INTRODUCTION
More than 40 years from its first description in the late 1970s[1], occult hepatitis B virus (HBV) infection (OBI) continues to be one of the most challenging topics in the field of viral hepatitis. OBI represents an important public health problem because of its many implications.
Since the introduction of hepatitis B surface antigen (HBsAg) testing in the routine screening of blood donors in the early 1970s, the incidence of transfusion-transmitted hepatitis has been dramatically reduced; however, it did not eliminate this unwanted event, and neither associating alanine aminotransferase level measurement nor anti HBc testing did. The majority of transmissions is attributable to occult hepatitis B. HBV remains the most frequent transfusion-transmitted viral infection[2].
OBI may also be the cause of HBV transmission in organ transplants or can represent a problem in patients receiving immunosuppressive therapy for various conditions, chemotherapy, or anti-CD-20 therapy because of the risk of reactivation.
The persistence of OBI may lead to the development of liver cirrhosis and, eventually, to hepatocellular carcinoma (HCC)[3]. OBI can be one of the possible causes of cryptogenic liver disease.
In people co-infected with hepatitis C virus (HCV) who have OBI, curing HCV with current direct-acting antiviral medication can lead to HBV reactivation, although this happens rarely in patients with OBI, mostly in those with overt HBV infection[4]. The risk of HBV reactivation is also documented in human immunodeficiency virus (HIV) co-infected patients, especially after withdrawal of antiretrovirals that are also active on HBV[5].
SOURCES AND SELECTION CRITERIA
We searched PubMed for studies published in English between January 1979 (when the first reference to OBI was considered to be found) and December 2019. We initially used the following search terms in combination with the term “HBV” or “hepatitis B”: “lifecycle,” “persistence,” “natural history,” “guidelines,” “cccDNA,” “integrated DNA,” “immunity,” “immune system,” “innate immunity,” “adaptive immunity,” “pathogenesis,” “physiopathology,” and then with “OBI HBV” or “Occult hepatitis” + “HBV”: “definition,” “cccDNA,” “mutations,” “HBV variants,” “immune system,” “innate immunity,” “adaptive immunity,” “physiopathology,” “pathogenesis,” “mechanism,” “viral factors,” “host factors,” “epigenetics,” “miRNA.”
We prioritized studies performed in humans, chimpanzees, or humanized chimeric mice, when available, but we also included those performed in other animal models, such as woodchucks and mice or in vitro primary hepatocytes or cell lines. We looked into the most recent sources first, but if data from older sources were still available, we also cited such data. For the best quality of clinical evidence, we prioritized guidelines, technical reviews, or high-quality prospective observational studies or their meta-analyses, when available.
DEFINITION OF OCCULT HEPATITIS B INFECTION
Over time, several definitions have been proposed for OBI.
The first article considered to refer to occult B hepatitis, although it did not identify it by this specific term, dates back to 1979. It retrospectively analyzed sera from 128 donors from 1971 to 1977, who were chosen because their blood recipients developed clinically recognizable posttransfusion hepatitis. “The detection of hepatitis B core antibody (anti-HBc) alone in nine of 29 implicated donors with HBV markers, suggests that some HBsAg-negative donors implicated in the transmission of hepatitis B may be low-level carriers potentially detectable using tests for anti-HBc. However, the total absence of HBV markers in many implicated donors probably indicates that such donors did not transmit HBV infection” was one of that study’s[1] conclusion, and today, we know that their assumptions were not completely correct, although they identified an important category of patients. When we refer to that study, we have to keep in mind that it was a retrospective study; furthermore, the sensitivity of the serological tests used at that time was low, and molecular biology testing was not available.
The definition that resulted from the first Taormina workshop on occult hepatitis B was as follows: "Presence of HBV DNA in the liver (with detectable or undetectable HBV DNA in the serum) of individuals testing HBsAg negative by currently available assays"[6]. The revision made 10 years later in the re-edited workshop was only slightly different: “the presence of replication-competent HBV DNA [i.e., episomal HBV covalently closed circular DNA (cccDNA)] in the liver and/or HBV DNA in the blood of people who test negative for HBsAg by currently available assays”; the latter emphasizes the importance of the fact that HBV DNA should be competent to replicate[7]. There is another change of optics between the two workshops; in the first one, patients with S gene mutations that make HBsAg not detectable by usual commercially available detection assays but with HBV DNA levels comparable to those of the overt infections were called false OBIs[6], whereas in the 2018 workshop, these patients were considered a subset of OBI.
OBI can be seropositive when either the anti-HBc and/or the hepatitis B surface antibody (anti-HBs) is positive (without prior hepatitis B vaccination) or seronegative and when both anti-HBc and anti-HBs are negative. Up to 20% of OBI are seronegative[3,8].
The latest issue of the Asian Pacific Association for the Study of the Liver clinical practice guidelines on the management of hepatitis B[8] and the latest European Association for the Study of the Liver clinical practice guidelines on the management of hepatitis B virus infection[9] recognize occult hepatitis B as a particular form of evolution of HBV infection, characterized by the absence of secreted HBsAg and the presence of HBV DNA either in the liver or in the blood of the patient. American Association for the Study of Liver Diseases (AASLD) guidelines identify a category of patients who test positive for anti-HBc antibodies but negative for HBsAg and, among them, a sub-category of patients who may or may not be HBV DNA positive - these groups may be at risk for reactivation or for developing HCC; however, AASLD guidelines do not define the category specifically as OBI[10].
TYPES OF OBIS
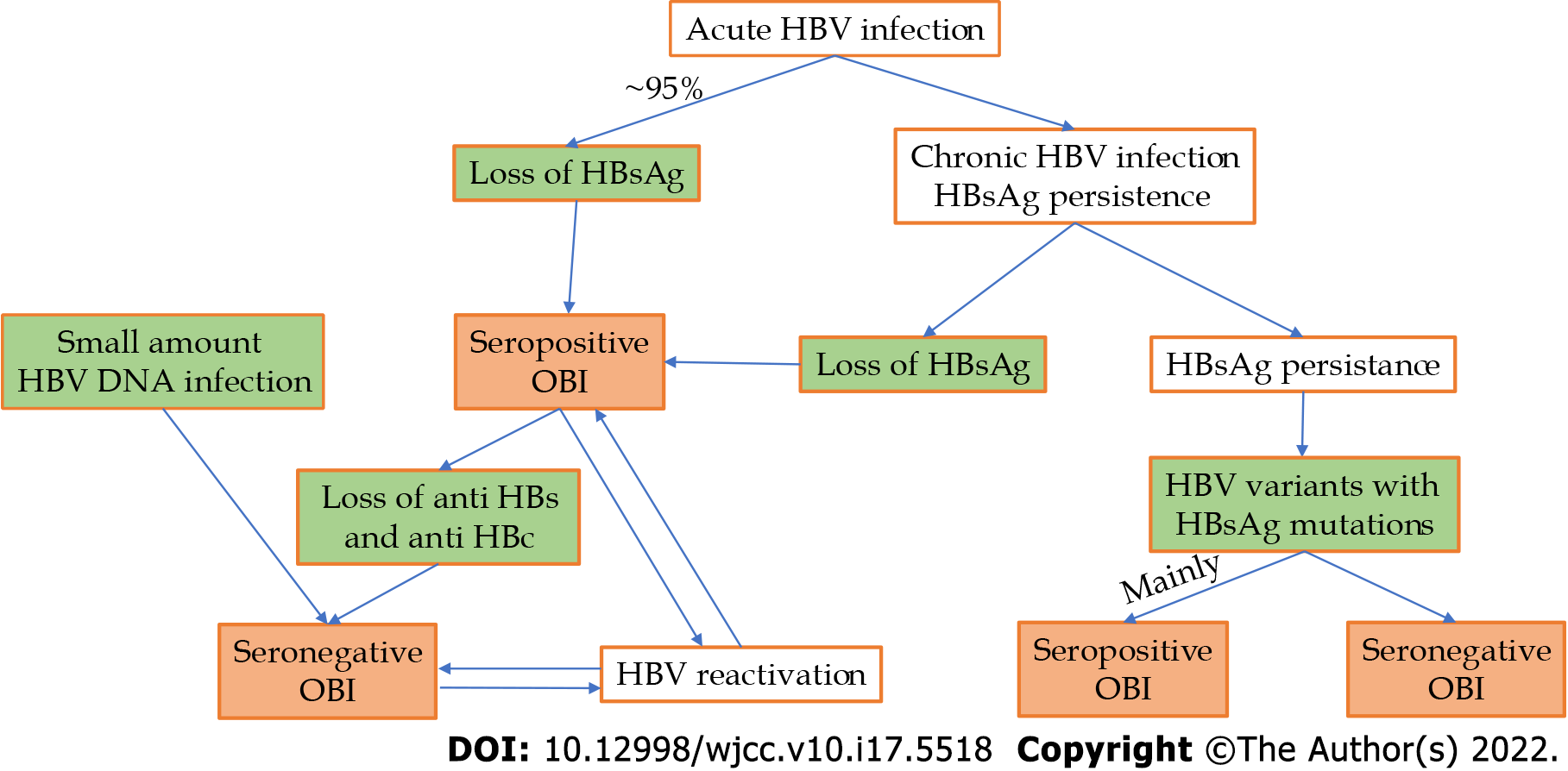
According to the 2019 Taormina workshop[7], OBIs can be categorized mainly into seropositive and seronegative. The seropositive status may be achieved either after the resolution of acute hepatitis (which is the case in more than 95% of immune-competent adults) or after a chronic HBV infection (with or without liver injury), either spontaneously or after antiviral treatment (that with actual substances rarely achieves this functional cure). In these cases, we have to ensure that the most sensitive HBsAg kits are used to rule out false OBIs. HBV DNA can be found intermittently in the blood of these patients, usually at levels below 200 IU/mL[3,7]. The seronegative status may be the consequence of an OBI that progressively lost anti-HBc and anti-HBs antibodies or might be negative from the beginning (a situation called primary occult infection and demonstrated in woodchucks infected with small amounts of viral particles[11]).
A special category of OBI is represented by HBV genetic variants in which the HBs antigen is not recognized by available assays. The main cause of this situation is a mutation in the S-gene (S-escape mutants), but a mutation in the S-gene promoter or a splice variant can also be considered[7]. This type of OBI is mainly seropositive; in a study regarding this issue, out of 99 patients with OBI and mutant HBV variants, only 3 patients were seronegative[3]. At the previous Taormina workshop, this type of OBI was considered false[6]. HBV DNA can have levels comparable with those of the overt HBV infection in this subtype of OBI, except in the one with splice variants, in which HBV DNA levels are low or undetectable[7]. The above pathways to OBI are summarized in Figure 1.
Figure 1 Pathways to different occult hepatitis B infection types.
OBI: Occult hepatitis B infection; HBV: Hepatitis B virus; HBsAg: Hepatitis B surface antigen.
The term “functional cure” refers to OBIs in which HBsAg is not detectable as a result of the immune system’s action and not because of the situations in which HBsAg is present, mutant, and not detected by conventional commercially available assays.
HEPATITIS B VIRUS LIFECYCLE AND PERSISTENCE
HBV is a member of the Hepadnaviridae family, and its complete HBV virion consists of an outer envelope, an inner nucleocapsid, and a 3.2 kb partially double-stranded DNA, known as relaxed circular DNA (RC-DNA), which is covalently connected with the DNA polymerase. The HBV genome contains four overlapping open reading frames, namely, preS1/S2/S, pre-core/core, polymerase, and X domains, which encode seven viral proteins[12]. Of these proteins, four are of major importance: (1) The viral polymerase, that has a role in viral replication and packaging; (2) The small (S), medium (M), and large (L) surface antigens, polypeptides that constitute the HBsAg, that is part of the viral envelope and play a major role in viral entry; (3) HBV core protein (HBc), part of the viral capsid (that play a role in viral replication and packaging); and (4) The X protein (HBx), which has various functions, one of them being the regulation of viral genome transcription. HBx functions may vary with the stage of the HBV infection[12-14].
HBV virions bind initially to hepatocytes by interacting with heparan sulfate proteoglycans for virus docking and subsequently with the recently discovered functional receptor - sodium taurocholate co-transporting polypeptide[13,15]. After endocytosis, the nucleocapsid is released in the cytoplasm and transported to the nucleus, where RC-DNA is released and converted (some say repaired) by host factors into cccDNA[12,14]. When RC-DNA is not completely converted to cccDNA, the aberrant double-stranded linear DNA of HBV can be used for viral integrations into the host genome[16]. The chromatinized cccDNA, which results after a complex multiple-step process, is a mini-chromosome that serves as a template for the pregenomic RNA and subgenomic RNA transcripts, encoding all viral proteins[12-14,17]. The pregenomic RNA is the template for the generation of the progeny HBV RC-DNA, and for this to occur, it has to interact with its own translation products, HBV polymerase, and the core protein, thus forming the nucleocapsid; the latter matures through complex processes and can either be enveloped to form HBV virions or can be re-imported into the nucleus to be converted into cccDNA from its RC-DNA in order to maintain a stable pool of cccDNA[12,14,17]. HBs-coated mature nucleocapsids containing RC-DNA are released from infected cells via host cellular multivesicular body function[14,18] and can infect other hepatocytes. Other sub-viral particles, such as HBsAg, which can be produced in excess, are released by similar pathways as subviral noninfectious HBsAg particles[13,14,18].
After exposure to HBV, over 95% of immune-competent adults can eliminate HBsAg and HBV DNA from circulation, in many cases with HBs and HBe antigen seroconversion (loss of these antigens with the appearance of corresponding antibodies); despite this, HBV DNA can persist in the liver in the form of cccDNA or be integrated in the genome for years or a lifetime. This is called a functional cure and is a type of OBI. By contrast, infections in newborns with HBeAg expressing HBV strains lead to chronic overt infections in over 90% of cases.
The persistence of viable HBV virus particles is maintained through cccDNA or genomic material persistence (integrated HBV DNA fragments). We should keep in mind that laboratory techniques that are used to characterize HBV persistence are not perfect; they lack sensitivity, they may fail in recognizing HBsAg mutant variants, and they cannot distinguish the origin of HBsAg - whether it is from cccDNA or integrated HBV DNA.
HOST IMMUNE RESPONSE TO HBV
Both innate and adaptive immune responses are important in the course of HBV infection.
Innate immunity represents the first line in host defense, playing an important role in the resolution of a viral infection either through its direct activity or by initiating and regulating adaptive immunity. HBV, similar to many other pathogens, is recognized through germline-encoded pattern recognition receptors that are present either on the cell surface or within some intracellular compartments. The activation of these receptors normally triggers the recruitment of different types of adaptor molecules that eventually activate the signaling pathways of nuclear factor-kb and interferon (IFN) regulatory factors. Finally, this leads to the production of interferon-stimulated genes (ISGs) and different inflammatory cytokines, interferons (type I/III IFNs), and chemokines[19,20]. Studies on experimentally infected chimpanzees and human patients with acute infection showed that HBV does not induce type I/III IFNs and also does not significantly increase ISGs, suggesting that the receptors are unable to recognize HBV or that HBV can actively block these pathways[21,22]. In vitro studies (on HBV-infected cultures of human hepatocytes) or studies on liver specimens collected by biopsy from patients with chronic HBV infection showed similar results - ISG levels are not increased compared with those of the controls[23,24]. Therefore, the term stealth virus was coined for HBV[22,23].
Natural killer (NK) cells are also important parts of the innate immune system, and they normally provide a rapid response when viral invasion is recognized. Their activity also seems to be impaired in HBV-infected hosts, and the mechanisms that could be involved include the reduction of the expression of NK group receptors 2D (NKG2D) and 2B4 (NKG2B4) (activating receptors), which consequently reduces NK cells’ capacity to produce INFs and mediate cytotoxicity; the suppression of the expression of the major complexes of histocompatibility class I-related molecules A and B; and the increased expression of T cell immunoglobulin- and mucin-domain-containing molecule-3 in circulating NKs[25,26].
Nevertheless, the innate immune system eventually responds to the presence of HBV through both its components (circulating and intra-hepatic) and its cells; macrophages, monocytes, NK cells, dendritic cells (DC), myeloid-derived suppressor cells, and innate lymphoid cells start producing different signals that will lead to the activation of the adaptive immune system[27]. The adaptive immune system acts mainly through specially developed subsets of T and B cells that are created to recognize and destroy HBV-infected hepatocytes. Prolonged exposure to viral components, such as HBsAg, HBeAg, and HBxAg, leads to immune system exhaustion and downregulation of host response[28].
Regulatory T cells (Tregs) normally play an immune suppressive role by suppressing DCs, NK cells, CD4 cells, and CD8 T cells[27,29]. They perform their role by producing immunosuppressive mediators, IL-10 and TGF-β, and through direct contact[27]. During acute HBV infection, Tregs protect the liver from exceedingly severe immune-mediated liver damage[30]. On the other hand, the same Tregs seem to play a role in promoting chronic infection, as they are found in larger amounts in patients with chronic hepatitis B than in those with acute hepatitis B or HBe-negative HBV infection (formerly inactive carriers).
CD4 T cells play a central role in HBV infection management by manifesting their functions, including the activation of innate immune cells, B cells, and cytotoxic T cells. They promote antibody production and generate signals to attract neutrophiles at the site of infection[31]. CD4 T cells also contribute to the selection and maintenance of HBV-specific CD8 T cells[32]. On the other hand, besides the above-mentioned functions, CD4 T cells are involved in the pathogenesis of HBV chronic infection by producing and promoting inflammation and fibrosis[27]. CD8 T cells are the actual effectors that perform viral clearance by inhibiting viral replication and contributing to the apoptosis of infected liver cells. In patients who achieve a functional cure, a polyclonal and multi-specific HBV CD8 T cell response can be identified, whereas in those with chronic hepatitis B, CD8 T cells display a narrow spectrum of epitopes and a consequently weak response[27,33,34].
B cells are essentially seen as cells that produce antibodies and elements that can differentiate into plasma cells, providing long-term immunity. Lately, a new function of B cells has been identified - the regulatory function. This B subtype is called B regulatory cells (Bregs). Bregs produce IL-10, a mediator that serves as a downregulatory agent for other immune cells and promotes immune tolerance[35]. In HBV-infected patients, Bregs are considered to promote viral replication and liver fibrosis. They could also be responsible for HBV flares by suppressing CD8 cells[36]. The antibody production function of B cells is critical for the management of HBV infection; these cells can produce antibodies against different viral proteins, including but not limited to HBsAg, HBcAg, and HBeAg. HBs antibodies are critical for controlling or preventing viral infection. They are produced by B cells stimulated by specific T helper cells (follicular helpers) mainly through IL-21. Impairment of this chain at any of its links leads to persistent HBV infection[31].
Also of interest are human apolipoprotein B mRNA-editing catalytic polypeptide-like enzymes (APOBECs), essential components of our innate immune system, which can inhibit a wide range of viruses using mainly de-amination processes[37]. By exerting their activity, APOBECs can lead to the selection of HBsAg mutants (e.g., G145R), which are not detectable by some commercially available assays[38] and can produce replication-incompetent transcripts (splice variants), which can also elude some HBsAg detection kits[39]. Furthermore, APOBECs can inhibit DNA-RNA hybridization; they can increase susceptibility to nuclease digestion and decrease protein processing, leading to OBI[37,39].
The interaction of viral factors with the immune system of the host can lead to various outcomes, and the following are the five phases of the natural evolution of HBV infection[10,11]: (1) HBeAg-positive chronic infection (immune tolerance); (2) HBeAg-positive chronic hepatitis (immune clearance/chronic hepatitis); (3) HBeAg-negative chronic infection (inactive carriers); (4) HBeAg-negative chronic hepatitis (chronic hepatitis); and (5) HBsAg-negative phase (OBI).
MECHANISMS THAT LEAD TO OBI
In recent years, significant advances have been made in understanding the HBV lifecycle and the molecular mechanisms leading to the persistence of the virus in the occult state. These factors are mainly related to the host immune system and, to a smaller proportion, to the virus[40]. Some external factors can contribute to the appearance of OBI by interfering either with the host immune system or with the lifecycle of HBV. Of these, notable are HIV and HCV co-infections[3,40]. Furthermore, co-infection with Schistosoma mansoni was found to inhibit HBV replication[39,41].
Viral factors
The factors related to HBV that result in OBIs are mainly related to the situation in which HBsAg is not recognized by available kits because of various mutations of the virus.
HBV variants may show different types of mutations:
In the major hydrophilic region (MHR/aminoacids 99-169) of the S protein. G119R, Q129R, T140I, and D144A are the mutations of this region found in one study, affecting mainly the “a” determinant of MHR (aminoacids 124-147), which contains a cluster of B-cells epitopes[42]. P120T and S143L are other mutations associated with OBI[43]. Another study looked even further and differentiated between OBIs in different genotypes. In this case, sM103, sS113, sS114, sG130, sS132, and sK160 appear specific to OBI with genotype B; sD102 and sW165 appear specific to OBI with genotype C; and sT118, sP135, and sS154 appear common to both genotypes[12]. E2 mutations (E2G/A/V/D) can also influence the detection of HBsAg, leading to OBI[12,44].
In the T-cell epitopes. Positions 41, 44, 48, 93, 96, 97, 171, 175, 176, 178, 185, 190, 207, and 213 are affected and may generate immune-escape variants, with some not being recognized even by the host’s circulating HBs antibodies. These positions are outside the MHR, in the N-terminal and C-terminal regions of the S domain[40,45].
In the pre S1/S2 genomic region. The following pre-S1 mutations were found in one study: F25L, A28T, K57T, del 57-99, P65L, S78N, P89T, N98NK, N98NT, N98I, G102R; the following were found for pre S2: del 9-22, A11T, P36Q, and P54Q[42]. Mutations in this genomic region can affect antigenicity, immunogenicity, cell elimination, and/or expression of HBsAg, leading to the failure of its detection or reducing or even inhibiting the replication and/or secretion of virions and thus having a negative effect on HBsAg detection[46].
In virus regulatory elements. Gene promoter regions are essential sites in DNA recognized by proteins for the downstream processes of replication and transcription. Alterations in these regions can lead to the down- or upregulation of the respective genes[40]. One study showed that a 129 bp in-frame deletion in the S promoter region is associated with reduced levels of middle and small surface protein transcripts, resulting in a marked reduction in the expression of the two proteins. In infections with these mutants, a large amount of surface proteins accumulates inside the hepatocytes[47].
In the core protein. Generally, studies have focused on the mutations occurring in the S and pre-S regions, but at least two studies have shown that core protein mutations can also lead to occult HBV infection. The W62R mutation in the core protein significantly reduces HBcAg and HBeAg production during HBV replication, potentially contributing to the occurrence of OBI[48,49].
Mutations that affect the posttranslational production of virus envelope proteins. For example, N-glycosylation in the position N146 of the S domain in wild-type virus may lead to an escape variant[50].
Mutations that appear as a consequence of treatment with nucleotide/nucleoside analogs and may affect both viral polymerase and S protein[51]. Lamivudine-associated polymerase gene mutations M204I and L180M/M204I, corresponding to sI195M and sW196S in HBsAg, have been shown to be associated with reduced binding to HBs antibodies, and these mutants may not be correctly identified by HBsAg detection kits[39].
Although a greater genetic variability in the S gene of HBV isolated from OBIs was found compared with overt infection, it has also been proven that the majority of OBI patients are not infected with mutant variants, suggesting that the mechanisms of OBI are not mainly viral mutations[40]. MHR variants and, generally, S gene mutants may escape anti-HBs antibodies and may not be recognized by available kits, thus representing a serious health problem because they could infect even vaccinated persons[39]. The same mutants are implicated in reinfections following liver transplantation, despite correct Hepatitis B Immune Globulin (HBIG) prophylaxis. Stopping HBIG administration after the procedure allows the mutant HBV to revert into the wild type, suggesting that HBIG may favor the selection of MHR mutants[52]. In patients who present the reactivation of HBV infection from OBI during or after immunosuppressive therapy, the heterogeneity of reactivated HBV has been reported to be significantly lower than that from HBsAg-positive carriers, suggesting that OBI individuals are infected with HBV populations of low genomic heterogeneity in their liver[53].
RNA alternative splicing is an important posttranscriptional mechanism that enables single genes to produce multiple proteins. RNA splicing contributes to mRNA and protein diversities. It regulates gene expressions, providing an important causative relationship link between genetic variation and disease[54,55]. Splicing has been shown to have a significant effect on gene expression in HBV, and its implication in the occurrence of OBI has been claimed. A G-to-A mutation at position 458 of the surface gene altered the splicing of the S gene mRNA because nucleotide 458 is close to the 5’ splice site of S gene mRNA. The mutation prevents the splicing of the pre-S2/S mRNA from positions 458 to 1305, and the two analyzed patients did not express pre-S2/S mRNA and HBsAg[56]. Another group found another mutation mechanism based on splicing, which is specific to genotype D. They describe an evolutionary branch in which the acceptor site at nucleotide 202 and the donor site at nucleotide 2986 are involved in a splice event, resulting in the loss of the spacer region from the viral polymerase gene while retaining the original reading frame. As a result, polymerase functions are not affected, but the expression of the small, middle, and large surface proteins is. Reduced HBsAg expression in the infection with HBV with this mutation leads to OBI[57].
Despite all the above arguments, it is important to note that most OBI patients are not infected with specific mutants. Mutant populations, especially pre-S/S variants, can be found in people with overt infections. Occult HBV genotypes are more often perfectly able to replicate, and their heterogeneity is similar to those from overt infections. In vitro studies have shown that HBV taken from the host’s environment is going back to the wild type, being able to normally synthesize proteins and replicate. A similar situation is described above regarding post-liver transplantation from OBI donor reactivation of HBV in its wild type[40,51,52].
Host factors
Immune host factors: The first evidence, though indirect, that the host immune system is important in OBI is the possibility of HBV infection reactivation in patients subjected to immune suppression, regardless of the possible virus mutations.
A long-term follow-up study has shown that cytotoxic-T lymphocyte (CTL) response following an acute HBV infection persists for decades after serological recovery. CTL response is directly correlated with the presence of HBV DNA in the serum of these patients[58]. Therefore, it is possible to hypothesize that during the occult phase of the infection, HBV can still synthesize very small amounts of antigens that are not detectible by available kits but are sufficient to maintain an HBV-specific T cell response. This assumption is confirmed by the findings showing that, apart from HBV cccDNA molecules, all viral HBV transcripts (including the pregenomic RNA) can also be detected and quantified in the livers of OBI individuals[40,58]. Some other studies have reported similar vigorous T cell responses in OBI[59,60]. The presence or absence of serologic HBV markers defined two profiles of HBV-specific T-cell responses in occult infection. Anti-HBc-positive patients showed a T-cell response typical of protective memory, with robust in vitro expansion and IFN-γ production by HBV-specific T cells, suggesting that this condition represents a resolved infection with immune-mediated virus control. By contrast, HBV-specific T cells in anti-HBc-negative patients did not readily expand and produce interferon-gamma in vitro, suggesting the possibility of less complete maturation of protective memory[60]. It has been demonstrated that clearance of more than 90% of intrahepatic HBV DNA does not require lysis of HBV-infected hepatocytes, suggesting that some noncytolytic immune responses are critical in the clearance of acute HBV infection. It has also been shown that even HBV cccDNA is susceptible to these noncytolytic mechanisms. A noncytolytic HBsAg-specific T-cell response has been suggested as the potential mechanism for occult HBV infections associated with very low and undetectable levels of HBsAg[39].
One study that examined cytokine expression in OBI compared with chronic HBV hepatitis found that interleukin 2, interleukin 4, and IFN-β responses were low in both situations. The authors also found that significantly lower levels of the soluble form of the anti-apoptotic regulator Fas (sFas) were detected in occult HBV infection than in chronic HBV infection (P = 0.01)[61]. As a marker of apoptotic inhibition, decreased sFas during occult HBV infection would indicate that apoptosis occurs at higher rates in occult compared with chronic HBV infection and, therefore, may contribute to HBsAg clearance and HBV replication downregulation. Another study showed that reduced expression of CXCL12, a chemokine that modulates apoptosis, may play a role in occult HBV infection[62]. Increased apoptosis may thus play a role in the occurrence of OBI[39,61,62]. A more recent study found that in patients with OBI and chronic HCV infections compared with monoinfected (HCV) patients and healthy donors, the levels of TNF-α, IL-10, IL-6, IL-4, and IL-2 were increased[63]. Vitamin D3 and vitamin D receptor (VDR) regulate several cytokines and are important determinants of anti-HBV response. They also modulate HBV loads and HBV protein expression[64,65]. The polymorphisms in the T/T allele of exon 9 of VDR are possibly associated with OBI, and VDR and its functional polymorphisms are likely to be related to the occurrence of OBI in some patients[66].
With regard to antibodies, one study found that positive anti-HBs (≥ 10 mIU/mL) were more frequent in HBsAgNx [ARCHITECT HBsAg NEXT (sensitivity 0.005 IU/mL)]- negative than in HBsAgNx-positive nucleic acid testing yield samples (P = 0.0014), while there was no significant difference for the HBsAgNx-negative vs HBsAgNx-positive OBI samples (P = 0.0748)[42]. HBsAgNx is a “supersensitive” assay and its use in this study has been shown to improve HBsAg detection with 22.6% as compared to standard tests[42]. The masking of HBsAg by anti-HBs has been proposed as one reason for the lack of detection of OBI even if anti-HBs is undetectable[67]. Data from the first study[42] suggest that anti-HBs levels over 300 mIU/mL may affect the detection of samples with extremely low viral loads (median viral load: 4.42 IU/mL) and that the detection of such samples would require at least a 20000-fold excess of HBsAg to reach the detection limit of the HBsAgNx assay. Whether anti-HBs might be a consequence of vaccination in these cases or produced as a normal response to the immunogenic stimulus could be the subject of another discussion.
The physiological function of apolipoprotein B mRNA-editing enzyme catalytic polypeptides is cytidine deamination[68]. The expression of APOBEC3G in cells replicating HBV resulted in a 50-fold reduction in HBV DNA levels. Both deamination-dependent and deamination-independent mechanisms of inhibition of HBV replication have been reported for APOBECs[69]. Both mechanisms have also been implicated in the APOBEC-induced inhibition of HBV replication[70]. APOBEC deamination-dependent activity may lead to HBsAg mutants, as mentioned above[38,39]. IFN-alpha can upregulate APOBEC3A in HBV-infected cells in which HBV core protein mediates the interaction of APOBEC3A with HBV cccDNA, resulting in cytidine deamination, apurinic/apyrimidinic site formation, and, finally, cccDNA degradation[70]. Deamination-independent processes of APOBECs lead to decreased HBV DNA production and to a decrease in HBV protein synthesis[37,38].
Epigenetic host factors: Some of the mechanisms that control HBV transcription or replication can be influenced in some cases by the modification of gene expression rather than by the alteration of the DNA sequence itself. This is called epigenetic modification. Epigenetic modifications can alter the expression pattern of a gene without changing its nucleotide sequence. Many studies have revealed that epigenetic mechanisms are important for the occurrence of OBI[39,71,72].
HBV cccDNA minichromosomes are located in the nucleus of infected hepatocytes and can be associated with histones, such as H1, H2A, H2B, H3, and H4[72], or non-histone proteins (HBV core proteins)[73]. The acetylation status of cccDNA-bound histones H3 and H4 regulates HBV replication, while the recruitment of histone deacetylase 1 correlates with low HBV replication[74]. In the presence of histone deacetylase inhibitors (valproic acid or trichostatin A), high HBV transcript levels and increased HBV replication are correlated with an increase in acetylated histones bound to cccDNA[74]. IFN-α can inhibit cccDNA-based RNA transcription by inducing the hypoacetylation of cccDNA-bound histones. This mechanism could be implicated not only in OBI but also in an active epigenetic long-term control of cccDNA activity after IFN-α therapy[75,76].
Along with histones, HBx protein can be recruited to cccDNA, and an HBx mutant has been shown to induce rapid hypoacetylation of histones, thus reducing HBV pregenomic RNA and HBV regulation[77,78].
Besides acetylation, methylation is another epigenetic mechanism considered to be involved in OBI occurrence. Cytosine-guanine dinucleotide (CpG) methylation in a gene promoter region rich in CpGs (CpG island) acts like a switch, silencing the gene[79]. It was already demonstrated a long time ago that HBV DNA integrated into the host genome is methylated, leading to the loss of HBV core protein in PLC/PRF/5. Methylation of HBV DNA is an epigenetic mechanism that modifies HBV proteins, interferes HBV replication, and impairs HBV virion production, possibly leading to occult HBV infection[80]. Methylation of CpG island 2 in the HBV genome is frequently detected in occult HBV infection[81]. Hypermethylated HBV DNA sequences are often found in HCC patients with occult HBV infection[82].
Emerging data suggest that microRNAs (miRNAs) play vital roles in the occurrence and development of HBV infection, particularly in OBI occurrence. MiR-199a-3p and miR-210 were found to efficiently reduce HBsAg expression, and quantification of HBV DNA by real-time PCR showed that both miRNAs suppressed viral replication[83]. In another study, miR-125a-5p was found to interact with the viral sequence and to suppress HBsAg expression and release[84]. MiR-141 was identified to repress HBV expression, and synthetic miR-141 could also significantly suppress HBV expression and replication by targeting peroxisome proliferator-activated receptor alpha[85]. In a more recent study, miRNAs, including hsa-miR-25-3p, -486-5p, -92a-3p, and -1-3p, showed the ability to distinguish OBI from healthy controls efficiently, with an area under the curve value of 0.874, 0.776, 0.886, and 0.807, respectively. In total, 32 differentially expressed miRNAs were identified between OBI and the healthy controls by miRNA sequencing[86]. Compared with the case of the healthy controls, plasma miR-451a and miR-340-3p were significantly upregulated in OBI, making the authors propose these markers for distinguishing OBI from healthy donors[86].
The above depicted mechanisms that may lead to OBI are summarized in Figure 2.
Figure 2 Mechanisms that produce occult hepatitis B infection.
APOBECs: Apolipoprotein B mRNA-editing catalytic polypeptide-like enzymes; OBI: Occult hepatitis B infection; HBsAg: Hepatitis B surface antigen; cccDNA: Covalently closed circular DNA; miRNA: MicroRNAs.
CONCLUSION
OBI remains one of the most challenging problems in the hepatology field. It is a public health problem and a subject that needs further research in the future. OBI is currently diagnosed using PCR and real-time PCR assays. However, all efforts should be made to exclude false negative HBsAg, and new standardized methods must be developed to correctly identify OBI. Some of the studies mentioned above have found different markers (especially in the miRNA field) that could be used in the future for this purpose. Facts regarding OBI have become clearer in recent years; the factors that determine this outcome are now better understood, with host factors (immune or epigenetic) being identified as seemingly the main contributors. Viral factors are important but account for only a minority of OBIs. Some external factors can contribute to the appearance of OBI by interfering either with the host immune system or with the lifecycle of HBV. Of these, HIV and HCV co-infections are notable. Co-infection with Schistosoma mansoni was also found to inhibit HBV replication. Future research in this domain and increased awareness regarding this topic must be encouraged, as this particular form of evolution of HBV infection is still far from being completely understood and controlled.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: Romania
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): E
P-Reviewer: Gerlich W, Germany; Lee GH, Singapore; Mrzljak A, Croatia S-Editor: Gong ZM L-Editor: A P-Editor: Gong ZM