Published online Jan 14, 2022. doi: 10.12998/wjcc.v10.i2.448
Peer-review started: July 9, 2021
First decision: November 11, 2021
Revised: November 11, 2021
Accepted: December 7, 2021
Article in press: December 7, 2021
Published online: January 14, 2022
Processing time: 186 Days and 14.4 Hours
In recent years, the prevalence of Alzheimer’s disease (AD) has increased, which places a great burden on society and families and creates considerable challenges for medical services. N6-methyladenine (m6A) deoxyribonucleic acid (DNA) adenine methylation is a novel biomarker and is abundant in the brain, but less common in AD. We support to analyze the relationship between DNA m6A and cognition in patients with AD and normal controls (NCs) in China.
To analyze the relationship between the novel m6A DNA and cognition in patients with AD and NCs in China.
A total of 179 AD patients (mean age 71.60 ± 9.89 years; males: 91; females: 88) and 147 NCs (mean age 69.59 ± 11.22 years; males: 77; females: 70) who were age- and sex-matched were included in our study. All subjects underwent neuropsychological scale assessment and magnetic resonance imaging examination. Apolipoprotein E (APOE) genotypes were measured through agarose gel electrophoresis. Global m6A levels were evaluated by a MethylFlash m6A DNA Methylation ELISA Kit (colorimetric). Global m6A levels in total DNA from ten AD patients with 18F-AV-45 (florbetapir) positron emission tomography (PET) positivity and ten NCs with PET negativity were analyzed by dot blotting to determine the results.
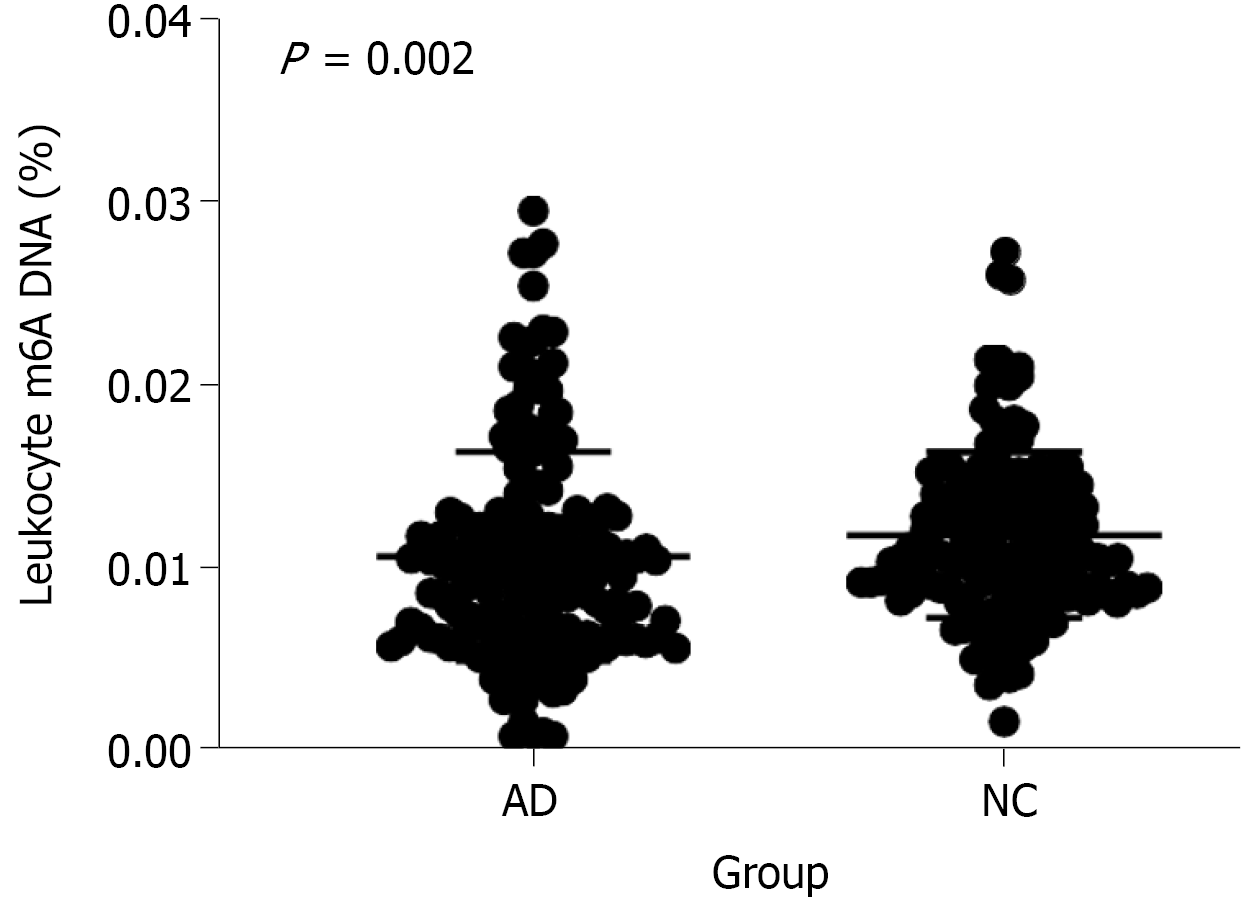
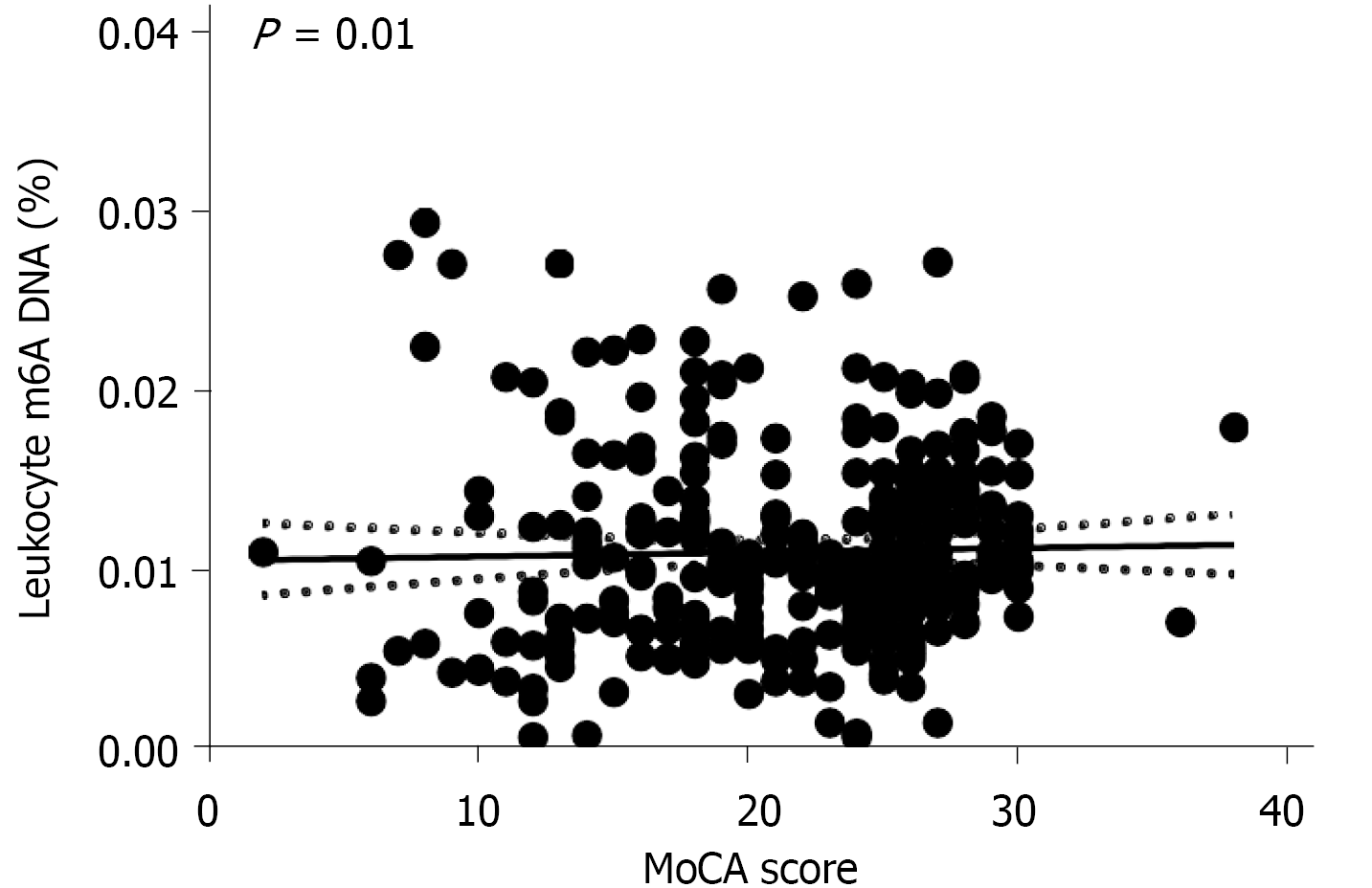
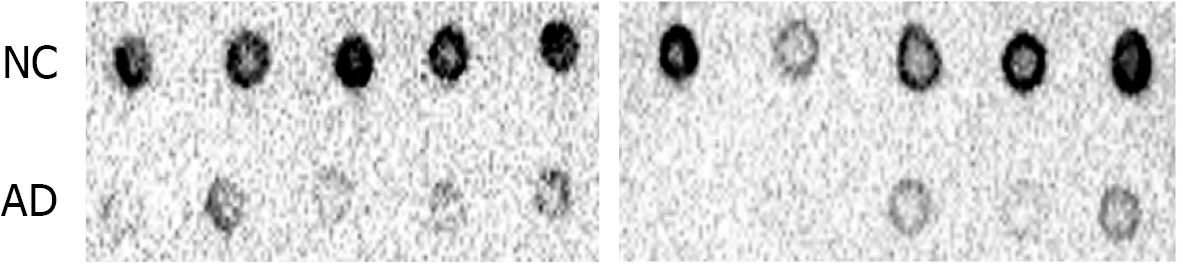
Our ELISA results showed that the global m6A DNA levels in peripheral blood were different between patients with AD and NCs (P = 0.002; < 0.05). And ten AD patients who were PET positive and ten NCs who were PET negative also showed the same results through dot blotting. There were significant differences between the two groups, which indicated that the leukocyte m6A DNA levels were different (P = 0.005; < 0.05). The m6A level was approximately 8.33% lower in AD patients than in NCs (mean 0.011 ± 0.006 vs 0.012 ± 0.005). A significant correlation was found between the Montreal Cognitive Assessment score and the peripheral blood m6A level in the tested population (r = 0.143, P = 0.01; < 0.05). However, no relationship was found with APOE ε4 (P = 0.633, > 0.05). Further studies should be performed to validate these findings.
Our results show that reduced global m6A DNA methylation levels are significantly lower in AD patients than in NCs by approximately 8.33% in China.
Core Tip: Although Alzheimer’s disease (AD) cannot be cured, early diagnosis and treatment can greatly improve the prognosis of AD patients. Thus, we aimed to identify biomarkers of AD that can be useful in the clinic. The diagnostic criteria for AD were strictly employed in the study. We found that N6-methyladenine (m6A) DNA adenine methylation may be a novel biomarker of AD. Twenty subjects underwent 18F-AV-45 (florbetapir) positron emission tomography to test this assertion. In addition, the global m6A DNA methylation level was also correlated with cognition level.
- Citation: Lv S, Zhou X, Li YM, Yang T, Zhang SJ, Wang Y, Jia SH, Peng DT. N6-methyladenine-modified DNA was decreased in Alzheimer’s disease patients. World J Clin Cases 2022; 10(2): 448-457
- URL: https://www.wjgnet.com/2307-8960/full/v10/i2/448.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i2.448
Alzheimer’s disease (AD) is the most common form of dementia. It is a progressive neurodegenerative disease with symptoms of initial memory impairment and cognitive decline. Usually, it affects patients’ behavior, speech, visuospatial orientation and motor system[1]. Pathological tau and amyloid-β (Aβ) deposition and neurodegeneration are biomarkers of AD. Studying the biological mechanisms of cognitive symptoms and trajectories of decline is important for clinicians to be able to determine prognosis and apply precision medicine in AD patients[2]. Although the incidence of AD is increasing, the treatment is still limited in preventing, slowing, and stopping the progression of the disease[3].
Cytosine deoxynucleotides in eukaryotic genomic DNA were first found to be methylated 60 years ago[4]. DNA methylation plays a crucial role in epigenetic mechanisms, including the regulation of gene expression, transposon suppression, and epigenetic memory maintenance[5]. Previous studies have shown that 5-methylcytosine DNA methylation is important in epigenetic mechanisms[6]. Because of technical limitations, the presence of N6-methyladenine (m6A) within DNA was not found in eukaryotes in earlier generations of studies, and as such, m6A was believed to be absent from eukaryotic genomes. However, recently, m6A was discovered in unicellular organisms, namely, Caenorhabditis elegans[7], Drosophila[8], zebrafish and mammals[9]. DNA methylation usually refers to the addition of a methyl group (CH3) to any of the four types of DNA nucleotides[10]. When methylations appear on the sixth position of the purine ring of adenine, the resulting modification is called m6A. m6A is abundant in the mammalian brain[11]. In the mammalian central nervous system, stimulus-dependent regulation of m6A was found in response to sensory experiences, learning and injury[12]. A recent study showed that m6A methylation mRNA was lower in 6-month-old familial Alzheimer’s disease mice[13]. However, the study of m6A DNA in AD patients has been less studied.
Apolipoprotein E (APOE) was first proposed as an Aβ-binding protein in the brain[14]. A study showed that normal elderly individuals with APOE ε4 homozygosity (ε4/ε4) and even the ε 4 allele have a very high risk of developing clinical AD[15,16]. APOE not only changes the protein codon but also changes the quantity of CpG dinucleotides, which are the primary sites for DNA methylation[17]. A previous study showed that DNA methylation may have a relationship with APOE and AD[18]. However, there has been no research on the relationship between m6A DNA levels and APOE. Here, we examined the relationship between m6A and APOE. The levels of m6A in patients with AD and normal controls (NCs) were determined to assess whether the m6A DNA level is a new marker of AD that can be used for early detection or diagnosis.
Participants from the Neurology Clinic of China-Japan Friendship Hospital were enrolled from March 2018 to February 2021. Before initiation, the trial was registered at http://www.chinadrugtrials.org.cn/index.html, Unique identifier: CTR20171631. This retrospective study received institutional review board approval (Ethics ID: 2017SY51), and all subjects signed an informed consent form. The participants were indepen
Among them, ten AD patients who were 18F-AV-45 (florbetapir) positron emission tomography (PET) positive (mean age 69.1 ± 10.16 years; males: 4; females: 6) and ten NCs (mean age 68.6 ± 7.95 years; males: 4; females: 6) who were PET negative were included in the subsequent analysis.
A 2-mL peripheral blood sample was obtained from each patient using a standard venipuncture technique. Each sample was centrifuged to separate the plasma and white blood cells. The white blood cells were rinsed with red blood cell lysis buffer (TAKARA, Japan) and then labeled with RNAlater (Thermo, United States). All the samples were stored at -80°C until the next test. According to the manufacturer’s instructions, DNA was isolated from white blood cells using the QIAamp DNA Blood Mini Kit (QIAGEN, Germany). An ND-1000 spectrophotometer (Nanodrop Technologies, Delaware) was used to quantify the DNA samples at 450 nm to ensure that the DNA quantity was sufficient for further experiments. The ratio of the absorbance at 260/280 nm was required to be 1.8-1.9 for the DNA samples. We determined the precise length of genomic DNA by gel electrophoresis using 1% agarose gels. The DNA concentration was corrected to 100 ng/μL, and DNA samples with concentrations less than 100 ng/μL were excluded. Then, the genotyping of the APOE SNPs rs7412 and rs429358 was performed by agarose gel electrophoresis[22].
Global m6A levels in total DNA were measured using the MethylFlash m6A DNA Methylation ELISA Kit (colorimetric) (Epigentek, United States) by adding 200 ng of DNA extracted from human peripheral blood. All the experimental details followed the manufacturer’s instructions. The absolute amount of m6A in each sample was calculated by using a standard curve generated by plots of the absorbance of the positive and negative controls. m6A% indicates the ratio of m6A to total DNA.
DNA that was previously corrected to 100 ng/μL before was spotted onto a nylon membrane (Bio-Rad, United States), with 1 μL of DNA in each sample, and allowed to air dry. DNA was ultraviolet (UV) crosslinked to the membrane, and the membranes were blocked for 1 h in 3% nonfat dry milk in 0.1% PBS (blocking buffer) at room temperature[23,24]. Then, the cells were washed with Tween-TBS (Solarbio, China) for 10 min three times. The membranes were detected by anti-m6A antibody (1:200 dilution, Abcam, United Kingdom) in 3% milk TBS at 4 °C overnight and washed three times with Tween-TBS for 10 min each time. The membranes were detected with anti-mouse IgG secondary antibodies (1:10000 dilution, Easybio, China) for 1 h at room temperature. The visual blots were finally captured using the ECL Imaging System (Merck Millipore, United States). The signals were analyzed with Fiji ImageJ software.
We first evaluated whether the data were normally distributed. Comparisons of two groups, such as the analysis of differences in baseline characteristics between the AD patients and NCs, involved independent-samples Mann-Whitney U-tests (unpaired). The data were expressed as the mean ± standard deviation (SD) if the variance between groups was similar. The analysis of the relationships with APOE genotypes was performed by the chi-square test. When the expected count was less than 5, the Fisher’s chi-square test was used instead of the chi-square test. Spearman analysis was used to assess correlations. The associations between clinical and biological characteristics and m6A DNA levels were evaluated through linear and multivariate regression analyses with adjustment for age and sex. Medians and interquartile ranges (IQRs) are reported for non-Gaussian distributed variables. All statistical analyses in our study were performed with Statistical Package for Social Sciences (SPSS) version 20 (Armonk, United States). Two-tailed P < 0.05 was considered to indicate a significant difference in all statistical analyses.
We determined the global m6A DNA level in peripheral blood samples from 179 AD patients and 147 NCs (shown in Figure 1). Our results showed no differences in terms of age, sex, education, body mass index, systolic blood pressure, diastolic blood pressure, smoking and drinking habits between the AD and NC groups. The raw data are shown in Table 1. Figure 1 shows that the leukocyte m6A levels were different in patients with AD and NCs. Our study showed that the m6A level was approximately 8.33% lower in the AD patients than in the NCs (mean 0.011 ± 0.006 vs 0.012 ± 0.005). Multivariate regression analysis further confirmed that the m6A level had a positive correlation with the occurrence of AD after adjustment for age and sex (P ≤ 0.01). Thus, we found that reduced leukocyte m6A DNA levels were associated with AD.
| Total (326) | AD (179) | NC (147) | P value | ||
| Age (yr) | 73 (64, 78) | 74 (68, 78) | 72 (62, 79) | 0.165 | |
| Sex (male/female) | 168/158 | 91/88 | 77/70 | 0.781 | |
| m6A% | 0.010% (0.008%, 0.014%) | 0.010% (0.006%, 0.013%) | 0.011% (0.009%, 0.014%) | 0.002a | |
| MoCA score | 24 (18, 26) | 18 (14, 21) | 26 (25, 28) | 0.001a | |
| Education (yr) | 12 (9, 15) | 12 (9, 15) | 11 (9, 15) | 0.435 | |
| BMI (kg/cm2) | 22.29 (20.16, 24.51) | 22.27 (20.03, 24.58) | 22.31 (20.32, 24.62) | 0.460 | |
| Smoking (%) | 69 (21.66) | 31 (17.32) | 38 (25.85) | 0.061 | |
| Alcohol (%) | 107 (32.82) | 56 (31.28) | 51 (34.69) | 0.514 | |
| SBP (mmHg) | 129 (115, 140) | 127 (117, 138) | 130 (113, 143) | 0.434 | |
| DBP (mmHg) | 77(69, 83) | 75 (68, 83) | 78 (69, 84) | 0.265 | |
| APOE (%) | ε2/2 | 1 (0.31) | 0 (0) | 1 (0.68) | |
| Allele (%) | ε2/3 | 26 (7.98) | 12 (6.70) | 14 (9.52) | |
| ε3/3 | 195 (59.82) | 98 (43.58) | 97 (65.99) | ||
| ε3/4 | 89 (27.30) | 57 (31.84) | 32 (21.77) | ||
| ε4/4 | 15 (4.60) | 12 (6.70) | 3 (2.04) | ||
We further verified the relationship between leukocyte m6A DNA levels and AD through dot blotting. Ten AD patients who were PET positive and ten NCs who were PET negative were age- and sex-matched. There were significant differences between the two groups, which indicated that the leukocyte m6A DNA levels were different (P = 0.005; < 0.05, n = 10 people per group) (shown in Figure 2).
In addition, we also analyzed the correlation between the MoCA score and peripheral blood m6A levels and found that there was a significant correlation between the two in the tested population (r = 0.143, P = 0.01; < 0.05) (shown in Figure 3). In addition, the linear regression analysis showed that the two were positively correlated, and a positive correlation still existed after adjustment for sex and age. Thus, the m6A DNA level is associated with cognition.
The APOE genotype was detected by agarose gel electrophoresis. The results for male patients were as follows: ε2/2 (1, 0.60%), ε2/3 (13, 7.74%), ε3/3 (103, 61.31%), ε3/4 (42, 25.00%) and ε4/4 (9, 5.36%); the results for female patients were as follows: ε2/3 (13, 8.23%), ε3/3 (92, 58.23%), ε3/4 (47, 29.75%), and ε4/4 (6, 3.80%). There was no significant difference between APOE ε4 and sex in our study (P = 0.537 > 0.05; 95%CI: 0.727, 1.845). The Kruskal-Wallis test showed that the leukocyte m6A DNA level was not associated with APOE carrying ε4 (including ε4/4 and ε3/4) or not carrying ε4 (including ε2/3 and ε3/3) (P = 0.633, > 0.05). It has been shown that APOE ε4 confers greater AD risk in females than in males[25]. Therefore, we studied APOE ε4 further in the female participants. In the female participants, the m6A level was also not associated with APOE in the participants carrying ε3 (including ε2/3 and ε3/3) or in the participants carrying ε4 (including ε4/4 and ε3/4) (P = 0.425, > 0.05). Thus, in our study, the m6A DNA level was not associated with APOE.
DNA methylation can affect many biological processes by changing DNA structure and topology. Recent studies have demonstrated that m6A, a novel modified form of adenine in DNA, may function as an epigenetic biomarker of DNA modification preserved in prokaryotes and eukaryotes[26]. m6A significantly affects DNA replication, repair, virulence, and gene regulation[27]. It can also be used to distinguish host DNA from foreign DNA and other foreign nucleic acid elements, which is important for prokaryotic immunity[28]. However, the occurrence and biological effects of m6A methylation are still poorly understood[29]. Therefore, we analyzed whether m6A had any effect in AD. Liu et al[9] showed that m6A accounts for up to 0.1%-0.2% of total adenines during early embryogenesis in zebrafish and pigs, but during embryo development, the m6A level is relatively low. Stephen J Mondo et al[30] showed that the high m6A level present in early-diverging fungal lineages is related to transcriptionally active genes, and the percentage of methylated adenines can be as high as 2.8% of all adenines. M6A is associated with not only nervous system development, but also neurodegenerative diseases. To our knowledge, no study has evaluated m6A DNA methylation between NCs and AD patients. In our study, we found that global m6A DNA methylation levels were higher in NCs than in AD patients. We demonstrated this result through not only a MethylFlash m6A DNA Methylation ELISA Kit but also dot blotting. The m6A level was significantly lower in the AD patients than in the NCs by approximately 8.33% (mean 0.011 ± 0.005 vs 0.012 ± 0.005). The dot blot results revealed that the number of NCs who were PET negative was significantly higher than the number of AD patients who were PET positive. Therefore, we speculate that m6A can be used as a new marker of AD for early detection and diagnosis.
Memory loss and cognitive impairment are the main clinical features of AD patients[1]. Next, we explored the relationship between the MoCA score and the m6A level because the MoCA is widely used to screen for dementia[31]. In clinical work, the MoCA is also used to assess the severity of cognitive impairment[32]. In our study, we showed that there was a positive correlation between the MoCA score and the m6A level, indicating that there may be a positive correlation between m6A and cognitive function. This result further validates our hypothesis that m6A is associated with AD. Chen et al[33] also suggested that m6A methylation may be associated with cognitive dysfunction. Deng et al[34] found that m6A reader protein (insulin-like growth factor 2 mRNA binding protein 2) was abnormally highly expressed in AD patients. The APOE ε4 allele is the best-characterized amyloid-β (Aβ) chaperone and is related to Aβ metabolism and tau phosphorylation[35]. ε4 carriers have brain structural and developmental abnormalities (e.g., lower cortical gray matter volume in regions particularly affected by AD) that, together with functional features (e.g., deficient neuronal maintenance and repair), increase their vulnerability to neuropathological changes and subsequent late-life cognitive decline. ε4 allele insertion in mice causes tau accumulation[36]. A randomized trial showed that the amelioration of cognitive function among people aged over 65 years may occur through reducing the Ca:Mg ratio, which is mediated by reductions in 5-mC levels in APOE[37]. However, the biological mechanisms through which the ε4 allele contributes to disease pathophysiology are incompletely understood. Therefore, we hypothesized that APOE would also be related to the m6A level. However, no relationship was found (P = 0.633; > 0.05). Another study showed that compared with males, females have a higher risk of AD[38]. Thus, we further assessed whether APOE allele status had any relationship with m6A levels in females. However, no relationship was found in the female subgroup or the total group. Some limitations of our study should be noted. First, the sample size of the study was small. In addition, we did not conduct a large sample size or conduct a multicenter study, which may have caused bias in the results, such as gender bias and age bias. We concluded that the m6A level was correlated with the overall level of cognition but did not further analyze the correlation between the m6A level and various aspects of cognition (e.g., memory, executive function, visual space). Further studies are required to validate these findings.
The above study and analysis showed that the m6A level was significantly correlated with the incidence of AD. We conducted a linear regression analysis to determine the relationship between the m6A level and AD, which showed a positive correlation. The m6A level was approximately 8.33% lower in AD patients than in NCs. We will further study the effect of the m6A level on the pathological mechanisms of AD to elucidate its role in the disease.
Alzheimer’s disease (AD) is the most common form of dementia and places a large burden on both society and family members. Extracellular senile plaques composed of amyloid-beta (Aβ) peptide and intracellular tau-containing neurofibrillary tangles in the brain are the classical view of AD pathogenesis.
Currently, targeting Aβ and tau-containing neurofibrillary tangles fails to stop the progression of AD. Studies have shown that early diagnosis and treatment are beneficial for improving the prognosis of AD patients. Thus, it is important to identify AD biomarkers.
This study aimed to determine the relationship between the novel m6A DNA and cognition in patients with AD and normal controls (NCs) in China. Complete the biomarkers of AD in clinical.
The study included 179 AD patients and 147 NCs who were age- and sex-matched. All of them underwent neuropsychological scale assessment and magnetic resonance imaging examination. Blood samples were obtained from each subject to analyze apolipoprotein E (APOE) genotypes and global m6A levels. Global m6A levels were evaluated by a MethylFlash m6A DNA ELISA Kit (colorimetric). In addition, m6A levels from ten AD patients with 18F-AV-45 (florbetapir) positron emission tomography (PET) positivity and ten NCs with PET negativity were analyzed by dot blotting.
The study showed that the m6A level was approximately 8.33% lower in AD patients than in NCs. Multivariate regression analysis further confirmed that the m6A level had a positive correlation with the occurrence of AD (P ≤ 0.01). The correlation between the MoCA score and peripheral blood m6A levels revealed that there was a significant correlation between the two in the tested population (r = 0.143, P = 0.01; < 0.05). However, m6A levels were not associated with APOE.
The study showed that leukocyte m6A DNA levels are associated with AD and MoCA scores. Global m6A DNA methylation levels are significantly lower in AD patients than in NCs.
We will further analyze the correlation between the m6A level and various aspects of cognition, such as memory and executive function. A further study will be performed to elucidate the effect of the m6A level on the pathological mechanisms of AD.
We thank our colleagues at Peking University, Graduate School of Peking Union Medical College and Chinese Academy of Medical Sciences, Capital Medical University and China-Japan Friendship Hospital. We thank all the staff who helped us during the study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Neurosciences
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sachu A, Toledano A S-Editor: Wu YXJ L-Editor: A P-Editor: Wu YXJ
| 1. | DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer's disease. Mol Neurodegener. 2019;14:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1846] [Cited by in RCA: 1893] [Article Influence: 315.5] [Reference Citation Analysis (0)] |
| 2. | Ten Kate M, Dicks E, Visser PJ, van der Flier WM, Teunissen CE, Barkhof F, Scheltens P, Tijms BM; Alzheimer’s Disease Neuroimaging Initiative. Atrophy subtypes in prodromal Alzheimer's disease are associated with cognitive decline. Brain. 2018;141:3443-3456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 111] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 3. | Im JJ, Jeong H, Bikson M, Woods AJ, Unal G, Oh JK, Na S, Park JS, Knotkova H, Song IU, Chung YA. Effects of 6-month at-home transcranial direct current stimulation on cognition and cerebral glucose metabolism in Alzheimer's disease. Brain Stimul. 2019;12:1222-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 116] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 4. | ARBER W, DUSSOIX D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J Mol Biol. 1962;5:18-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 308] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 5. | von Meyenn F, Iurlaro M, Habibi E, Liu NQ, Salehzadeh-Yazdi A, Santos F, Petrini E, Milagre I, Yu M, Xie Z, Kroeze LI, Nesterova TB, Jansen JH, Xie H, He C, Reik W, Stunnenberg HG. Impairment of DNA Methylation Maintenance Is the Main Cause of Global Demethylation in Naive Embryonic Stem Cells. Mol Cell. 2016;62:848-861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 151] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 6. | Saghafinia S, Mina M, Riggi N, Hanahan D, Ciriello G. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep. 2018;25:1066-1080.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 238] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 7. | Masiello I, Biggiogera M. Ultrastructural localization of 5-methylcytosine on DNA and RNA. Cell Mol Life Sci. 2017;74:3057-3064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 8. | Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizábal-Corrales D, Hsu CH, Aravind L, He C, Shi Y. DNA Methylation on N6-Adenine in C. elegans. Cell. 2015;161:868-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 503] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 9. | Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J, Li C, Liu B, Luo Y, Zhu Y, Zhang N, He S, He C, Wang H, Chen D. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161:893-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 466] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 10. | Liu J, Zhu Y, Luo GZ, Wang X, Yue Y, Zong X, Chen K, Yin H, Fu Y, Han D, Wang Y, Chen D, He C. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat Commun. 2016;7:13052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 184] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 11. | Liang Z, Shen L, Cui X, Bao S, Geng Y, Yu G, Liang F, Xie S, Lu T, Gu X, Yu H. DNA N6-Adenine Methylation in Arabidopsis thaliana. Dev Cell. 2018;45:406-416.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 171] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 12. | Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-1646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2265] [Cited by in RCA: 3191] [Article Influence: 245.5] [Reference Citation Analysis (0)] |
| 13. | Shafik AM, Zhang F, Guo Z, Dai Q, Pajdzik K, Li Y, Kang Y, Yao B, Wu H, He C, Allen EG, Duan R, Jin P. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer's disease. Genome Biol. 2021;22:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 170] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 14. | Widagdo J, Zhao QY, Kempen MJ, Tan MC, Ratnu VS, Wei W, Leighton L, Spadaro PA, Edson J, Anggono V, Bredy TW. Experience-Dependent Accumulation of N6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J Neurosci. 2016;36:6771-6777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 183] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 15. | Wisniewski T, Frangione B. Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci Lett. 1992;135:235-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 571] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 16. | Hanseeuw BJ, Betensky RA, Jacobs HIL, Schultz AP, Sepulcre J, Becker JA, Cosio DMO, Farrell M, Quiroz YT, Mormino EC, Buckley RF, Papp KV, Amariglio RA, Dewachter I, Ivanoiu A, Huijbers W, Hedden T, Marshall GA, Chhatwal JP, Rentz DM, Sperling RA, Johnson K. Association of Amyloid and Tau With Cognition in Preclinical Alzheimer Disease: A Longitudinal Study. JAMA Neurol. 2019;76:915-924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 312] [Cited by in RCA: 579] [Article Influence: 96.5] [Reference Citation Analysis (0)] |
| 17. | Foraker J, Millard SP, Leong L, Thomson Z, Chen S, Keene CD, Bekris LM, Yu CE. The APOE Gene is Differentially Methylated in Alzheimer's Disease. J Alzheimers Dis. 2015;48:745-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 18. | Jiang L, Lin H, Alzheimer's Disease Neuroimaging Initiative, Chen Y. Sex difference in the association of APOE4 with cerebral glucose metabolism in older adults reporting significant memory concern. Neurosci Lett. 2020;722:134824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Gamst A, Holtzman DM, Jagust WJ, Petersen RC, Snyder PJ, Carrillo MC, Thies B, Phelps CH. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270-279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8107] [Cited by in RCA: 7488] [Article Influence: 534.9] [Reference Citation Analysis (0)] |
| 20. | Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, Holtzman DM, Jagust W, Jessen F, Karlawish J, Liu E, Molinuevo JL, Montine T, Phelps C, Rankin KP, Rowe CC, Scheltens P, Siemers E, Snyder HM, Sperling R; Contributors. NIA-AA Research Framework: Toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535-562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3730] [Cited by in RCA: 6648] [Article Influence: 1108.0] [Reference Citation Analysis (1)] |
| 21. | Li Y, Kang M, Sheng C, Chen G, Li T, Wang J, Cai Y, Wang R, Han Y. Relationship between Urinary Alzheimer-Associated Neuronal Thread Protein and Apolipoprotein Epsilon 4 Allele in the Cognitively Normal Population. Neural Plast. 2020;2020:9742138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Sáiz PA, Morales B, G-Portilla MP, Alvarez V, Coto E, Fernández JM, Bousoño M, Bobes J. Apolipoprotein E genotype and schizophrenia: further negative evidence. Acta Psychiatr Scand. 2002;105:71-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 23. | Qian JY, Gao J, Sun X, Cao MD, Shi L, Xia TS, Zhou WB, Wang S, Ding Q, Wei JF. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene. 2019;38:6123-6141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 154] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 24. | Couturier M, Lindås AC. The DNA Methylome of the Hyperthermoacidophilic Crenarchaeon Sulfolobus acidocaldarius. Front Microbiol. 2018;9:137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 25. | Xiong J, Ye TT, Ma CJ, Cheng QY, Yuan BF, Feng YQ. N 6-Hydroxymethyladenine: a hydroxylation derivative of N6-methyladenine in genomic DNA of mammals. Nucleic Acids Res. 2019;47:1268-1277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 26. | Koh CWQ, Goh YT, Toh JDW, Neo SP, Ng SB, Gunaratne J, Gao YG, Quake SR, Burkholder WF, Goh WSS. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 2018;46:11659-11670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 27. | HOTCHKISS RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175:315-332. [PubMed] |
| 28. | Chen H, Shu H, Wang L, Zhang F, Li X, Ochola SO, Mao F, Ma H, Ye W, Gu T, Jiang L, Wu Y, Wang Y, Kamoun S, Dong S. Phytophthora methylomes are modulated by 6mA methyltransferases and associated with adaptive genome regions. Genome Biol. 2018;19:181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 29. | Mondo SJ, Dannebaum RO, Kuo RC, Louie KB, Bewick AJ, LaButti K, Haridas S, Kuo A, Salamov A, Ahrendt SR, Lau R, Bowen BP, Lipzen A, Sullivan W, Andreopoulos BB, Clum A, Lindquist E, Daum C, Northen TR, Kunde-Ramamoorthy G, Schmitz RJ, Gryganskyi A, Culley D, Magnuson J, James TY, O'Malley MA, Stajich JE, Spatafora JW, Visel A, Grigoriev IV. Widespread adenine N6-methylation of active genes in fungi. Nat Genet. 2017;49:964-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 240] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 30. | Folstein MF, Folstein SE, McHugh PR. "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56757] [Cited by in RCA: 60706] [Article Influence: 1214.1] [Reference Citation Analysis (0)] |
| 31. | Jiao F, Yi F, Wang Y, Zhang S, Guo Y, Du W, Gao Y, Ren J, Zhang H, Liu L, Song H, Wang L. The Validation of Multifactor Model of Plasma Aβ 42 and Total-Tau in Combination With MoCA for Diagnosing Probable Alzheimer Disease. Front Aging Neurosci. 2020;12:212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 32. | Zukotynski K, Gaudet V, Kuo PH, Adamo S, Goubran M, Scott CJM, Bocti C, Borrie M, Chertkow H, Frayne R, Hsiung R, Laforce R Jr, Noseworthy MD, Prato FS, Sahlas DJ, Smith EE, Sossi V, Thiel A, Soucy JP, Tardif JC, Black SE. The Use of Random Forests to Identify Brain Regions on Amyloid and FDG PET Associated With MoCA Score. Clin Nucl Med. 2020;45:427-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Chen H, Gao S, Liu W, Wong CC, Wu J, Liu D, Gou H, Kang W, Zhai J, Li C, Su H, Wang S, Soares F, Han J, He HH, Yu J. RNA N6-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m6A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology. 2021;160:1284-1300.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 196] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 34. | Deng Y, Zhu H, Xiao L, Liu C, Liu YL, Gao W. Identification of the function and mechanism of m6A reader IGF2BP2 in Alzheimer's disease. Aging (Albany NY). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 35. | Bennett RE, Esparza TJ, Lewis HA, Kim E, Mac Donald CL, Sullivan PM, Brody DL. Human apolipoprotein E4 worsens acute axonal pathology but not amyloid-β immunoreactivity after traumatic brain injury in 3xTG-AD mice. J Neuropathol Exp Neurol. 2013;72:396-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Zhu X, Borenstein AR, Zheng Y, Zhang W, Seidner DL, Ness R, Murff HJ, Li B, Shrubsole MJ, Yu C, Hou L, Dai Q. Ca:Mg Ratio, APOE Cytosine Modifications, and Cognitive Function: Results from a Randomized Trial. J Alzheimers Dis. 2020;75:85-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 37. | Arnold M, Nho K, Kueider-Paisley A, Massaro T, Huynh K, Brauner B, MahmoudianDehkordi S, Louie G, Moseley MA, Thompson JW, John-Williams LS, Tenenbaum JD, Blach C, Chang R, Brinton RD, Baillie R, Han X, Trojanowski JQ, Shaw LM, Martins R, Weiner MW, Trushina E, Toledo JB, Meikle PJ, Bennett DA, Krumsiek J, Doraiswamy PM, Saykin AJ, Kaddurah-Daouk R, Kastenmüller G. Sex and APOE ε4 genotype modify the Alzheimer's disease serum metabolome. Nat Commun. 2020;11:1148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 132] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 38. | Shao Y, Shaw M, Todd K, Khrestian M, D'Aleo G, Barnard PJ, Zahratka J, Pillai J, Yu CE, Keene CD, Leverenz JB, Bekris LM. DNA methylation of TOMM40-APOE-APOC2 in Alzheimer's disease. J Hum Genet. 2018;63:459-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |