Published online Jan 14, 2022. doi: 10.12998/wjcc.v10.i2.607
Peer-review started: January 31, 2021
First decision: July 16, 2021
Revised: July 19, 2021
Accepted: December 9, 2021
Article in press: December 9, 2021
Published online: January 14, 2022
Processing time: 345 Days and 23.5 Hours
Lissencephaly (LIS) is a malformation of cortical development with broad gyri, shallow sulci and thickened cortex characterized by developmental delays and seizures. Currently, 20 genes have been implicated in LIS. However, GRP56-related LIS has never been reported. GRP56 is considered one of the causative genes for bilateral frontoparietal polymicrogyria. Here, we report a twin infant with LIS and review the relevant literature. The twins both carried the novel compound heterozygous GPR56 mutations.
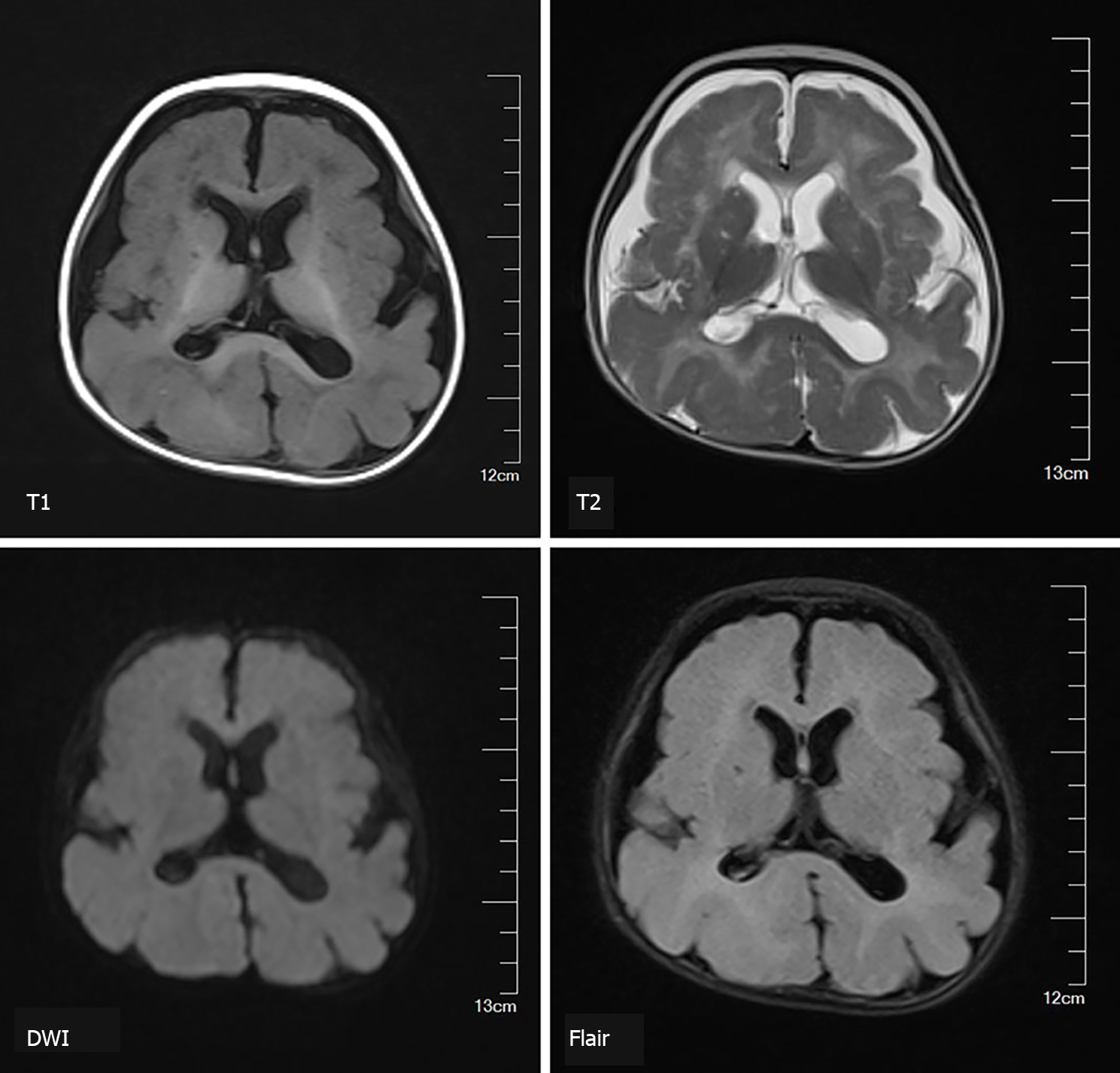
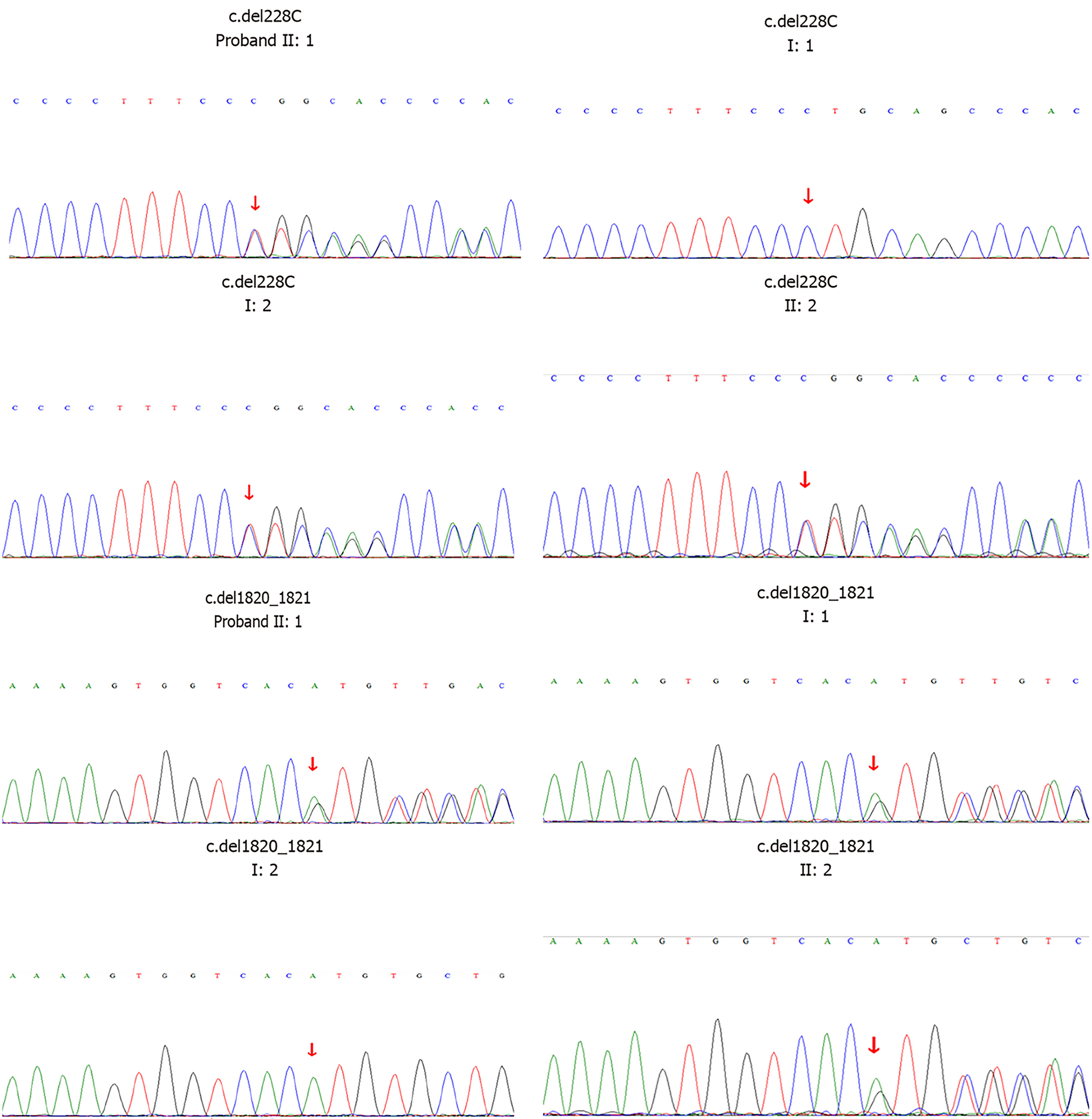
A 5-mo-old female infant was hospitalized due to repeated convulsions for 1 d. The patient had a flat head deformity that manifested as developmental delays and a sudden onset of generalized tonic-clonic seizures at 5 mo without any causes. The electroencephalography was normal. Brain magnetic resonance imaging revealed a simple brain structure with widened and thickened gyri and shallow sulci. The white matter of the brain was significantly reduced. Patchy long T1 and T2 signals could be seen around the ventricles, which were expanded, and the extracerebral space was widened. Genetic testing confirmed that the patient carried the GPR56 gene compound heterozygous mutations c.228delC (p.F76fs) and c.1820_1821delAT (p.H607fs). The unaffected father carried a heterozygous c.1820_1821delAT mutation, and the unaffected mother carried a heterozygous c.228delC mutation. The twin sister carried the same mutations as the proband. The patient was diagnosed with LIS.
This is the first case report of LIS that is likely caused by mutations of the GPR56 gene.
Core Tip: We report a twin infant with lissencephaly (LIS). The twins both carried the novel compound heterozygous GPR56 mutations, p.F76fs and p.H607fs, which have not been reported in the Human Gene Mutation Database. To our knowledge, this is the first case of GRP56-related LIS. Therefore, GPR56 gene mutations may lead to LIS.
- Citation: Lin WX, Chai YY, Huang TT, Zhang X, Zheng G, Zhang G, Peng F, Huang YJ. Novel compound heterozygous GPR56 gene mutation in a twin with lissencephaly: A case report. World J Clin Cases 2022; 10(2): 607-617
- URL: https://www.wjgnet.com/2307-8960/full/v10/i2/607.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i2.607
Lissencephaly (LIS) is a group of abnormal cerebral cortical dysplasias caused by the defective migration of neurons. It can be diagnosed clinically by neuroimaging. It is characterized by thickening of the cerebral cortex, widening of the gyri, and disappearance or shallowness of the sulci. The complete disappearance of the sulci and gyri showing smooth surface of the brain is called agyria and is seen in severe cases[1]. According to the neuroimaging, LIS is divided into six grades, ranging from severe agyria (grade 1) to mild subcortical band heterotopias (grade 6). The severity of nerve damage is closely related to the grade of LIS and cortical thickening, and the mortality rate of severe LIS is high[2]. In the early stages, patients often exhibit developmental delays and hypotonia, followed by seizures, and a severe intellectual disability eventually. Although a LIS patient may develop normally in the neonatal period, many neonates suffer from persistent feeding problems and different types of epilepsy, which are difficult to cure[3]. An individual with mild LIS and normal intelligence has been reported[4]. Currently, 20 genes have been implicated in LIS. Many of these genes are microtubule genes[5,6].
GPR56 (OMIM#606854, NM_0001145773) encodes an orphan G protein-coupled receptor (GPCR) that is extensively expressed in the nervous system and is essential for the normal development of the cerebral cortex and cerebellar morphology[7-9]. The reported mutations of the GPR56 gene have been confirmed to be related to bilateral frontoparietal polymicrogyria (BFPP)[10].
Herein, we report a twin infant with LIS who came from a nonconsanguineous family. The twins both carried a novel compound heterozygous GPR56 mutation. To our knowledge, this is the first case of GRP56-related LIS.
A 5-mo-old female infant was hospitalized due to repeated convulsions for 1 d.
The patient was admitted to the Children’s Hospital of Nanjing Medical University due to repeated convulsions. The patient had a sudden onset of generalized tonic-clonic seizures without any causes. In addition, she had a flat head deformity and developmental delays.
The patient had no history of past illness.
The patient was the first child of nonconsanguineous Chinese parents. She was delivered by cesarean section due to twin pregnancy at 32 wk of gestation, with a birth weight of 2.6 kg. No intrauterine distress or postnatal asphyxia had occurred. She had a twin sister with LIS.
The patient showed a flat head deformity. The neurological examination was normal. There were no other abnormal signs.
The electroencephalography and laboratory findings (full blood count, liver, kidney and thyroid function tests, creatine kinase, uric acid, metabolic study and chromosome karyotyping) were normal.
Brain magnetic resonance imaging (MRI) revealed a simple brain structure, with widened and thickened gyri and shallow sulci. The white matter of the brain was significantly reduced. The patchy long T1 and long T2 signals could be seen around the ventricles, which were expanded, and the extracerebral space was widened (Figure 1).
According to the clinical characteristics, imaging and genetic test findings (Figure 2), the infant was diagnosed with LIS.
During the hospital stay, the patient had no epileptic seizures. She received rehabilitation, but anti-epileptic treatment was refused.
The patient experienced repeated convulsions after she was discharged from hospital. The convulsions occurred once a day to more than ten times a day without any causes, each episode lasting several minutes. She died 3 mo later.
The GPR56 gene spans 45 kb and consists of 14 exons encoding an orphan GPCR of 693 amino acids[7,11]. GPR56 is a member of the adhesion GPCR family, which has an N- and a C-terminal fragment and a GPCR proteolytic site[12]. In the central nervous system, GPR56 plays an important role in the normal development of the cerebral cortex and cerebellar morphogenesis[8]. In the peripheral nervous system, GPR56 can regulate the formation and maintenance of myelin sheaths[13]. Therefore, the normal expression of GPR56 is essential for the function of the nervous system.
It is known that mutations of the GPR56 gene are related to BFPP (Table 1). The clinical manifestations of BFPP are overall growth retardation and seizures. MRI shows symmetrical polygyria (the frontal parietal area is the most serious part), ventricular enlargement, and bilateral white matter changes. Twenty-eight pathogenic GPR56 mutations related to the BFPP phenotype have been reported[11,14]. The affected individuals inherit the mutants in an autosomal recessive mode. The majority of missense mutations resulted in similar clinical symptoms, indicating that the similar phenotype might be caused by the same mechanism. However, the mechanism remains unclear, although it may involve GPR56 trafficking and a decrease in receptor levels at the cell membrane[15-17]. GPR56 knockdown did not affect the migration of neural progenitor cells, while GPR56 overexpression inhibited the migration of neural progenitor cells. This mechanism might occur through the reorganization of cerebral cortex actin to change the cell morphology and regulate neural progenitor cell behavior[8]. LIS is caused by premature stop of neuronal migration, which might explain the mechanism of the GPR56 mutations causing LIS in the present case.
| Ref. | Mutation | Exon/intron | Case number | Ethnicity | Consanguinity | Motor delay | Cognitive delay | Seizure | MRI | ||
| Gyri | White matter abnormalities | Brainstem/cerebellum | |||||||||
| Piao et al[10,11], 2004 and 2005 | c.112C>T (p.R38W) | Exon 3 | 2 | Arabic (Qatar) | First cousin | + | Moderate | GTC, myoclonic | BFPP | Patchy signal change | Small brainstem |
| + | NA | + | |||||||||
| 1 | Arabic (UAE) | First cousin | + | + | NA | BFPP | Reduced volume, patchy signal change | Slightly small pons and vermis | |||
| c.113G>A (p.R38Q) | Exon 3 | 1 | Turkish | First cousin | + | + | + | BFPP | Severely reduced volume, patchy signal change | Small pons and vermis | |
| c.263A>G (p.Y88C) | Exon 3 | 2 | French Canadian | N | + | + | NA | BFPP | Reduced volume, patchy signal change | Small pons, small/dysplastic cerebellum | |
| c.739-746 delCAGGACC (p.Q246Tfx*72) | Exon 5 | 2 | Indian | N | + | + | Blank episodes | BFPP | Reduced volume, patchy signal change | Slightly small pons and vermis | |
| + | + | AS | |||||||||
| 1 | Pakistani | First cousin | Severe | Severe | Generalized | BFPP | Patchy radiolucency | Small cerebellum | |||
| 1 | Afghani | First cousin | Moderate | + | NA | BFPP | Reduced volume, patchy signal change | Small pons and superior vermis | |||
| c.E5- 1G>C (NA) | Exon 5 | 2 | Palestinian | N | + | + | Episodes of startles | BFPP | Reduced volume, periventricular signal change | Small pons and superior vermis | |
| c.1036T>A(p.C346S) | Exon 8 | 2 | Palestinian | First cousin | + | + | NA | BFPP | Reduced volume, patchy signal change | Small pons and cerebellum | |
| 1 | Palestinian | First cousin | + | Severe | + | BFPP | Reduced volume, frontal subcortical signal change | Small brainstem and cerebellum | |||
| c.1046G>C(p.W349S) | Exon 8 | 2 | Israeli Jewish | First cousin | + | + | GTC | BFPP | Reduced volume, patchy signal change | Small pons and vermis | |
| + | + | Myoclonic | BFPP | ||||||||
| 1 | Israeli Jewish | N | + | Severe | + | BFPP | Patchy signal change | Small vermis | |||
| c.IVS9+3G>C (NA) | Intron 9 | 3 | Palestinian | First cousin | + | + | FS, atonic-drop | BFPP | Patchy signal change | Slightly small pons and superior vermis | |
| + | Moderate | GTC, AS | BFPP | ||||||||
| Severe | Severe | FS, GTC | BFPP | ||||||||
| 2 | Palestinian | First cousin | + | Severe | GTC, atonic | BFPP | Patchy signal change | Small pons and superior vermis | |||
| + | Severe | No | BFPP | ||||||||
| c.1693C>T (p.R565W) | Exon 13 | 3 | Arabic (Bedouin) | C | + | Severe | GTC, myoclonic | BFPP | Reduced volume, patchy signal change | Small vermis | |
| 1 | Italian | Second cousin | + | + | + | BFPP | Reduced volume, patchy signal change | Slightly small vermis | |||
| c.1919T>G (p.L640R) | Exon 13 | 1 | Hispanic | N | + | + | + | BFPP | Mildly reduced volume, patchy signal change | Slightly small cerebellar hemispheres | |
| Parrini et al[18], 2009 | c.97C>G (p.R33P) | Exon 2 | 2 | Turkish | C | + | Severe | Atypical absences, GTC, tonic | BFPP | NA | NA |
| + | Severe | Tonic, atypical absences, recurrent nonconvulsive status epilepticus | BFPP | Patchy signal change | NA | ||||||
| c.235C>T (R79X) | Exon 2 | 1 | Italian | C | + | Severe | Infantile spasms, tonic and atonic seizures | BFPP | Patchy signal change | NA | |
| c.1693C>T (p.R565W) | Exon 13 | 1 | Italian | C | + | Severe | Tonic atonic GTC, atypical absences, recurrent nonconvulsive statusepilepticus | BFPP | Patchy signal change | Slightly small vermis | |
| Bahi-Buisson et al[19], 2010 | c.174-175insC (p.E59Rfs*24) | Exon 3 | 2 | NA | C | NA | Severe | + | NA | NA | NA |
| Walking at 4 yr | Severe | Focal seizures | BFPP | Patchy periventricular predominance | Hypoplastic pons | ||||||
| c.272G>A (p.C91Y) | Exon 3 | 2 | NA | C | Walking at 2 yr | Severe | NA | BFPP | Patchy | Hypoplastic pons | |
| Walking at 2 yr | Severe | GTC/atypical absence, atonic seizures | BFPP | Patchy periventricular and frontal predominance | Hypoplastic pons, Cyst in the ventral pons | ||||||
| c.367C>T (p.Q123X) | Exon 3 | 1 | NA | C | + | Severe | Focal seizures, GTC | BFPP | Patchy periventricular and frontal predominance | Hypoplastic pons, Cyst in the ventral pons | |
| c.671delA (p.D224Wfs*96) | Exon 5 | 3 | NA | C | Walking at 4 yr | Severe | GTC | BFPP | Patchy periventricular and frontal predominance | Hypoplastic pons | |
| Walking at 18 mo | Severe | GTC | BFPP | ||||||||
| Sitting without support | Severe | GTC | BFPP | Diffuse | Hypoplastic pons | ||||||
| c.1215-1216delC (p.L406S406fs*41) | Exon 10 | 1 | NA | C | Walking acquired but subsequently lost (11 yr) | Severe | + | BFPP | Patchy | Hypoplastic pons | |
| c.1254C>G (p.C418W) | Exon 10 | 3 | Pakistani | First cousin | Walking at 5 yr | Severe | GTC | BFPP | Diffuse | Hypoplastic pons | |
| Walking at 5 yr | Severe | GTC | BFPP | Patchy with subcortical and frontal predominance, reduced volume | Severely hypoplastic pons with posterior concavity, cyst in the ventral pons | ||||||
| NA | NA | NA | NA | NA | NA | ||||||
| c.1345delCTG (p.L449del) | Exon 11 | 1 | NA | C | Walking at 3 yr | Severe | Atypical absence | BFPP | Patchy with subcortical predominance | Severely hypoplastic pons with posterior concavity | |
| c.1453C>T (p.S485P) | Exon 11 | 2 | NA | C | Walking at 18 mo | Severe | Focal seizures, generalized tonic seizures | BFPP | Patchy with subcortical and frontal predominance | Hypoplastic pons | |
| Walking at 18 mo | Severe | Focal seizures | BFPP | ||||||||
| Luo et al[20], 2011 | c.1486G>A (p.E496K) | NA | 1 | Yemeni | First cousin | Walking | Severe | Tonic-clonic seizures | BFPP | Asymmetric areas of abnormal signal in the white matter of both cerebral hemispheres | Mild hypoplasia of the inferior cerebellar vermis and pons |
| Quattrocchi et al[16], 2013 | c.105C>A (p.C35X) | Exon 2 | 1 | NA | NA | Ataxic gait | Severe | Focal seizures, myoclonic | BFPP | Patchy subcortical and periventricular white matter abnormalities | Mildly hypoplastic cerebellar vermis, flattening of the ventral aspect of the pons, hemispheric cerebellar cysts, vermian cysts |
| c.429G>A (p.W143X) | Exon 2 | 1 | NA | NA | Ataxic gait | Moderate | No | BFPP | Patchy subcortical and periventricular white matter abnormalities | Mildly hypoplastic cerebellar vermis, flattening of the ventral aspect of the pons, hemispheric cerebellar cysts, vermian cysts | |
| c.1453C>T (p.S485P) | Exon 11 | 2 | NA | NA | Walking at 18 mo | Severe | GTS, focal seizures | BFPP | Patchy subcortical and periventricular white matter abnormalities | Hypoplastic pons and superior vermis, hemispheric cerebellar cysts, vermian cysts | |
| Walking at 22 mo | Severe | Focal seizures | BFPP | Patchy subcortical and periventricular white matter abnormalities | Hypoplastic pons and superior vermis, hemispheric cerebellar cysts, vermian cysts | ||||||
| c.1796-1801delTGCGCC/insAGATCCTGTGGGCAGAT (premature stop codon at position 614) | Exon 12 | 1 | NA | NA | Ataxic gait | Moderate | No | BFPP | Patchy subcortical and periventricular white matter abnormalities | Flattening of the ventral aspect of the pons, hemispheric cerebellar cysts | |
| Fujii et al[21], 2014 | c.107G>A and c.113G>A(p.S36N and p.R38Q) | Exon 2 | 1 | Japanese | N | Able to walk with help | Severe | Complex partial seizures, tonic seizures, epileptic spasms | BFPP | Patchy high signals in the frontal subcortical | Hypoplastic pons |
| Desai et al[22], 2015 | c.113G>A (p.R38Q) | Exon 3 | 1 | Indian (Marathi) | C | Mode-rate | Moderate | Complex febrile seizures | BFPP | Diffuse | Mild thinning and cerebellar cysts |
| c.739–746 delCAGGACC (p.Q246Tfx*72) | Exon 4 | 1 | Indian (Punjabi) | N | Severe | Mild | No | BFPP | Frontal and periventricular | Mild thinning and cerebellar cysts | |
| c.739–746 delCAGGACC (p.Q246Tfx*72) | Exon 4 | 1 | Indian (Sindhi) | N | Severe | Moderate | No | BFPP | Frontal and periventricular | Inferior vermian hypoplasia; cerebellar cyst | |
| c.1426 C>T (p.R476X) | Exon 12 | 1 | Indian (Gujarati) | C | Severe | Severe | Generalized seizures | BFPP | Diffuse | Mild thinning and cerebellar cysts | |
| Santos-Silva et al[17], 2015 | 811C > T (R271X) | Exon 6 | 1 | Caucasian | N | Severe | Severe | Hot water epilepsy | BFPP | Reduced volume, patchy signal change | Hypoplasia of the pons and cerebellar vermis |
| Öncü-Öner et al[14], 2018 | 811C > T (R271X) | Exon 6 | 1 | NA | C | Severe | Severe | Focal onset bilateral tonic-clonic seizure | BFPP | Yes | Thin brainstem and normal cerebellar structure |
| Current report | c.228delC and c.1820-1821del AT (p.F76fs and p.H607fs) | Exon 6 and Exon 13 | 2 | Chinese | N | + | Severe | GTC | LIS | Reduced volume, patchy signal change | Normal |
| + | + | No | LIS | NA | NA | ||||||
The development of the brain is a delicate and complex physiological process, and the proper migration of neurons is one of the most critical steps. LIS is brain dysplasia caused by the premature stop of neuronal migration. Type I LIS is characterized by a thickened cerebral cortex (10-20 mm, whereas normal is 4 mm), but no other brain development malformations, such as severe congenital microcephaly, corpus callosum hypoplasia, or cerebellar hypoplasia[2]. Microscopically, the cerebral cortex in LIS is divided into four thick and dysplastic layers: The molecular layer, the superficial cellular layer, the cell spare layer, and the deeper cellular layer; the normal cerebral cortex has six layers[1].
Currently, 20 genes have been reported to be associated with LIS, and many of them are microtubule genes[5,6]. In a cohort study of 811 patients with LIS, the overall mutation frequency of the entire cohort was 81%, of which LIS1 accounted for 40%, followed by DCX (23%), TUBA1A (5%), and DYNC1H1 (3%). Other genes accounted for 1% or less. Interestingly, the cause of LIS in 19% of the patients was unknown, which indicates that additional genes are involved and need to be discovered[6]. There have been no other reports of LIS caused by GPR56 gene mutations. Therefore, the relationship between LIS and GPR56 still needs further research.
There is no specific treatment method for LIS. Current treatments typically involve symptomatic relief, such as anti-epileptic treatment and rehabilitation training. Studies in animal models have shown that it might be possible to restart neuronal migration by re-expressing the missing/nonfunctional genes after birth[2]. Even if the degree of cortical deformity is partially improved, it may significantly decrease seizure frequency and clinical severity[2]. Therefore, with the advances in genetic testing and medical technology, the diagnosis and treatment of LIS will continue to be improved and optimized.
The compound mutations in the GPR56 gene identified in the twin sisters with LIS were novel and unreported mutations. This finding has broadened our knowledge of the clinical manifestations of LIS and increased our understanding of GPR56. Genetic testing is necessary when patients suffer from LIS symptoms.
We sincerely appreciate the patients and their parents for their help and willingness in this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Pediatrics
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Idiculla PS S-Editor: Zhang H L-Editor: A P-Editor: Zhang H
| 1. | Guerrini R, Parrini E. Neuronal migration disorders. Neurobiol Dis. 2010;38:154-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 167] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 2. | Fry AE, Cushion TD, Pilz DT. The genetics of lissencephaly. Am J Med Genet C Semin Med Genet. 2014;166C:198-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 3. | Parrini E, Conti V, Dobyns WB, Guerrini R. Genetic Basis of Brain Malformations. Mol Syndromol. 2016;7:220-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 144] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 4. | Leventer RJ, Cardoso C, Ledbetter DH, Dobyns WB. LIS1 missense mutations cause milder lissencephaly phenotypes including a child with normal IQ. Neurology. 2001;57:416-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Di Donato N, Chiari S, Mirzaa GM, Aldinger K, Parrini E, Olds C, Barkovich AJ, Guerrini R, Dobyns WB. Lissencephaly: Expanded imaging and clinical classification. Am J Med Genet A. 2017;173:1473-1488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 6. | Di Donato N, Timms AE, Aldinger KA, Mirzaa GM, Bennett JT, Collins S, Olds C, Mei D, Chiari S, Carvill G, Myers CT, Rivière JB, Zaki MS; University of Washington Center for Mendelian Genomics, Gleeson JG, Rump A, Conti V, Parrini E, Ross ME, Ledbetter DH, Guerrini R, Dobyns WB. Analysis of 17 genes detects mutations in 81% of 811 patients with lissencephaly. Genet Med. 2018;20:1354-1364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 7. | Ke N, Ma H, Diedrich G, Chionis J, Liu G, Yu DH, Wong-Staal F, Li QX. Biochemical characterization of genetic mutations of GPR56 in patients with bilateral frontoparietal polymicrogyria (BFPP). Biochem Biophys Res Commun. 2008;366:314-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Iguchi T, Sakata K, Yoshizaki K, Tago K, Mizuno N, Itoh H. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem. 2008;283:14469-14478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 163] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 9. | Chiang NY, Hsiao CC, Huang YS, Chen HY, Hsieh IJ, Chang GW, Lin HH. Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J Biol Chem. 2011;286:14215-14225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 10. | Piao X, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, Voit T, Ross ME, Michaud JL, Déscarie JC, Barkovich AJ, Walsh CA. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033-2036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 408] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 11. | Piao X, Chang BS, Bodell A, Woods K, Benzeev B, Topcu M, Guerrini R, Goldberg-Stern H, Sztriha L, Dobyns WB, Barkovich AJ, Walsh CA. Genotype-phenotype analysis of human frontoparietal polymicrogyria syndromes. Ann Neurol. 2005;58:680-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Cauley ES, Hamed A, Mohamed IN, Elseed M, Martinez S, Yahia A, Abozar F, Abubakr R, Koko M, Elsayed L, Piao X, Salih MA, Manzini MC. Overlap of polymicrogyria, hydrocephalus, and Joubert syndrome in a family with novel truncating mutations in ADGRG1/GPR56 and KIAA0556. Neurogenetics. 2019;20:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Ackerman SD, Luo R, Poitelon Y, Mogha A, Harty BL, D'Rozario M, Sanchez NE, Lakkaraju AKK, Gamble P, Li J, Qu J, MacEwan MR, Ray WZ, Aguzzi A, Feltri ML, Piao X, Monk KR. GPR56/ADGRG1 regulates development and maintenance of peripheral myelin. J Exp Med. 2018;215:941-961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 14. | Öncü-Öner T, Ünalp A, Porsuk-Doru İ, Ağılkaya S, Güleryüz H, Saraç A, Ergüner B, Yüksel B, Hız-Kurul S, Cingöz S. GPR56 homozygous nonsense mutation p.R271* associated with phenotypic variability in bilateral frontoparietal polymicrogyria. Turk J Pediatr. 2018;60:229-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Luo R, Jin Z, Deng Y, Strokes N, Piao X. Disease-associated mutations prevent GPR56-collagen III interaction. PLoS One. 2012;7:e29818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 16. | Quattrocchi CC, Zanni G, Napolitano A, Longo D, Cordelli DM, Barresi S, Randisi F, Valente EM, Verdolotti T, Genovese E, Specchio N, Vitiello G, Spiegel R, Bertini E, Bernardi B. Conventional magnetic resonance imaging and diffusion tensor imaging studies in children with novel GPR56 mutations: further delineation of a cobblestone-like phenotype. Neurogenetics. 2013;14:77-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Santos-Silva R, Passas A, Rocha C, Figueiredo R, Mendes-Ribeiro J, Fernandes S, Biskup S, Leão M. Bilateral frontoparietal polymicrogyria: a novel GPR56 mutation and an unusual phenotype. Neuropediatrics. 2015;46:134-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Parrini E, Ferrari AR, Dorn T, Walsh CA, Guerrini R. Bilateral frontoparietal polymicrogyria, Lennox-Gastaut syndrome, and GPR56 gene mutations. Epilepsia. 2009;50:1344-1353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 19. | Bahi-Buisson N, Poirier K, Boddaert N, Fallet-Bianco C, Specchio N, Bertini E, Caglayan O, Lascelles K, Elie C, Rambaud J, Baulac M, An I, Dias P, des Portes V, Moutard ML, Soufflet C, El Maleh M, Beldjord C, Villard L, Chelly J. GPR56-related bilateral frontoparietal polymicrogyria: further evidence for an overlap with the cobblestone complex. Brain. 2010;133:3194-3209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 20. | Luo R, Yang HM, Jin Z, Halley DJ, Chang BS, MacPherson L, Brueton L, Piao X. A novel GPR56 mutation causes bilateral frontoparietal polymicrogyria. Pediatr Neurol. 2011;45:49-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Fujii Y, Ishikawa N, Kobayashi Y, Kobayashi M, Kato M. Compound heterozygosity in GPR56 with bilateral frontoparietal polymicrogyria. Brain Dev. 2014;36:528-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Desai NA, Udani V. GPR56-Related Polymicrogyria: Clinicoradiologic Profile of 4 Patients. J Child Neurol. 2015;30:1819-1823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |