 | Kanzonol F | 32.47 | 0.89 |
| Gancao | MOL004820 | Kanzonols W | 50.48 | 0.52 |
| Gancao | MOL005003 | Licoagrocarpin | 58.81 | 0.58 |
| Gancao | MOL005012 | Licoagroisoflavone | 57.28 | 0.49 |
| Gancao | MOL000497 | Licochalcone a | 40.79 | 0.29 |
| Gancao | MOL004841 | Licochalcone B | 76.76 | 0.19 |
| Gancao | MOL004848 | Licochalcone G | 49.25 | 0.32 |
| Gancao | MOL004882 | Licocoumarone | 33.21 | 0.36 |
| Gancao | MOL004885 | Licoisoflavanone | 52.47 | 0.54 |
| Gancao | MOL004883 | Licoisoflavone | 41.61 | 0.42 |
| Gancao | MOL004884 | Licoisoflavone B | 38.93 | 0.55 |
| Gancao | MOL004904 | Licopyranocoumarin | 80.36 | 0.65 |
| Gancao | MOL004860 | Licorice glycoside E | 32.89 | 0.27 |
| Gancao | MOL004855 | Licoricone | 63.58 | 0.47 |
| Gancao | MOL004903 | Liquiritin | 65.69 | 0.74 |
| Gancao | MOL003656 | Lupiwighteone | 51.64 | 0.37 |
| Gancao | MOL002565 | Medicarpin | 49.22 | 0.34 |
| Gancao | MOL004328 | Naringenin | 59.29 | 0.21 |
| Gancao | MOL005016 | Odoratin | 49.95 | 0.3 |
| Gancao | MOL005017 | Phaseol | 78.77 | 0.58 |
| Gancao | MOL004833 | Phaseolinisoflavan | 32.01 | 0.45 |
| Gancao | MOL004961 | Quercetin der. | 46.45 | 0.33 |
| Gancao | MOL004827 | Semilicoisoflavone B | 48.78 | 0.55 |
| Gancao | MOL004891 | Shinpterocarpin | 80.3 | 0.73 |
| Gancao | MOL004935 | Sigmoidin-B | 34.88 | 0.41 |
| Gancao | MOL000359 | Sitosterol (Zicao) | 36.91 | 0.75 |
| Gancao | MOL000500 | Vestitol | 74.66 | 0.21 |
| Gancao | MOL005018 | Xambioona | 54.85 | 0.87 |
| Zicao | MOL002372 | (6Z, 10E, 14E, 18E)-2, 6, 10, 15, 19, 23-hexamethyltetracosa-2, 6, 10, 14, 18, 22-hexaene | 33.55 | 0.42 |
| Zicao | MOL007715 | [(1R)-1-(5, 8-dihydroxy-1, 4-dioxo-2-naphthyl)-4-methyl-pent-3-enyl] propanoate | 54.64 | 0.29 |
| Zicao | MOL007714 | 1-methoxyacetylshikonin | 73.09 | 0.29 |
| Zicao | MOL007734 | 5-[(E)-5-(3-furyl)-2-methyl-pent-2-enyl]-2, 3-dimethoxy-p-benzoquinone | 61.8 | 0.24 |
| Zicao | MOL007716 | Acetylshikonin | 62.39 | 0.27 |
| Zicao | MOL007735 | Des-O-methyllasiodiplodin | 30.12 | 0.2 |
| Zicao | MOL002883 | Ethyl oleate (NF) | 32.4 | 0.19 |
| Zicao | MOL007722 | Isoarnebin 4 | 64.79 | 0.2 |
| Zicao | MOL007728 | Lithospermidin A | 75.08 | 0.38 |
| Zicao | MOL007736 | Lithospermidin B | 60.48 | 0.39 |
| Zicao | MOL001494 | Mandenol | 42 | 0.19 |
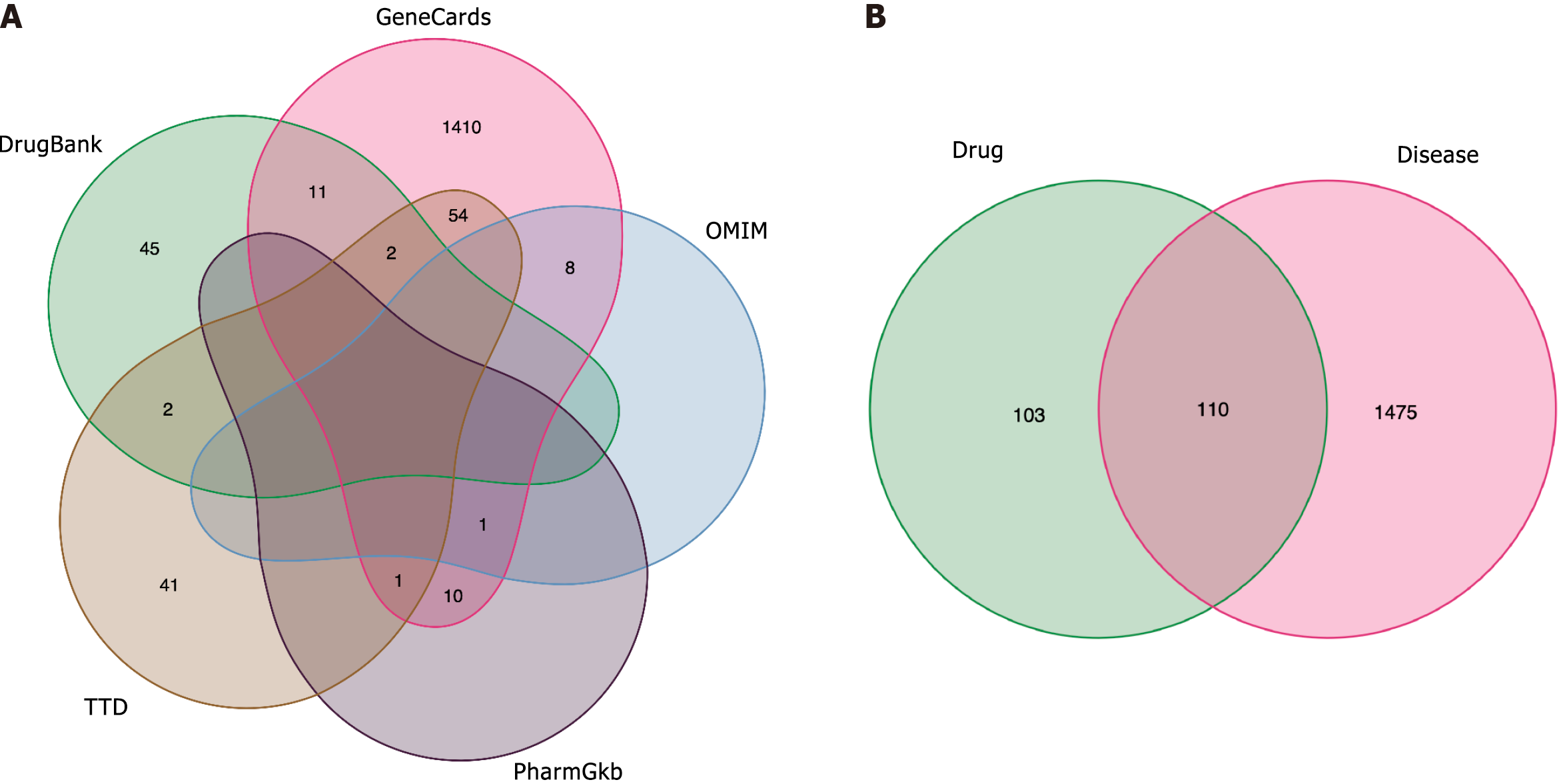
We used “Psoriasis” as a keyword to search GeneCards, OMIM, PharmGkb, TTD, and DrugBank databases. With a relevance score > 1.0, genes from the GeneCards database were filtered. Overall, 1700 PSO-related genes were retrieved (GeneCards, 1497; OMIM, 13; PharmGkb, 12; TTD, 100; and DrugBank, 78). After deduplication of the retrieved genes, a Venn diagram of the identified genes was drawn (Figure 1A), yielding 1585 PSO gene targets.
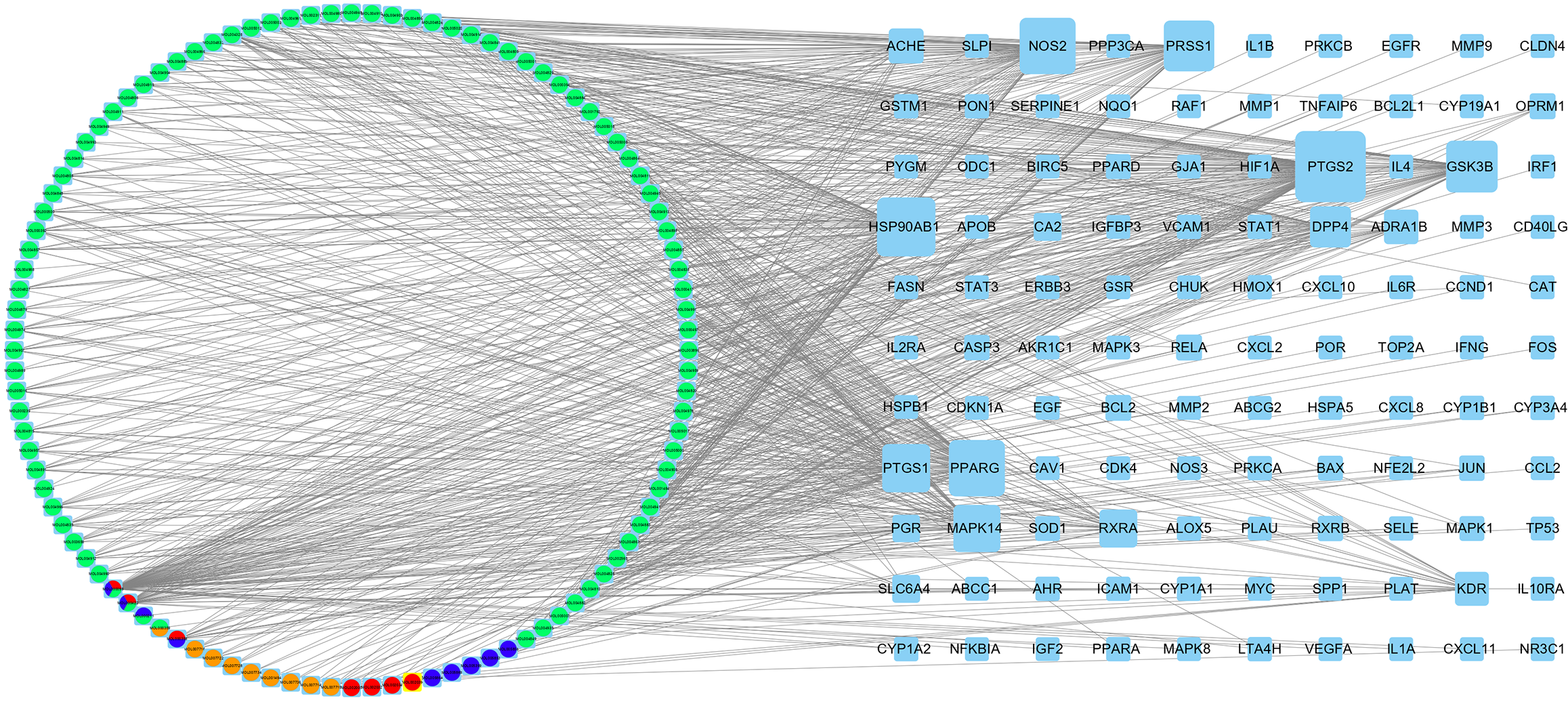
The intersection of 1585 PSO-related genes and the 213 predicted BYT targets yielded a total of 110 BYT-PSO genes, and a Venn diagram was drawn (Figure 1B). MAPK8, NFKBIA, STAT3, TNFAIP6, IFNG, ICAM1, IL1A, IL2RA, IL10RA, VCAM1, EGFR, VEGFA, MMP2, and other genes associated with PSO were included in this intersection. The target gene mapping rate for BYT in PSO was 51.6%, indicating some specificity (Figure 2).
Cytoscape software was used to construct a BYT active ingredient/PSO gene target network (Figure 2). The network comprised 110 potential target genes and 104 active ingredients. The larger the metric value of the target, the more the target and drug components constituted the events, implying a greater role in the network. The same ingredient could be derived from multiple drugs simultaneously. Each potential target gene interacted with multiple active ingredients, which indicated that BYT has effects on PSO via multiple pathways, multiple components, and multiple targets.
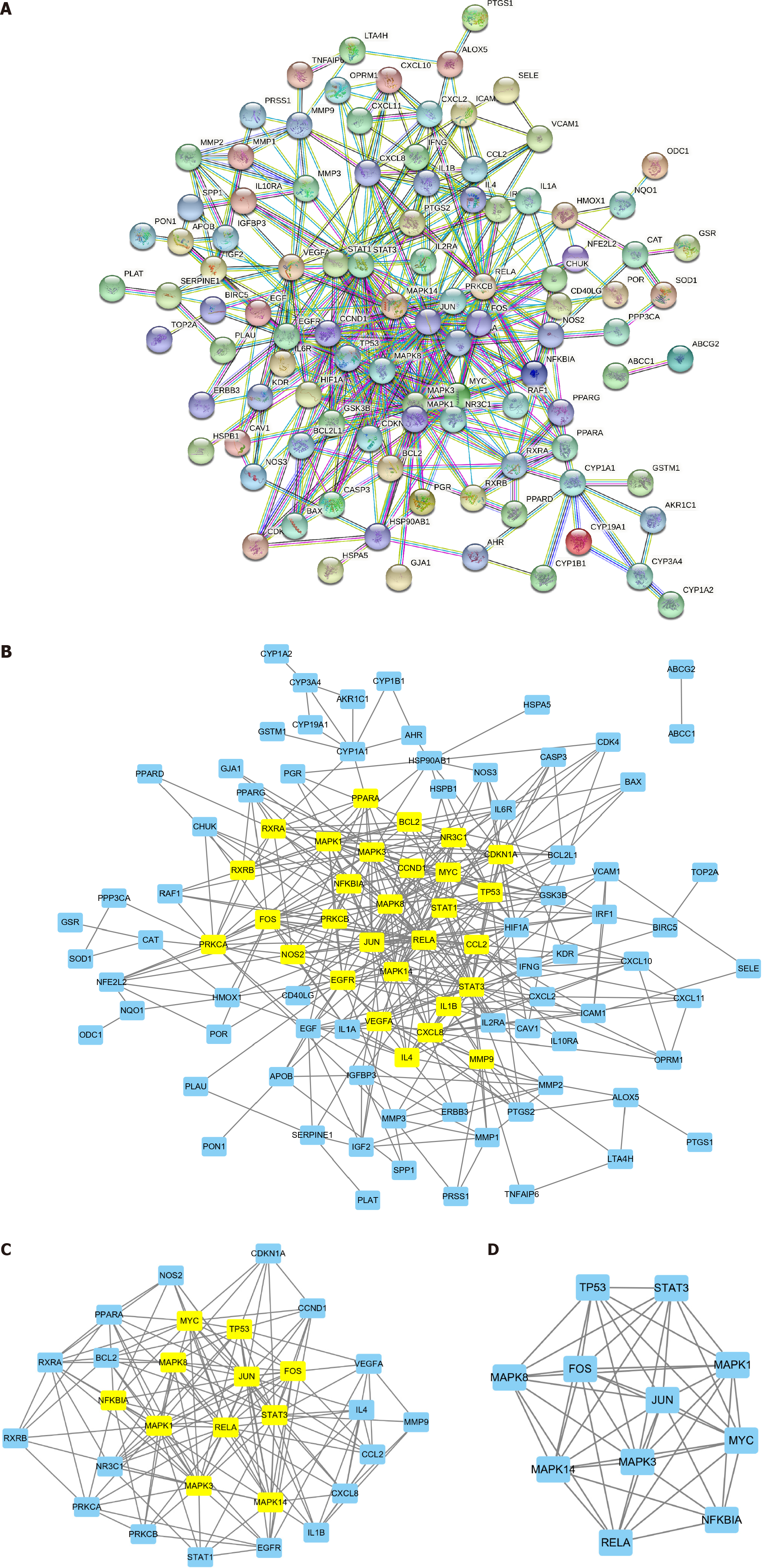
We imported the 110 BYT-PSO intersection genes into the STRING database (species: Homo sapiens; filter: Highest confidence, 0.900) and removed free nodes to obtain the BYT-PSO PPI network (Figure 3A). The network comprised 101 nodes and 399 edges, with lines of different colors representing different ways of proving the protein interaction relationship between nodes.
We imported the obtained PPI network diagram into Cytoscape software (Figure 3B) and used the CytoNCA plug-in to score nodes for BC, CC, DC, EC, LAC, and NC. The nodes for which all values exceeded the median were retained in the PPI network diagram (Figure 3C). The filter conditions were as follows: BC > 55.53, CC > 0.21, DC > 6, EC > 0.05, LAC > 2.55, and NC > 3.5. This secondary PPI network contained 29 nodes and 157 edges. The calculation was repeated with the following filter conditions: BC > 7.18, CC > 0.58, DC > 9, EC > 0.15, LAC > 5.45, and NC > 7.18. The obtained core PPI network comprised 11 nodes and 47 edges (Figure 3D), with an average degree of 8.55. Based on the degree value, the order of the targets that have a major role in BYT treatment of PSO was as follows: MAPK3, JUN, MAPK1, FOS, MAPK14, MYC, MAPK8, TP53, RELA, STAT3, and NFKBIA (Table 2).
| Name | Betweenness | Closeness | Degree | Eigenvector | LAC | Network |
| MAPK8 | 0.793650794 | 0.833333333 | 8 | 0.287379682 | 6.25 | 7.142857143 |
| TP53 | 0.472222222 | 0.833333333 | 8 | 0.290343016 | 6.5 | 7.428571429 |
| RELA | 1.205555556 | 0.833333333 | 8 | 0.280704081 | 6 | 7.142857143 |
| NFKBIA | 0.222222222 | 0.714285714 | 6 | 0.217291564 | 4.666666667 | 5.6 |
| STAT3 | 0.472222222 | 0.833333333 | 8 | 0.290343136 | 6.5 | 7.428571429 |
| MAPK3 | 2.576984127 | 1 | 10 | 0.341228724 | 7.4 | 10 |
| JUN | 2.576984127 | 1 | 10 | 0.341228724 | 7.4 | 10 |
| MAPK1 | 2.354761905 | 0.909090909 | 9 | 0.30924511 | 6.444444444 | 7.978571429 |
| FOS | 1.043650794 | 0.909090909 | 9 | 0.31924358 | 7.111111111 | 8.482142857 |
| MAPK14 | 2.354761905 | 0.909090909 | 9 | 0.30924511 | 6.444444444 | 7.978571429 |
| MYC | 1.926984127 | 0.909090909 | 9 | 0.311044633 | 6.666666667 | 8.196428571 |
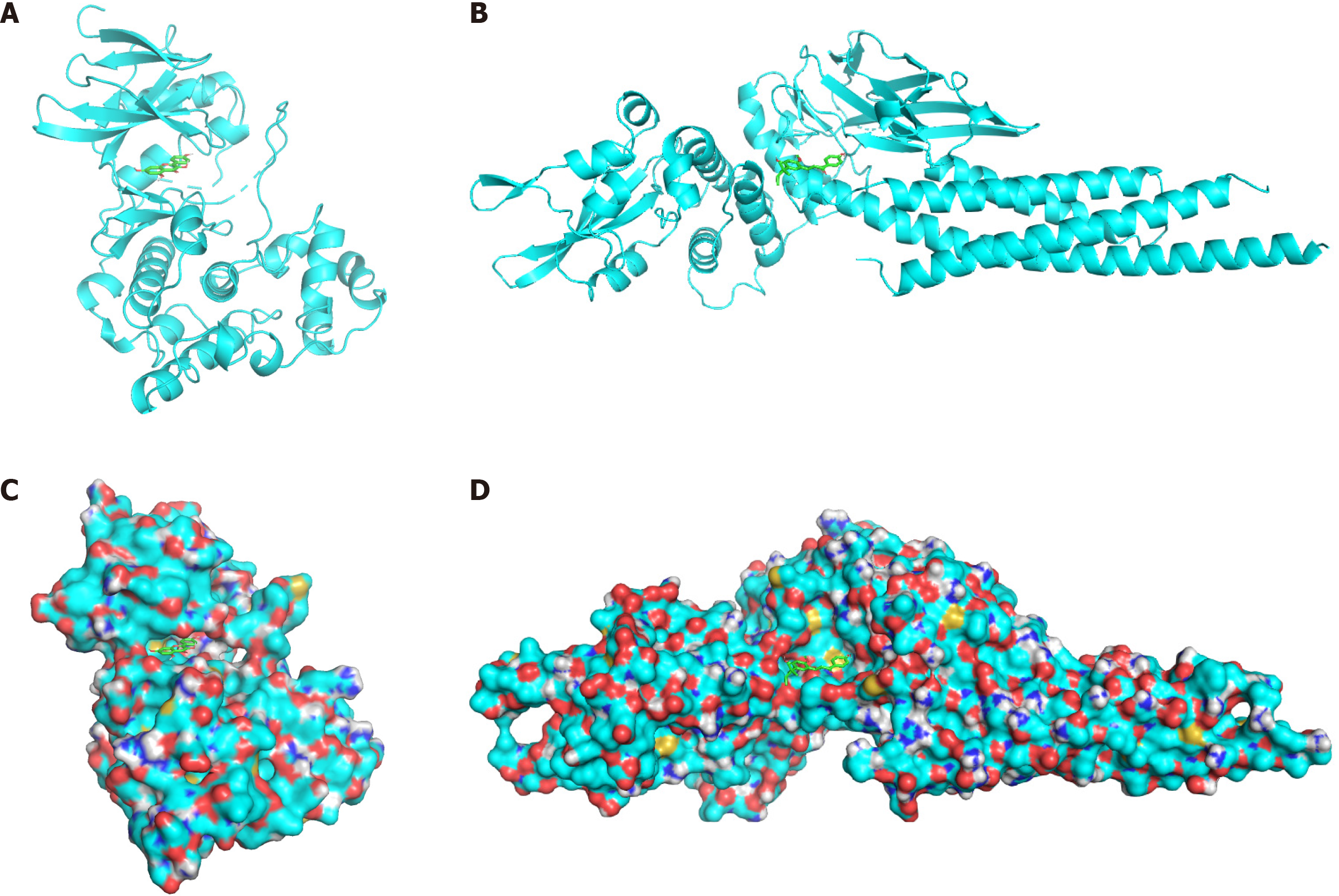
We searched the literature for PSO gene research and selected two core targets from the PPI core network: MAPK8 and STAT3. MAPK8 and STAT3 proteins (3ELJ and 6NJS, respectively) were retrieved from the PDB database. We pretreated the receptor protein macromolecules by removing water molecules and small ligand molecules and adding polar hydrogen; then, we set the active pocket search range. Based on the BYT active ingredient/PSO gene target network, the active ingredients kaempferol (for MAPK8) and licochalcone A (for STAT3) were selected.
Vina software was used to verify molecular docking between the receptor macromolecule and Chinese medicine small molecule component. The 20 main interaction methods were identified by calculating binding energies from small to large (Table 3). The model with the smallest binding energy was selected, and PyMOL software was used for visualization (Figure 4). The smaller the binding energy, the stronger the bond between the ligand and receptor. When affinity is less than -4.25 kcal/mol, the ligand shows certain binding to the receptor. Affinity less than -5.0 kcal/mol indicates good binding activity, and affinity less than -7.0 kcal/mol indicates strong binding activity. The energy of the binding of BYT core targets in PSO treatment with active ingredients was between -5.0 kcal/mol and -8.7 kcal/mol, indicating that the main active ingredients of BYT demonstrate good binding activity with the core gene targets of the disease with relatively reliable prediction.
| Number | MAPK8/kaempferol Affinity (kcal/mol) | STAT3/licochalcone A Affinity (kcal/mol) |
| 1 | -8.7 | -6.6 |
| 2 | -8.7 | -6.2 |
| 3 | -8.6 | -6 |
| 4 | -8.6 | -5.8 |
| 5 | -8.5 | -5.7 |
| 6 | -8.4 | -5.7 |
| 7 | -8.2 | -5.5 |
| 8 | -8 | -5.5 |
| 9 | -8 | -5.5 |
| 10 | -7.9 | -5.4 |
| 11 | -7.8 | -5.3 |
| 12 | -7.7 | -5.3 |
| 13 | -7.7 | -5.3 |
| 14 | -7.6 | -5.3 |
| 15 | -7.5 | -5.3 |
| 16 | -7.4 | -5.2 |
| 17 | -7.4 | -5.2 |
| 18 | -7.3 | -5.1 |
| 19 | -7.2 | -5.1 |
| 20 | -7.1 | -5 |
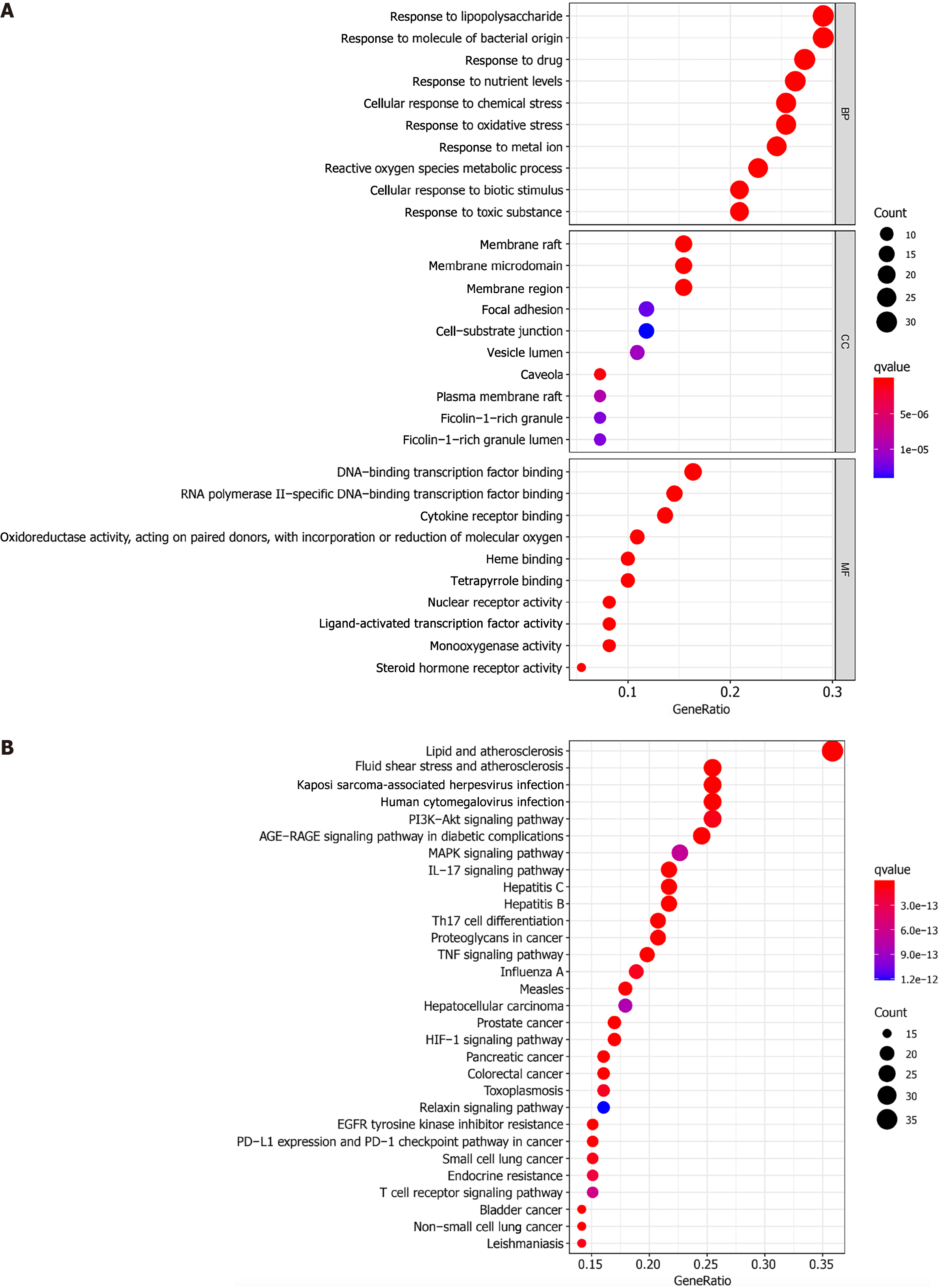
GO enrichment of the BYT-PSO intersection genes indicated 2083 types of BP, 42 types of CC, and 150 types of MF. We selected the first 30 with significant enrichment to draw images (Figure 5A). In terms of BP, these target genes were mainly involved in biological reactions, such as the response to lipopolysaccharides, response to molecules of bacterial origin, and response to oxidative stress. In terms of CC, target genes were mainly distributed in cell parts, such as membrane rafts, membrane microdomains, and membrane regions. In terms of MF, the target genes were significantly associated with DNA binding of transcription factors, RNA polymerase II-specific DNA binding of transcription factors, and cytokine receptor binding.
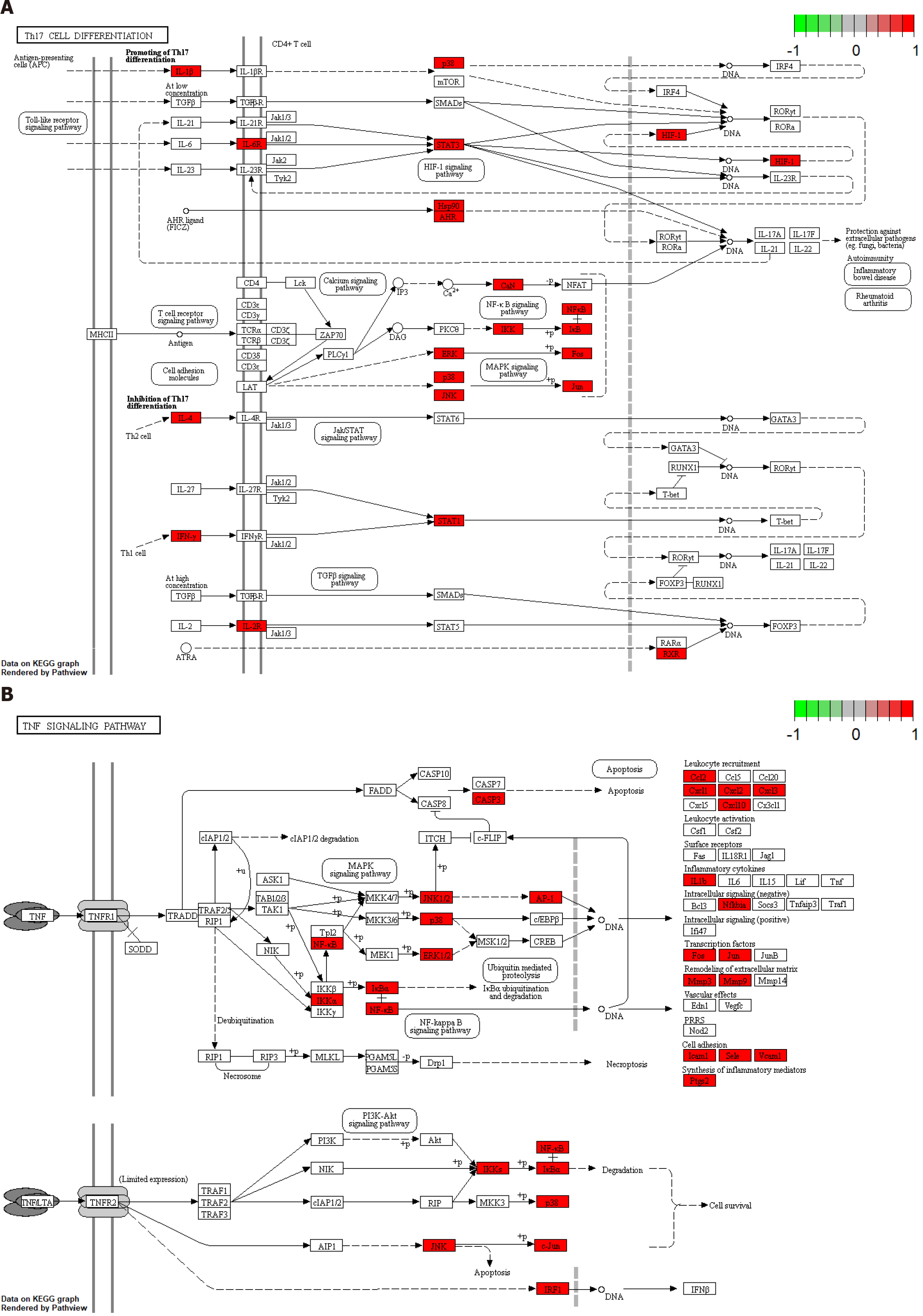
Overall, 170 signal pathways were obtained by KEGG pathway enrichment analysis. The first 30 signal pathways were screened out by statistical significance (P < 0.05; if not, then the q-value correction was used) for visual analysis (Figure 5B). The target genes had a role in multiple classic PSO-related pathways, such as the IL-17 signaling pathway, Th17 cell differentiation, TNF signaling pathway, T-cell receptor signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway, and PI3K-Akt signaling pathway. Therefore, BYT acts synergistically on PSO through multiple pathways. The more relevant Th17 cell differentiation and TNF signaling pathways were selected to draw a pathway diagram (Figure 6).
In this study, we explored the mechanism of BYT in PSO treatment using a network pharmacology and molecular docking study. We screened 117 major active ingredients, including quercetin, kaempferol, naringenin, and acetyl-shikonin, and identified 213 gene targets, including MAPK3, JUN, FOS, MYC, MAPK8, STAT3, and NFKBIA. Molecular docking analysis revealed that the main active ingredients demonstrated good binding to the core targets. The GO analysis showed that these ingredients were significantly associated with biological activities, such as DNA binding of transcription factors and cytokine receptor binding, whereas the KEGG analysis showed that the decoction ingredients act on Th17 cell differentiation and the TNF, MAPK, and other important PSO signaling pathways.
PSO pathogenesis has not been fully elucidated[16]. Excessive activation of the adaptive immune system is considered to be at the core of PSO[17]. Early during PSO, various cell types secrete cytokines to activate myeloid dendritic cells. When activated, these cells secrete IL-12 and IL-23[18] . IL-23-mediated activation of the Th17 pathway is considered the main pathway that guides the transcription of key inflammatory mediators via the Tyk2-Jak2 and STAT3 pathways[19]. These cytokines cause abnormal proliferation of downstream keratinocytes, increase the expression of vascular endothelial growth factor (VEGF) and vascular cell adhesion molecules, and cause immune cells to continue to infiltrate the affected skin[20].
Because of limited therapeutic effects or unavoidable side effects, the long-term use of modern medical traditional therapies for PSO remains a challenge. Complementary and alternative therapies, such as traditional Chinese medicine (TCM), are widely used in East Asia. TCM formulations are complex drug systems. The complex interactions between their multiple components and multiple targets can lead to the regulation of various pathways and biological processes, thus providing treatment for numerous complex diseases. TCM treatment of PSO has a therapeutic effect that is not inferior to the therapeutic effects of modern conventional medical therapies[21]; furthermore, it could possibly strengthen the therapeutic effects of modern conventional medical therapies[22]. Because of the unique theory regarding their clinical use and successful application as PSO treatment agents, we believe that the development of new small-molecule drugs based on TCM will be a promising strategy to provide safe, effective, and less costly treatment for patients with PSO. Therefore, we used a network pharmacology research method to explore the mechanism by which BYT can treat PSO from the perspective of active ingredients, target genes, and signaling pathways.
Most of the BYT ingredients have good oral bioavailability and drug-like properties. The 124 active ingredients screened out during this study have affinity for 213 cell targets, thus providing a pharmacodynamic basis for the diversity and effectiveness of TCM formulations. In the BYTPSO network, there were 104 active ingredients of BYT. Among them, quercetin, kaempferol, beta-sitosterol, naringenin, acetyl-shikonin, and other compounds have affinity for multiple gene targets. Quercetin is a polyflavonoid compound in the human diet that has biological activity. It regulates many intracellular and extracellular signaling pathways related to disease progression. It is considered a promising natural compound for the prevention and treatment of diseases[23]. The anti-psoriatic mechanism of quercetin may be related to its antioxidant and anti-inflammatory effects, inhibition of nuclear factor (NF)-κB signaling pathway activation, and downregulation of miR-155, which is highly expressed in PSO[24,25]. Kaempferol is a natural flavanol found in many plants that has powerful anti-inflammatory, antioxidant, and anticancer properties. Kaempferol can protect mice from PSO-like skin damage caused by topical imiquimod[26]. It can also improve imiquimod-induced psoriatic lesions, mainly by reducing CD3+ T cell infiltration and the expression of proinflammatory cytokine genes (including IL-6, IL-17A, and TNF-α) in psoriatic lesions, downregulating NF-κB signal transduction in the skin, and reducing the percentage of IL-17A+ CD4+ T cells in the spleen and lymph nodes of the PSO mice model. Kaempferol can also inhibit T cell proliferation and mTOR signal transduction, which is associated with PSO, in vitro. Naringenin is a citrus flavonoid with various pharmacological properties. It demonstrates anti-inflammatory activity by inhibiting TNF-α and IL-1β[27]. Hepatotoxicity is an adverse side effect of methotrexate, which is used for the treatment of malignant tumors, PSO, and rheumatoid arthritis. Naringenin may inhibit methotrexate-related hepatotoxicity by scavenging active free radicals and exerting anti-inflammatory effects[28]. Naringenin significantly improved the changes in methotrexate-related biochemical markers and liver histopathology. Shikonin is the main active compound of Zicao and has been widely proven to inhibit PSO-like inflammation. Shikonin can reduce the overexpression of IL-17-related VEGF and K17 proteins by blocking the JAK2/STAT3 pathway. Additionally, shikonin can significantly inhibit the proliferation of HaCaT cells and induce apoptosis. However, the relationship of acetyl-shikonin in Zicao with PSO requires further research[29,30].
During this study, 11 target genes that are closely related to PSO, such as MAPK3, JUN, FOS, MYC, MAPK8, STAT3, and NFKBIA, were obtained through the PPI core network diagram. With PSO and related inflammatory skin diseases, innate and/or adaptive immune cells are activated and recruited to the site of inflammation, thus promoting further inflammation[31]. Continuous recruitment and activation of immune cells are regulated by a combination of cytokines and chemokines, which is, in turn, regulated by transcription factors, such as AP-1 (FOS/JUN), NF-κB, STATs, and MAPK[32]. MYC—a proto-oncogene involved in a variety of cellular processes, including cell growth, proliferation, and apoptosis—can play an important role in angiogenesis through a VEGF-dependent mechanism[33], is often amplified with skin cancer[34], and can stimulate the abnormal proliferation of normal keratinocytes. These effects of MYC are consistent with the pathological characteristics of abnormal proliferation and incomplete differentiation of psoriatic epithelial cells and abnormal dermal vascular proliferation. These core genes require further study to clarify their mechanisms of action and to provide new insights into the prevention and treatment of PSO.
We obtained low binding energies (-8.7 kcal/mol and -6.6 kcal/mol, respectively) for the molecular docking of MAPK8 with Kaempferol and STAT3 with licochalcone A, which show that the selected drug molecule-gene target combinations have a good binding activity. These results suggest that these drug molecules may directly bind to the corresponding core targets with high and stable affinity and provide evidence for the key role of these drug components in psoriasis treatment. The docking results confirm the accuracy of the PPI network analysis in identifying the core targets.
Based on the KEGG pathway enrichment analysis, BYT has a wide range of effects on the IL-17 signaling pathway, Th17 cell differentiation, TNF signaling pathway, PI3K-Akt signaling pathway, T cell receptor signaling pathway, MAPK signaling pathway, and other signaling pathways directly related to the pathological mechanism of PSO. The TNFα/IL-23/Th17 inflammatory pathway is a characteristic pathway of PSO. To date, clinically relevant signaling pathways in PSO have mainly been shown to be mediated by IL-17, which leads to the activation of a series of intracellular kinases (such as extracellular signal-regulated kinase, p38 MAPK, transforming growth factor-β-activated kinase 1, and others)[31]. These internal kinases enhance signaling pathways, such as the NF-κB pathway, thus promoting the expression of proinflammatory cytokines, chemokines, and antimicrobial peptides. Cytokines produced by Th1 and Th2 have a role through the JAK-STAT signaling pathway, whereas NF-κB can mediate the Th17 response[6]. Fluid shear stress and atherosclerosis affect hemodynamics by regulating the expression of genes and proteins in endothelial cells and changing the differentiation, morphology, and permeability of blood vessel walls of vascular endothelial cells[35]. It is inferred that this pathway may be related to the proliferation and expansion of blood vessels in the dermis, leading to persistent abnormalities in local skin function. Signaling pathways related to PSO comorbidities, such as the advanced glycation end products (AGEs)/receptor of AGE (RAGE) signaling pathway in diabetic complications, have been identified as well. AGEs and their receptors in the AGE-RAGE signal transduction pathway have been a focus of research recently. This pathway activates the NF-κB pathway and induces oxidative and inflammatory reactions by expressing and releasing VEGF, transforming growth factor-β1, nicotinamide adenine dinucleotide phosphate, and other factors, which ultimately cause cell and tissue damage[36,37].
TCM network pharmacology is the network biology basis for understanding complex diseases, TCM syndromes, and TCM treatment, reflecting the new trend of combining computation, experimentation, and clinical practices[38]. In this study, we used network pharmacology to establish drug target pathways and networks and predicted the possible protein targets of BYT in the treatment of psoriasis and the main interactions between these targets and drugs. Nonetheless, this study has some limitations. First, all data were from public databases and public data, and multi-faceted analysis was performed using bioinformatics. The management of public databases is heterogeneous and beyond our control; thus, the possibility of selection bias cannot be ruled out. Second, considering the multiple components, proteins, genes, and pathways involved in TCM, greater algorithm support is needed for a deeper understanding of the correlation between different data to reveal the internal biological network regulation mechanisms of TCM. Finally, this study lacks experimental validation, but it provides a possible direction for further in vitro and in vivo experimental studies to explore the detailed mechanism between BYT and psoriasis.
Using a network pharmacology approach, this study combined bioinformatics technology with TCM theory to analyze the chemical components, effective active ingredients, and targets of BYT compounds as TCM and the target genes in PSO. Relevant software was used to analyze and construct the target network and cell pathways targeted by BYT. The main active ingredients identified include quercetin, kaempferol, beta-sitosterol, naringenin, and acetyl-shikonin. Target genes include MAPK3, JUN, FOS, MYC, MAPK8, STAT3, and NFKBIA, which can regulate the inflammatory state mediated by PSO immune cells and mediate the expression of factors that adjust local skin inflammation. Our results confirm that BYT can work on PSO through multi-component, multi-target, and multi-channel synergy and provide a basis for further in-depth clinical research of BYT treatment for PSO.
Psoriasis (PSO) is a major public health challenge, causing great physical, psychological and economic burden. Biyu Decoction (BYT) has clear clinical efficiency in treating PSO.
The molecular mechanism of BYT treatment for PSO is still unclear.
To identify the targets and pathways through which BYT interferes with PSO.
Therapeutic targets for BYT were predicted by network pharmacology and validated by molecular docking.
A total of 213 gene targets corresponding to 117 active components of BYT can intervene in PSO through a variety of biological pathways highly related to PSO, such as Th17 cell differentiation and TNF signaling Pathway. The stability of these biological reactions was verified by molecular docking.
BYT can the pathogenesis of PSO with multiple targets.
The results of this study could serve as a fundamental basis for the further exploration in the in vitro and in vivo experiments for clinical promotions.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C, C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Apiratwarakul K, Thailand; Batool SN, Pakistan; Di Meglio L, Italy; Govindarajan KK, India; Hasabo EA, Sudan A-Editor: Zhu JQ, United States S-Editor: Wang LL L-Editor: A P-Editor: Chen YX
| 1. | Wei JC, Chang YJ, Wang YH, Yeh CJ. The Risk of Gout in Patients with Psoriasis: A Population-Based Cohort Study in Taiwan. Clin Epidemiol. 2022;14:265-273. [PubMed] |
| 2. | Ding X, Wang T, Shen Y, Wang X, Zhou C, Tian S, Liu Y, Peng G, Zhou J, Xue S, Wang R, Tang Y, Meng X, Pei G, Bai Y, Liu Q, Li H, Zhang J. Prevalence of psoriasis in China: a population-based study in six cities. Eur J Dermatol. 2012;22:663-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 167] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 3. | Wei JC, Shi LH, Huang JY, Wu XF, Wu R, Chiou JY. Epidemiology and Medication Pattern Change of Psoriatic Diseases in Taiwan from 2000 to 2013: A Nationwide, Population-based Cohort Study. J Rheumatol. 2018;45:385-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 4. | Rapp SR, Feldman SR, Exum ML, Fleischer AB Jr, Reboussin DM. Psoriasis causes as much disability as other major medical diseases. J Am Acad Dermatol. 1999;41:401-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1001] [Cited by in RCA: 999] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 5. | Dubertret L, Mrowietz U, Ranki A, van de Kerkhof PC, Chimenti S, Lotti T, Schäfer G; EUROPSO Patient Survey Group. European patient perspectives on the impact of psoriasis: the EUROPSO patient membership survey. Br J Dermatol. 2006;155:729-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 226] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 6. | Langley RG, Krueger GG, Griffiths CE. Psoriasis: epidemiology, clinical features, and quality of life. Ann Rheum Dis. 2005;64 Suppl 2:ii18-23; discussion ii24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 520] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 7. | Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker JNWN. Psoriasis. Lancet. 2021;397:1301-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 1252] [Article Influence: 313.0] [Reference Citation Analysis (0)] |
| 8. | Christophers E, van de Kerkhof PCM. Severity, heterogeneity and systemic inflammation in psoriasis. J Eur Acad Dermatol Venereol. 2019;33:643-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 9. | Dai D, Wu H, He C, Wang X, Luo Y, Song P. Evidence and potential mechanisms of traditional Chinese medicine for the treatment of psoriasis vulgaris: a systematic review and meta-analysis. J Dermatolog Treat. 2020;1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 10. | Chen X, Zhang R, Duan X, Xue M, Qu T, Li L. Effectiveness of Xiaoyin Jiedu granules in the treatment of psoriasis vulgaris in patients with blood-heat symptom patterns in terms of Traditional Chinese Medicine. J Tradit Chin Med. 2020;40:863-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 11. | Lan XO, Wang HX, Qi RQ, Xu YY, Yu YJ, Yang Y, Guo H, Gao XH, Geng L. Shikonin inhibits CEBPD downregulation in IL17treated HaCaT cells and in an imiquimodinduced psoriasis model. Mol Med Rep. 2020;22:2263-2272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Zhang CS, Yu JJ, Parker S, Zhang AL, May B, Lu C, Xue CC. Oral Chinese herbal medicine combined with pharmacotherapy for psoriasis vulgaris: a systematic review. Int J Dermatol. 2014;53:1305-1318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Jo S, Ryu J, Kim H, Kim M, Ryu MH, Cho SI. Anti-inflammatory Effects of Sanguisorbae Radix on Contact Dermatitis Induced by Dinitrofluorobenzene in Mice. Chin J Integr Med. 2020;26:688-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 14. | Gan DL, Yao Y, Su HW, Huang YY, Shi JF, Liu XB, Xiang MX. Volatile Oil of Platycladus Orientalis (L.) Franco Leaves Exerts Strong Anti-inflammatory Effects via Inhibiting the IκB/NF-κB Pathway. Curr Med Sci. 2021;41:180-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Li P, Li Y, Jiang H, Xu Y, Liu X, Che B, Tang J, Liu G, Tang Y, Zhou W, Zhang L, Dong C, Chen H, Zhang K, Du Z. Glabridin, an isoflavan from licorice root, ameliorates imiquimod-induced psoriasis-like inflammation of BALB/c mice. Int Immunopharmacol. 2018;59:243-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 16. | Hawkes JE, Chan TC, Krueger JG. Psoriasis pathogenesis and the development of novel targeted immune therapies. J Allergy Clin Immunol. 2017;140:645-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 672] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 17. | Luque-Martin R, Angell DC, Kalxdorf M, Bernard S, Thompson W, Eberl HC, Ashby C, Freudenberg J, Sharp C, Van den Bossche J, de Jonge WJ, Rioja I, Prinjha RK, Neele AE, de Winther MPJ, Mander PK. IFN-γ Drives Human Monocyte Differentiation into Highly Proinflammatory Macrophages That Resemble a Phenotype Relevant to Psoriasis. J Immunol. 2021;207:555-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 18. | Boehncke WH, Schön MP. Psoriasis. Lancet. 2015;386:983-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1395] [Cited by in RCA: 1822] [Article Influence: 182.2] [Reference Citation Analysis (0)] |
| 19. | Sun W, Gao Y, Yu X, Yuan Y, Yi J, Zhang Z, Cheng Y, Li Y, Peng X, Cha X. 'Psoriasis 1' reduces psoriasislike skin inflammation by inhibiting the VDRmediated nuclear NFκB and STAT signaling pathways. Mol Med Rep. 2018;18:2733-2743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Luengas-Martinez A, Paus R, Young HS. Antivascular endothelial growth factor-A therapy: a novel personalized treatment approach for psoriasis. Br J Dermatol. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 21. | Zhang CS, Yang L, Zhang AL, May BH, Yu JJ, Guo X, Lu C, Xue CC. Is Oral Chinese Herbal Medicine Beneficial for Psoriasis Vulgaris? J Altern Complement Med. 2016;22:174-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 22. | Sun X, Zhou X, Wei Y, Yang W, Huang N, Ding Y, Hu R, Guo S, Yang C, Weng H, Zhang Y. Our Choice: study protocol for a randomized controlled trial for optimal implementation of psoriasis treatment by the integration of Chinese and western medicine. Trials. 2020;21:299. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 23. | D'Andrea G. Quercetin: A flavonol with multifaceted therapeutic applications? Fitoterapia. 2015;106:256-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 484] [Reference Citation Analysis (0)] |
| 24. | Chen H, Lu C, Liu H, Wang M, Zhao H, Yan Y, Han L. Quercetin ameliorates imiquimod-induced psoriasis-like skin inflammation in mice via the NF-κB pathway. Int Immunopharmacol. 2017;48:110-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 114] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 25. | Kocic H, Damiani G, Stamenkovic B, Tirant M, Jovic A, Tiodorovic D, Peris K. Dietary compounds as potential modulators of microRNA expression in psoriasis. Ther Adv Chronic Dis. 2019;10:2040622319864805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 26. | Liu C, Liu H, Lu C, Deng J, Yan Y, Chen H, Wang Y, Liang CL, Wei J, Han L, Dai Z. Kaempferol attenuates imiquimod-induced psoriatic skin inflammation in a mouse model. Clin Exp Immunol. 2019;198:403-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 27. | Gaggeri R, Rossi D, Daglia M, Leoni F, Avanzini MA, Mantelli M, Juza M, Collina S. An eco-friendly enantioselective access to (R)-naringenin as inhibitor of proinflammatory cytokine release. Chem Biodivers. 2013;10:1531-1538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Malayeri A, Badparva R, Mombeini MA, Khorsandi L, Goudarzi M. Naringenin: a potential natural remedy against methotrexate-induced hepatotoxicity in rats. Drug Chem Toxicol. 2022;45:491-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Xu Y, Xu X, Gao X, Chen H, Geng L. Shikonin suppresses IL-17-induced VEGF expression via blockage of JAK2/STAT3 pathway. Int Immunopharmacol. 2014;19:327-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 30. | Yu YJ, Xu YY, Lan XO, Liu XY, Zhang XL, Gao XH, Geng L. Shikonin induces apoptosis and suppresses growth in keratinocytes via CEBP-δ upregulation. Int Immunopharmacol. 2019;72:511-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 31. | Rendon A, Schäkel K. Psoriasis Pathogenesis and Treatment. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 498] [Cited by in RCA: 1167] [Article Influence: 194.5] [Reference Citation Analysis (0)] |
| 32. | Helliwell PS. Assessment of disease activity in psoriatic arthritis. Clin Exp Rheumatol. 2015;33:S44-S47. [PubMed] |
| 33. | Feller JK, Mahalingam M. c-myc and cutaneous vascular neoplasms. Am J Dermatopathol. 2013;35:364-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Brandl L, Hartmann D, Kirchner T, Menssen A. Expression of n-MYC, NAMPT and SIRT1 in Basal Cell Carcinomas and their Cells of Origin. Acta Derm Venereol. 2019;99:63-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 35. | Akbari E, Spychalski GB, Rangharajan KK, Prakash S, Song JW. Competing Fluid Forces Control Endothelial Sprouting in a 3-D Microfluidic Vessel Bifurcation Model. Micromachines (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 36. | Li JH, Wang W, Huang XR, Oldfield M, Schmidt AM, Cooper ME, Lan HY. Advanced glycation end products induce tubular epithelial-myofibroblast transition through the RAGE-ERK1/2 MAP kinase signaling pathway. Am J Pathol. 2004;164:1389-1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 172] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 37. | Xu Y, Nie L, Yin YG, Tang JL, Zhou JY, Li DD, Zhou SW. Resveratrol protects against hyperglycemia-induced oxidative damage to mitochondria by activating SIRT1 in rat mesangial cells. Toxicol Appl Pharmacol. 2012;259:395-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 115] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 38. | Wang X, Wang ZY, Zheng JH, Li S. TCM network pharmacology: A new trend towards combining computational, experimental and clinical approaches. Chin J Nat Med. 2021;19:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 187] [Article Influence: 46.8] [Reference Citation Analysis (0)] |