Published online Jan 7, 2022. doi: 10.12998/wjcc.v10.i1.205
Peer-review started: June 17, 2021
First decision: July 16, 2021
Revised: July 23, 2021
Accepted: November 29, 2021
Article in press: November 29, 2021
Published online: January 7, 2022
Processing time: 195 Days and 18.3 Hours
Herein, we report the genetic, clinical, molecular and biochemical features of two Han Chinese pedigrees with suggested maternally transmitted non-syndromic hearing loss.
To investigate the pathophysiology of hearing loss associated with mitochondrial tRNA mutations.
Sixteen subjects from two Chinese families with hearing loss underwent clinical, genetic, molecular, and biochemical evaluations. Biochemical characterizations included the measurements of tRNA levels using lymphoblastoid cell lines derived from five affected matrilineal relatives of these families and three control subjects.
Three of the 16 matrilineal relatives in these families exhibited a variable seriousness and age-at-onset (8 years) of deafness. Analysis of mtDNA mutation identified the novel homoplasmic tRNAIle 4268T>C mutation in two families both belonging to haplogroup D4j. The 4268T>C mutation is located in a highly conserved base pairing (6U–67A) of tRNAIle. The elimination of 6U–67A base-pairing may change the tRNAIle metabolism. Functional mutation was supported by an approximately 64.6% reduction in the level of tRNAIle observed in the lymphoblastoid cell lines with the 4268T>C mutation, in contrast to the wild-type cell lines. The reduced level of tRNA was below the proposed threshold for normal respiration in lymphoblastoid cells. However, genotyping analysis did not detect any mutations in the prominent deafness-causing gene GJB2 in any members of the family.
These data show that the novel tRNAIle 4268T>C mutation was involved in maternally transmitted deafness. However, epigenetic, other genetic, or environmental factors may be attributed to the phenotypic variability. These findings will be useful for understanding families with maternally inherited deafness.
Core Tip: The 4268T>C mutation is located in a relatively highly conserved base-pairing (6U–67A) of tRNAIle. The elimination of 6U–67A base-pairing may change tRNAIle metabolism. Moreover, the reduced tRNA level is inferior to the proposed threshold to maintain normal respiration in lymphoblastoid cells.
- Citation: Zhao LJ, Zhang ZL, Fu Y. Novel m.4268T>C mutation in the mitochondrial tRNAIle gene is associated with hearing loss in two Chinese families. World J Clin Cases 2022; 10(1): 205-216
- URL: https://www.wjgnet.com/2307-8960/full/v10/i1/205.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i1.205
Hearing loss is a very important health problem, influencing nearly 1 in 700 newborns[1,2]. Loss of hearing may be associated with gene modifications or environmental factors, which include ototoxic drugs such as aminoglycoside antibiotics[3,4]. Mutations of mitochondrial rRNA and tRNA genes have been accredited as one of the most important reasons for sensorineural hearing loss[5-7]. Therefore, the 12S rRNA 1555A>G and 1494C>T mutations account for both aminoglycoside-induced and non-syndromic deafness in a large number of families worldwide. In addition, the 7472insC, 7510T>C, 7511T>C, 7445A>G and 7445A>C mutations in the tRNASer (UCN) gene have been associated with non-syndromic deafness[5,8-11]. The mtDNA mutations such as 1555A>G and 7445A>G usually exhibit incomplete penetrance, as some individuals carrying the mutation (s) have normal hearing[10,11]. In fact, mutations in the GJB2 gene encoding connexin 26 (Cx26) are a common reason for non-syndromic deafness worldwide[12]. There are more than 150 different GJB2 variants that have been identified, which include missense, nonsense and frameshift mutations (The Connexin-deafness homepage, http://davinci.crg.es/deafness). The range of GJB2 mutations vary in different race groups[12,13]. Of those variants, c.35delG may be the most common GJB2-deafness-causing mutation in European people, while c.235delC and c.167delT are the most common variations in East Asian people and Ashkenazi Jewish families, respectively[14,15]. In addition, they exhibit different age-at-onset, severity, and penetrance of hearing impairment for matrilineal intrafamily or interfamily relatives, carrying the same deafness-associated mtDNA mutation (s). Thus, the phenotypic variability and penetrance of deafness associated with these mtDNA mutations may be modulated by other modifying factors, such as environmental factors, nuclear modifier genes and mitochondrial haplotypes.
Mutations such as 7471InsG, 7510T>C, 7445AN>G, 7505T>C and 7511T>C in the tRNASer (UCN) gene, 4295A>G mutation in the tRNAIlet gene, and 12201T>C mutation in the tRNAHis gene[5,8-11], may change tRNA structure and functions, including modification of specific nucleotides, the processing of RNA precursors, amino
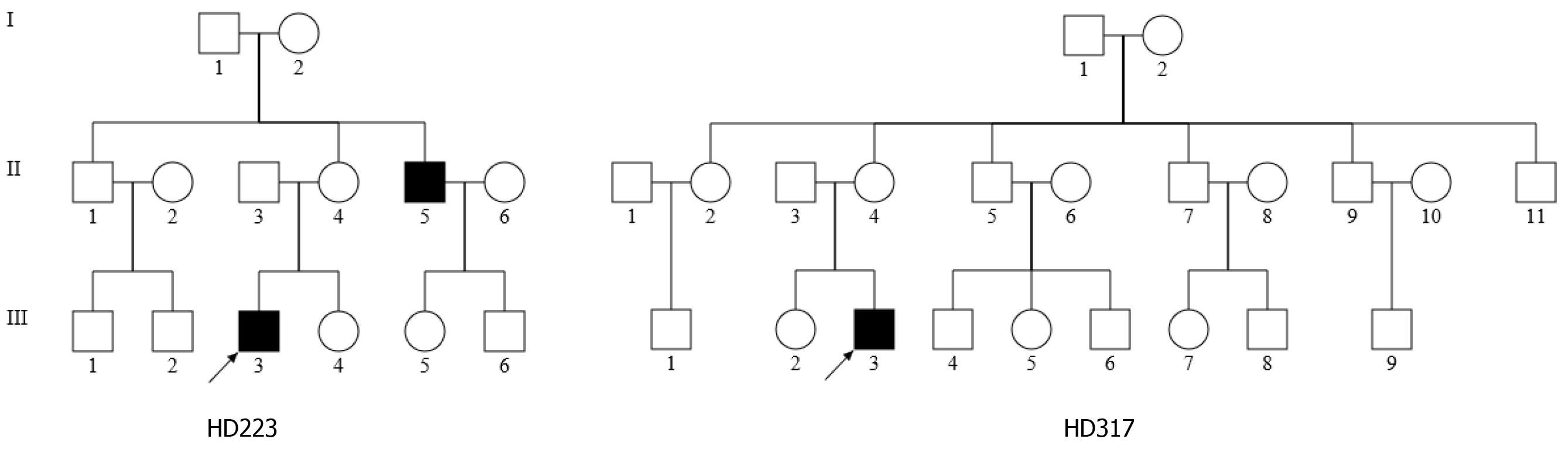
As part of the genetic screening program for hearing impairment, the two Han Chinese families, as shown in Figure 1, were examined at the Otology Department of the First Affiliated Hospital, Zhejiang University School of Medicine. A comprehensive history and physical examinations were performed to identify any syndromic findings, the history of the use of aminoglycosides, and genetic factors related to hearing impairment in members of this pedigree. An age-appropriate audiological examination was performed, which included pure-tone audiometry (PTA) and/or auditory brainstem response (ABR), immittance testing and distortion product otoacoustic emissions (DPOAE). The PTA was calculated from the sum of the audiometric thresholds at 500, 1000, 2000, 4000 and 8000 Hz. The severity of hearing impairment was classified into five grades: Normal < 26 decibel (dB); mild = 26-40 dB; moderate = 41-70 dB; severe = 71-90 dB; and profound > 90 dB. One hundred control DNAs were obtained from a panel of unaffected subjects of Han Chinese ancestry from the same region. Informed consent, blood samples, and clinical evaluations were obtained from all 36 subjects in these two families and 100 controls according to protocols approved by the Ethics Committee of Zhejiang University School of Medicine.
The Puregene DNA isolation kit (Biomega) was used to isolate total genomic DNA from the study participants, after which mitochondrial genomic DNA was assessed via Southern blotting as previously described[16]. A total of 24 overlapping PCR fragments were generated and amplified in order to provide full coverage of the mitochondrial genome, using appropriate pairs of light/heavy strand primers used in previous studies[17]. An ABI 3700 automated DNA sequencer was employed to sequence each of these fragments following purification with a Big Dye Terminator Cycle sequencing reaction kit. The revised Cambridge consensus sequence (GenBank accession number: NC_012920) was used for alignment of the sequenced fragments[16]. Detection of the 4268T>C mutation in family members and other controls was performed as previously described[18].
The DNA fragments spanning the entire coding region of GJB2 gene were amplified by PCR using the following oligodeoxynucleotides: forward: 5′-TATGACACT CCCCAGCACAG-3′ and reverse: 5′-GGGCAATGCTTAAACTGGC-3′. PCR amplification and subsequent sequencing analysis were performed as previously described[18]. The results were compared with the wild-type GJB2 sequence (GenBank Accession No. M86849) to identify the mutations.
The Epstein-Barr virus was used to generate immortalized cell lines from the proband patient (HD223-III-3) bearing the 4268T>C mutation, as well as from a control individual (C2). These cells were cultured in RPMI 1640 containing 10% FBS. Cybrid cells were generated using previous protocols[19]. Briefly, bromodeoxyuridine (BrdU)-resistant 143B.TK− cells were cultured in DMEM containing 5% FBS, and the ρo 206 cell line lacking mtDNA derived from these cells was also grown under these conditions in the presence of 50 μg uridine/mL. The patient and control cell lines were then enucleated and fused with the ρo 206 cells. The resultant cybrid cell lines were selected in uridine-free DMEM supplemented with BrdU, allowing for donor-derived cybrid lines that could then be assessed for the 4268T>C mutation, amounts of mtDNA, and other cellular genetic features. The resultant cybrid lines were maintained in DMEM containing 10% FBS.
TRIzol was used to isolate total mitochondrial RNA from knockdown or control cell lines, as previously described[20]. Briefly, 2 μg of total mitochondrial RNA was electrophoresed through a 10% polyacrylamide/7 M urea gel in Tris–borate–Ethylenediaminetetraacetic acid (EDTA) buffer (after heating the sample at 65 °C for 10 min), and then electroblotted onto a positively charged nylon membrane for the hybridization analysis with oligodeoxynucleotide probes. This was followed by transfer onto a positively charged membrane, which was then combined with appropriate DIG oligodeoxynucleoside probes as previously described[20], using tRNAIle, tRNAThr, tRNATyr, and tRNAAla utilized in previous studies[21].
The MitoSOX Red Mitochondrial Superoxide Indicator (Invitrogen, M36008) was used to assess reactive oxygen species (ROS) production in live cells based on a previously described protocol[22]. Briefly, approximately 2 × 106 cells of each cell line were harvested, resuspended in PBS, supplemented with 100 μM of MitoSOX, followed by incubation at 37 °C for 20 min. The cells were then resuspended. Samples were analyzed by flow cytometer, with an excitation at 488 nm and emission at 529 nm, and 10000 events were analyzed in each sample.
Statistical analysis was performed using the Student’s unpaired, two-tailed t-test in the SPSS version 17.0 program. Unless indicated otherwise, a P value < 0.05 was considered statistically significant.
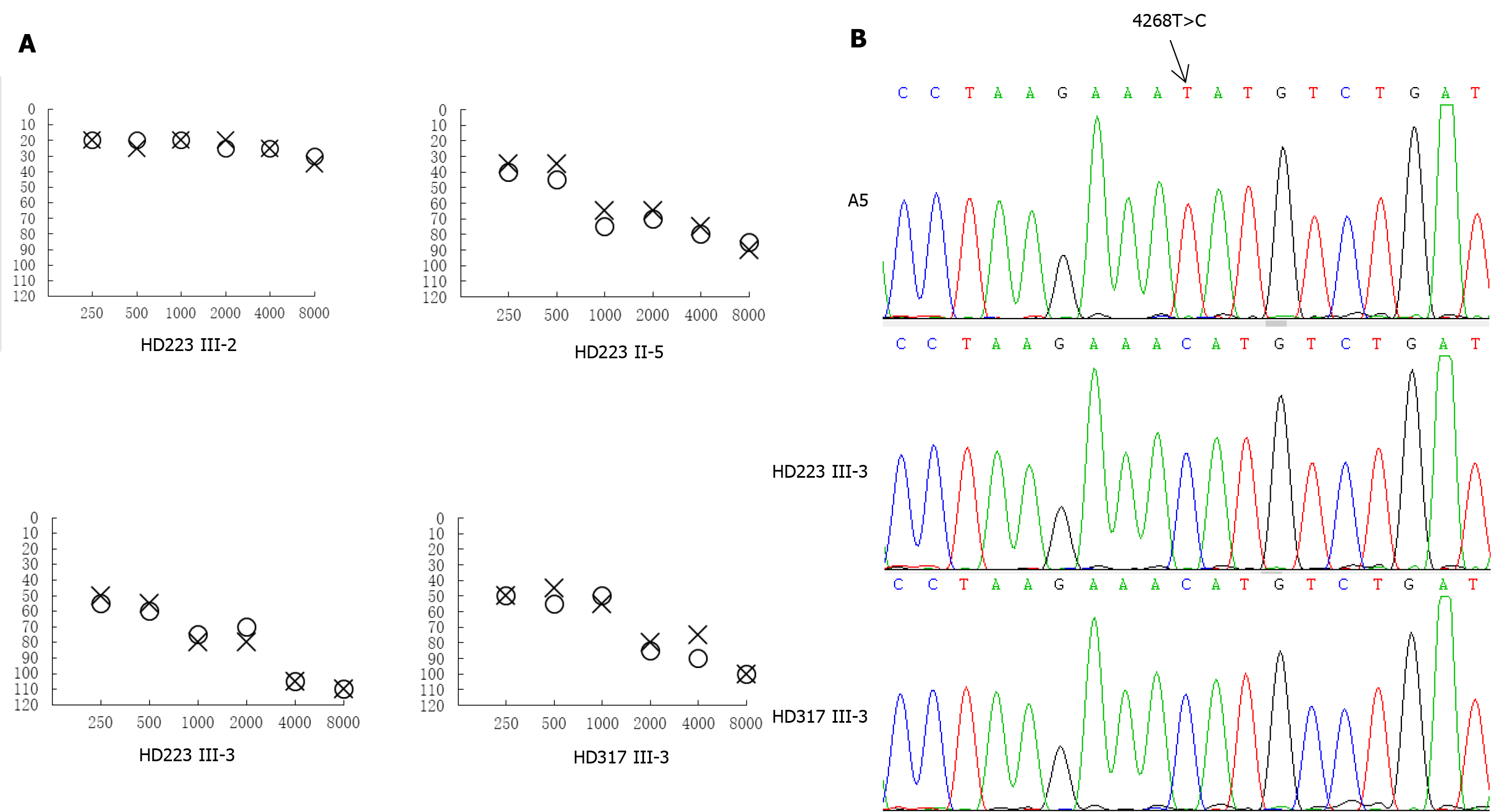
In the family HD223, the female proband (III-3) visited the otology clinic at the age of 28 years. As illustrated in Figure 2, auditory assessment showed that she had moderate-severe hearing loss (71.7 dB in the right ear and 67.5 dB in the left ear), with a sloped-shaped pattern. Moreover, her 48-year-old uncle (II-5) exhibited moderate hearing loss (66 dB and 61 dB in the right and left ears, respectively). Family history and clinical evaluations showed that the other members of this family had no hearing damage.
For the family HD317, however, the proband III-3, was a 22-year-old man, who suffered from bilateral hearing damage since birth. As shown in Figure 2, auditory evaluation showed that he had severe deafness (79.2 dB in the right ear and 80 dB in the left ear), with a sloped-shaped pattern. Other members of this family showed normal hearing.
We sequenced the mitochondrial genome of these two probands to explore potential mitochondrial mutations linked with deafness. Altogether there were 34 evident mutations in the mitochondrial genome compared with the revised Cambridge consensus sequence (NC_012920). The mitochondrial haplotypes of these two patients belonged to the same D4j. These variants were evaluated for pathogenicity using the following criteria: (1) Potential structural and functional alterations; (2) Absence in the controls; (3) Conservation index (CI) from other 16 vertebrates > 75%; (4) Missense mutation; (5) Pedigree analysis. As shown in Table 1, 12 of 34 variants were known silent variants, seven of them were known D-loop variants, nine of them were known missense mutations affecting protein-coding genes, four of them were known 12S rRNA variants, one of them was a known 16S rRNA variant, and one was a novel homoplasmy 4268T>C mutation in the tRNAIle gene (Figure 2). Currently, the detected missense mutations were as follows: 4048G>A (Val260Ile) in the ND1 gene, 8860A>G (Thr112Ala) in the ATP6 gene, 9612G>C (Val136Leu) in the mitochondrial CO3 gene, 11696G>A (Val313Ile) in the mitochondrial ND4 gene, 13708G>A (Ala458Thr), 14766C>T (Thr7Ile) and m.15326A>G (Thr>Ala) in the mitochondrial CYTB gene, 13928G>C (Ser531Thr) and 14002A>G (Thr556Ala) in the ND5 gene. The difference in these mutated RNA residues was compared across 16 different primate species, which showed that the tRNAIle 4268T > C mutation had a 100% CI across species, increasing its likelihood of functional significance after mutation, as shown in the patient. Moreover, we did not detect this mutation when 20 unrelated other patients and 100 Chinese control subjects were assessed.
| Gene | Position | Replacement | AA Position | Conservation (H/B/M/X)a | CRS | A23 | A27 | Previously reportedb |
| D-loop | 73 | A to G | A | G | G | Yes | ||
| 195 | T to C | C | C | C | Yes | |||
| 249 | Del A | A | Del A | Del A | Yes | |||
| 263 | A to G | A | G | G | Yes | |||
| 309 | C to CC | C | CC | CC | Yes | |||
| 310 | T to CTC | T | CTC | CTC | Yes | |||
| 16304 | T to C | T | C | C | Yes | |||
| 12S rRNA | 750 | A to G | A/A/A/- | A | G | G | Yes | |
| 1005 | T to C | T/T/T/T | T | C | Yes | |||
| 1438 | A to G | A/A/A/G | A | G | G | Yes | ||
| 1824 | T to C | T/A/A/A | T | C | C | Yes | ||
| 16S rRNA | 2706 | A to G | A/G/A/A | A | G | G | Yes | |
| ND1 | 3970 | C to T | C | T | T | Yes | ||
| 4084 | G to A | Val260Ile | V/M/T/M | G | A | A | Yes | |
| 4216 | G to A | G | A | Yes | ||||
| tRNAIle | 4268 | T to C | T/T/T/T | T | C | C | No | |
| ND2 | 4769 | A to G | A | G | G | Yes | ||
| 6392 | T to C | T | C | C | Yes | |||
| CO1 | 7028 | C to T | C | T | T | Yes | ||
| 7828 | A to G | A | G | G | Yes | |||
| A6 | 8860 | A to G | Thr112Ala | T/A/A/T | A | G | G | Yes |
| CO3 | 9612 | G to C | Val136Leu | V/V/V/V | G | C | C | Yes |
| ND3 | 10310 | G to A | G | A | A | Yes | ||
| ND4L | 10535 | T to C | T | C | C | Yes | ||
| 10586 | G to A | G | A | A | Yes | |||
| ND4 | 11696 | G to A | Val313Ile | V/T/T/M | G | A | A | Yes |
| 11719 | G to A | G | A | A | Yes | |||
| ND5 | 13708 | G to A | Ala458Thr | S/T/S/T | G | A | A | Yes |
| 13928 | G to C | Ser531Thr | L/S/N/T | G | C | C | Yes | |
| 14002 | A to G | Thr556Ala | T/I/I/S | A | G | G | Yes | |
| ND6 | 14560 | G to A | G | A | Yes | |||
| Cytb | 14766 | C to T | Thr7Ile | T/S/T/S | C | T | T | Yes |
| 15326 | A to G | Thr194Ala | T/M/I/I | A | G | G | Yes | |
| 15862 | T to C | T | C | Yes |
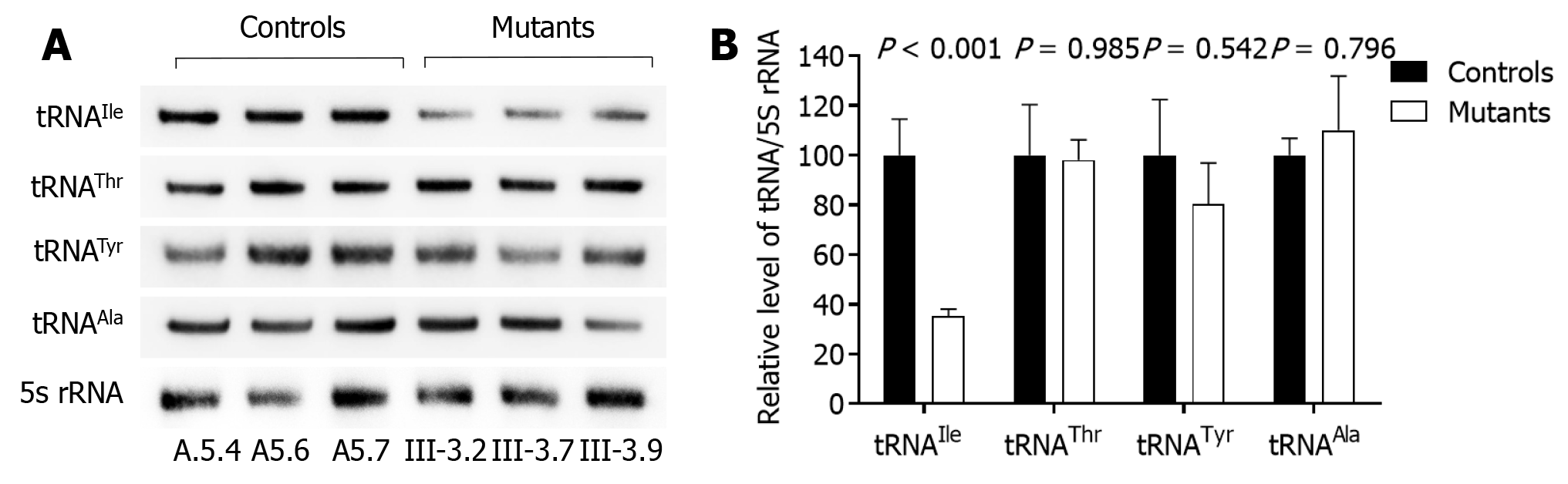
We examined how the 4268T>C mutation changed the metabolism of tRNAIle, which bears this mitochondrial mutation by subjecting cybrid cell lines to Northern blotting with probes specific to it and three other tRNAs. As shown in Figure 3, tRNAIle levels in the mutant cybrid lines were obviously reduced compared with control wild-type cells, with baseline tRNAIle levels in the mutant cells about 64.6% compared to those in control cells. The 5S RNA was applied for normalization. As a contrast, baseline tRNAIle, tRNATyr, tRNAThr, and tRNAAla levels in the mutant cell lines were consistent with control cells (98.0, 80.4, and 110.1%, respectively).
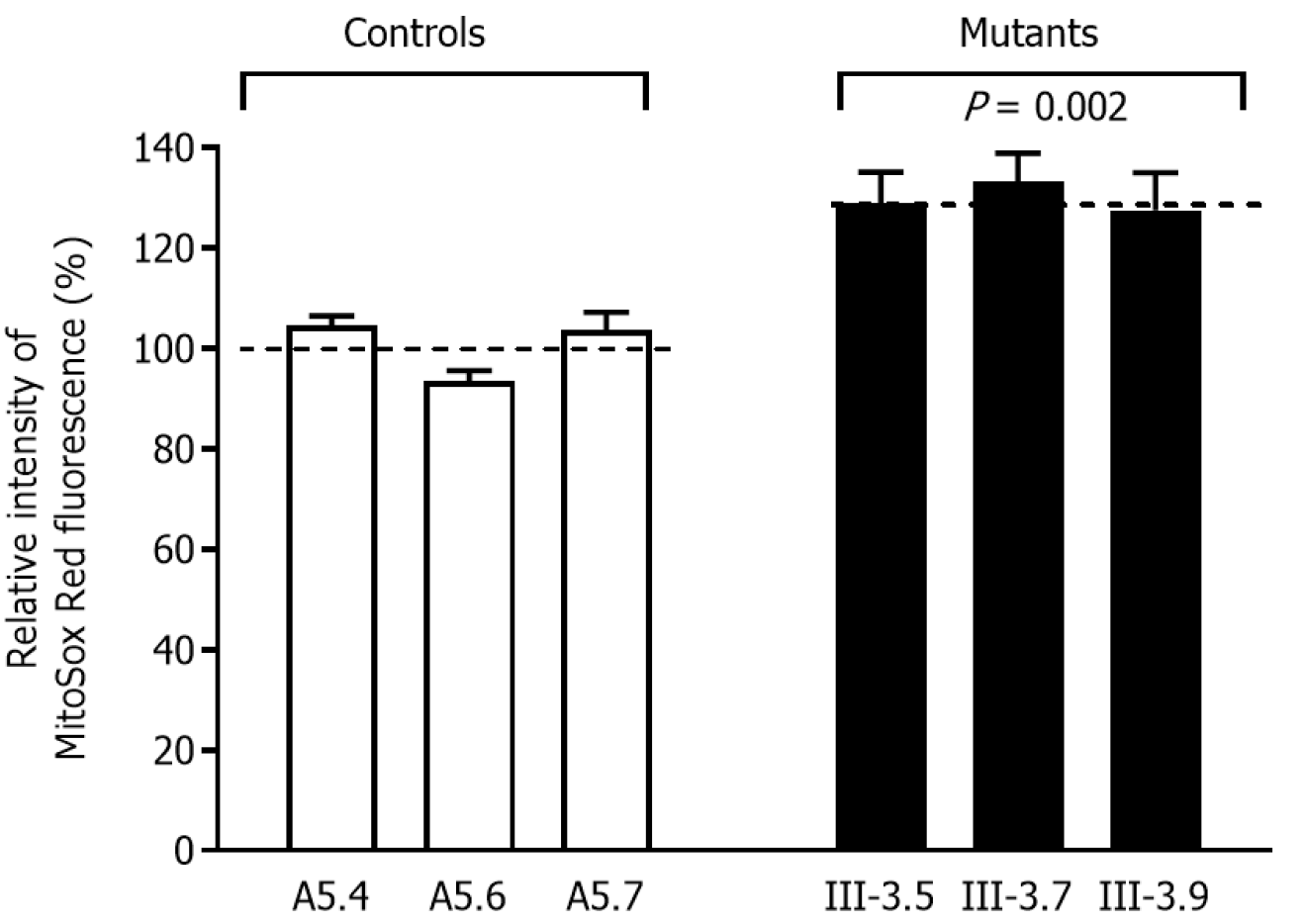
In order to obtain a ratio corresponding to ROS generation, we assessed ROS production at the mutant cybrid cell lines via flow cytometry, in contrast to baseline staining intensity for every cell line after oxidative stress. As shown in Figure 4, slightly increased ROS generation was observed in the mutant cybrid cell lines with the 4268T>C mutation, and with ROS production 127.6–130.1% (average: 130.0%) for control cells.
We conducted mutational screening of the GJB2 gene in seven affected and two unaffected matrilineal relatives of the Chinese family to assess the role of GJB2 gene in phenotypic expression of 4268T>C mutation. We found none of the variants in GJB2 gene in these matrilineal relatives of the Chinese pedigree. We found that the GJB2 gene could not be a modifier of the phenotypic effects of the 4268T>C mutation in the subjects, as none of the GJB2 gene variants in the subjects with hearing loss was verified.
Hearing loss is one of the major sensory disabilities worldwide, which is often due to the damage of spiral ganglion neurons or sensory hair cells in the inner ear[23-25]. Hearing loss can be caused by gene alterations, aging and environmental factors, with chronic cochlear infections, noise exposure, ototoxic drugs, and genetic factors accounting for more than 60% of the cases with hearing loss[26-31]. Numerous genes are known to cause syndromic hearing loss[32-37]. In the present study, we examined the clinical, genetic, biochemical and molecular characterization of two Chinese families with non-syndromic deafness. Only two pedigrees from the members had hearing loss with a single clinical phenotype. There was also a possible maternal inheritance of deafness in the families, except for two possible etiological types. The tRNAIle 4268T>C mutation was identified to belong to the Asian haplogroup D4j through the mutational analysis of mtDNA from the families. The 4268T>C mutation resided at position 6 of the ACC stem in the tRNAIle gene, and this position is very important for the structure and function of tRNA. A relationship was found between T>C transition at position 6 of tRNAGly (9996T>C), tRNACys (5821G>A) and human diseases[5,6]. It is known that 7472insC, 7510T>C, 7511T>C, 7445A>G and 7445A>C mutations in the tRNA gene are associated with hearing loss[5,8-11]. Similarly, m.5587T>C mutation in the tRNAAla gene was associated with Leber’s hereditary optic neuropathy[38]. The altered secondary tRNAIle structure caused by the 4268T>C mutation was thought to have led to a failure in tRNA metabolism. In the present study, an approximately 64.6% decrease in the tRNAIle level observed in cell lines with the mutation was consistent with our previous investigations, which showed the 4263A>G mutation at the ACC stem of tRNAIle in the reduced levels of tRNAIle in the cell lines. The mutant tRNAIle may be less stable metabolically and easier to degrade, which can lower the level of tRNAs.
The involvement of nuclear genes in the phenotypic manifestation was reflected by the phenotypic variability of matrilineal relatives amongst families. To date, over 140 loci have been mapped for non-syndromic deafness, and 82 deafness-causing genes have been identified (The Hereditary Hearing loss homepage. http://hereditaryhearingloss.org). GJB2 is one of the most common disease-causing genes in hereditary deafness worldwide, and the coding region comprises the majority of known GJB2 sequence variations (The Connexin-deafness homepage. http://davinci.crg.es/deafness). Nevertheless, other nuclear modifier genes may have contributed to the phenotypic variability and tissue-specific effect in this Chinese family in view of the absence of GJB2 mutation.
ROS are mainly generated from leaking electrons in the mitochondrial electron transport chain. Under normal conditions, the balance between production and scavenging of ROS is subtly and dynamically maintained. Previous studies have reported that many stress stimulators such as excessive and persistent noise exposure, ototoxic drugs including cisplatin and aminoglycosides, and mitochondrial dysfunction can increase ROS production in hair cells[35, 39-41], and eventually lead to hair cell apoptosis [42-47]. In this study, consistent with previous reports, we found that cybrids expressing the 4268T>C mutation elevated ROS production. Cellular macromolecules such as DNA and proteins can be disrupted by ROS production, which potentially lead to cellular dysfunction or asbestosis. As previously described for deafness-associated mitochondrial 7511A>G mutation, it could also potentially be attributed to the known deafness phenotype[48].
These two Chinese families carrying the 4268T>C mutation with the maternal lineages had wide phenotypic variability. There was 33.3% and 10% penetrance of deafness in these Chinese families, respectively, which was similar to the Scottish family carrying the m.7445A>G mutation and some reported Chinese families with the m.1555A>G mutation with low penetrance of deafness[49, 50]. The average age at deafness was 32.6 years (22-45 years) for matrilineal relatives with previous features. As a comparison, the average age was approximately 15 and 20 years old for 69 Chinese families and 19 Spanish families carrying the 1555A>G mutation[51,52], respectively, for those deaf individuals without aminoglycoside exposure, while some matrilineal relatives in a large Arab-Israeli family exhibited congenital profound deafness. Environmental factors, nuclear and mitochondrial genetic backgrounds and other modifiers may participate in slight mitochondrial dysfunction, which leads to late-onset hearing damage, similar to the homoplasmic nature of 4268T>C mutation in the phenotypic manifestation. Mitochondrial haplogroup did not play an important role in the phenotypic expression of functionally significant variants in the mtDNA. In addition, the phenotypic variability and tissue-specific effect may be caused by the nuclear modifier genes that were in synergy with the 4268T>C mutation. Alternatively, the deafness phenotype may develop due to tissue-specific disparity in tRNA metabolism.
The current study provides clinical, genetic, molecular and biochemical evidence that the tRNAIle 4268T>C mutation may be related to late-onset deafness. Nevertheless, nuclear modifier genes or tissue-specific tRNA metabolism could cause the tissue-specificity of this variant. Also, the future molecular diagnosis of deafness could include the inherited factors of RNAIle 4268T>C mutation, which may provide a new perspective for understanding the pathophysiology, management and treatment of maternally inherited deafness. Future research should include emerging linkages and their cause-effect relationship between deafness and mitochondrial dysfunction.
This study had several limitations. First, this investigation was limited to a small number of patients from the same hospital. Second, the 4268T>C mutation from these Chinese families may be an inherited risk factor for the development of deafness, but may not lead to a clinical phenotype. The slight mitochondrial dysfunction, hom
We identified the novel homoplasmic tRNAIle 4268T>C mutation in two families, which was located at a highly conserved base-pairing (6U–67A) of tRNAIle. The abolishment of 6U–67A base-pairing likely changes tRNAIle metabolism. Moreover, the proposed threshold of normal respiration in lymphoblastoid cells was more than the reduced tRNA level.
Mutations in mitochondrial rRNA and tRNA genes have been identified as one of the most common reasons for sensorineural hearing loss. In this investigation, two Han Chinese pedigrees with maternally transmitted non-syndromic deafness were studied and the clinical, molecular and genetic characterization was investigated, which suggested maternally transmitted non-syndromic hearing impairment.
In the present study, two Han Chinese pedigrees with maternally transmitted non-syndromic deafness were studied and the clinical, molecular and genetic characterization was investigated. Mutational analysis of the entire mtDNA identified the novel homoplasmic tRNAIle 4268T>C mutation, which disrupts a highly conservative base-pairing (6U–67A) on the ACC stem of tRNAIle. The steady-state levels of mitochondrial tRNA were detected with cybrid cell lines for functional significance of the 4268T>C mutation.
To investigate the pathophysiology of hearing loss associated with mitochondrial tRNA mutations.
Sixteen subjects from two Chinese families with deafness were assessed using clinical, genetic, molecular, and biochemical techniques. We measured the tRNA levels in lymphoblastoid cell lines derived from three control subjects and five affected matrilineal relatives of the families.
Variable severity and age-at-onset (8 years) of deafness were exhibited by three of the 16 matrilineal relatives from the families. The novel homoplasmic tRNAIle 4268T>C mutation was identified by mutational analysis of mtDNA in two families that belonged to haplogroup D4j. The base-pairing (6U–67A) of tRNAIle was a highly conserved gene where the 4268T>C mutation was located. The metabolism of tRNAIle could be changed by the abolishment of 6U–67A base-pairing. Compared with the wild-type cell lines, the lymphoblastoid cell lines with the 4268T>C mutation were observed at the level of tRNAIle with an approximately 64.6% reduction. Normal respiration in lymphoblastoid cells requires a higher threshold level than this reduced tRNA level. However, genotyping analysis did not detect any mutations in the prominent deafness-causing gene GJB2 in any members from the family.
The novel tRNAIle 4268T>C mutation may lead to maternally transmitted deafness. However, the phenotypic variability in the family may also be derived from epigenetic, other genetic, or environmental factors. These results may be of value in counseling, especially for families with maternally inherited deafness.
Future molecular diagnosis of deafness can include the tRNAIle 4268T>C mutation as one of the inherited risk factors. Thus, the results may have important value in deafness counseling, especially for families with maternally inherited deafness.
We thank all the people who participated in the study, including members of the two families.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Guo Guan S-Editor: Wang LL L-Editor: Webster JR P-Editor: Wang LL
| 1. | Morton CC. Genetics, genomics and gene discovery in the auditory system. Hum Mol Genet. 2002;11:1229-1240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 91] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 2. | Fitzpatrick EM, Whittingham J, Durieux-Smith A. Mild bilateral and unilateral hearing loss in childhood: a 20-year view of hearing characteristics, and audiologic practices before and after newborn hearing screening. Ear Hear. 2014;35:10-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 3. | Tsang SH, Aycinena ARP, Sharma T. Mitochondrial Disorder: Maternally Inherited Diabetes and Deafness. Adv Exp Med Biol. 2018;1085:163-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 4. | Meena R, Ayub M. Genetics Of Human Hereditary Hearing Impairment. J Ayub Med Coll Abbottabad. 2017;29:671-676. [PubMed] |
| 5. | Zhang M, Han Y, Zhang F, Bai X, Wang H. Mutation spectrum and hotspots of the common deafness genes in 314 patients with nonsyndromic hearing loss in Heze area, China. Acta Otolaryngol. 2019;139:612-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Ding Y, Teng YS, Zhuo GC, Xia BH, Leng JH. The Mitochondrial tRNAHis G12192A Mutation May Modulate the Clinical Expression of Deafness-Associated tRNAThr G15927A Mutation in a Chinese Pedigree. Curr Mol Med. 2019;19:136-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 7. | Xiang YB, Tang SH, Li HZ, Xu CY, Chen C, Xu YZ, Ding LR, Xu XQ. Mutation analysis of common deafness-causing genes among 506 patients with nonsyndromic hearing loss from Wenzhou city, China. Int J Pediatr Otorhinolaryngol. 2019;122:185-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Lévêque M, Marlin S, Jonard L, Procaccio V, Reynier P, Amati-Bonneau P, Baulande S, Pierron D, Lacombe D, Duriez F, Francannet C, Mom T, Journel H, Catros H, Drouin-Garraud V, Obstoy MF, Dollfus H, Eliot MM, Faivre L, Duvillard C, Couderc R, Garabedian EN, Petit C, Feldmann D, Denoyelle F. Whole mitochondrial genome screening in maternally inherited non-syndromic hearing impairment using a microarray resequencing mitochondrial DNA chip. Eur J Hum Genet. 2007;15:1145-1155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Labay V, Garrido G, Madeo AC, Nance WE, Friedman TB, Friedman PL, Del Castillo I, Griffith AJ. Haplogroup analysis supports a pathogenic role for the 7510T>C mutation of mitochondrial tRNA (Ser (UCN)) in sensorineural hearing loss. Clin Genet. 2008;73:50-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Tang X, Zheng J, Ying Z, Cai Z, Gao Y, He Z, Yu H, Yao J, Yang Y, Wang H, Chen Y, Guan MX. Mitochondrial tRNA (Ser (UCN)) variants in 2651 Han Chinese subjects with hearing loss. Mitochondrion. 2015;23:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 11. | Van Camp G, Smith RJ. Maternally inherited hearing impairment. Clin Genet. 2000;57:409-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 12. | Chan DK, Chang KW. GJB2-associated hearing loss: systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope. 2014;124:E34-E53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 224] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 13. | Kenneson A, Van Naarden Braun K, Boyle C. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet Med. 2002;4:258-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 289] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 14. | Lucotte G, Diéterlen F. The 35delG mutation in the connexin 26 gene (GJB2) associated with congenital deafness: European carrier frequencies and evidence for its origin in ancient Greece. Genet Test. 2005;9:20-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Yao J, Lu Y, Wei Q, Cao X, Xing G. A systematic review and meta-analysis of 235delC mutation of GJB2 gene. J Transl Med. 2012;10:136. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 16. | Jiang P, Ling Y, Zhu T, Luo X, Tao Y, Meng F, Cheng W, Ji Y. Mitochondrial tRNA mutations in Chinese Children with Tic Disorders. Biosci Rep. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Xie S, Zhang J, Sun J, Zhang M, Zhao F, Wei QP, Tong Y, Liu X, Zhou X, Jiang P, Ji Y, Guan MX. Mitochondrial haplogroup D4j specific variant m.11696G > a (MT-ND4) may increase the penetrance and expressivity of the LHON-associated m.11778G > a mutation in Chinese pedigrees. Mitochondrial DNA A DNA Mapp Seq Anal. 2017;28:434-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Wang J, Zhao N, Mao X, Meng F, Huang K, Dong G, Ji Y, Fu J. Obesity associated with a novel mitochondrial tRNACys 5802A>G mutation in a Chinese family. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Ji Y, Zhang J, Lu Y, Yi Q, Chen M, Xie S, Mao X, Xiao Y, Meng F, Zhang M, Yang R, Guan MX. Complex I mutations synergize to worsen the phenotypic expression of Leber's hereditary optic neuropathy. J Biol Chem. 2020;295:13224-13238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 20. | Meng F, Zhou M, Xiao Y, Mao X, Zheng J, Lin J, Lin T, Ye Z, Cang X, Fu Y, Wang M, Guan MX. A deafness-associated tRNA mutation caused pleiotropic effects on the m1G37 modification, processing, stability and aminoacylation of tRNAIle and mitochondrial translation. Nucleic Acids Res. 2021;49:1075-1093. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 21. | Li K, Wu L, Liu J, Lin W, Qi Q, Zhao T. Maternally Inherited Diabetes Mellitus Associated with a Novel m.15897G>A Mutation in Mitochondrial tRNAThr Gene. J Diabetes Res. 2020;2020:2057187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Ji Y, Nie Z, Meng F, Hu C, Chen H, Jin L, Chen M, Zhang M, Zhang J, Liang M, Wang M, Guan MX. Mechanistic insights into mitochondrial tRNAAla 3'-end metabolism deficiency. J Biol Chem. 2021;297:100816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Guo R, Zhang S, Xiao M, Qian F, He Z, Li D, Zhang X, Li H, Yang X, Wang M, Chai R, Tang M. Accelerating bioelectric functional development of neural stem cells by graphene coupling: Implications for neural interfacing with conductive materials. Biomaterials. 2016;106:193-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 105] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 24. | Völkening B, Schönig K, Kronenberg G, Bartsch D, Weber T. Type-1 astrocyte-like stem cells harboring Cacna1d gene deletion exhibit reduced proliferation and decreased neuronal fate choice. Hippocampus. 2018;28:97-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 25. | Guo R, Li J, Chen C, Xiao M, Liao M, Hu Y, Liu Y, Li D, Zou J, Sun D, Torre V, Zhang Q, Chai R, Tang M. Biomimetic 3D bacterial cellulose-graphene foam hybrid scaffold regulates neural stem cell proliferation and differentiation. Colloids Surf B Biointerfaces. 2021;200:111590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 26. | Tan F, Chu C, Qi J, Li W, You D, Li K, Chen X, Zhao W, Cheng C, Liu X, Qiao Y, Su B, He S, Zhong C, Li H, Chai R, Zhong G. AAV-ie enables safe and efficient gene transfer to inner ear cells. Nat Commun. 2019;10:3733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 168] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 27. | Zhang S, Zhang Y, Dong Y, Guo L, Zhang Z, Shao B, Qi J, Zhou H, Zhu W, Yan X, Hong G, Zhang L, Zhang X, Tang M, Zhao C, Gao X, Chai R. Knockdown of Foxg1 in supporting cells increases the trans-differentiation of supporting cells into hair cells in the neonatal mouse cochlea. Cell Mol Life Sci. 2020;77:1401-1419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 28. | Han S, Xu Y, Sun J, Liu Y, Zhao Y, Tao W, Chai R. Isolation and analysis of extracellular vesicles in a Morpho butterfly wing-integrated microvortex biochip. Biosens Bioelectron. 2020;154:112073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 29. | Cheng C, Wang Y, Guo L, Lu X, Zhu W, Muhammad W, Zhang L, Lu L, Gao J, Tang M, Chen F, Gao X, Li H, Chai R. Age-related transcriptome changes in Sox2+ supporting cells in the mouse cochlea. Stem Cell Res Ther. 2019;10:365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 30. | He Z, Guo L, Shu Y, Fang Q, Zhou H, Liu Y, Liu D, Lu L, Zhang X, Ding X, Tang M, Kong W, Sha S, Li H, Gao X, Chai R. Autophagy protects auditory hair cells against neomycin-induced damage. Autophagy. 2017;13:1884-1904. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 204] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 31. | Michel V, Booth KT, Patni P, Cortese M, Azaiez H, Bahloul A, Kahrizi K, Labbé M, Emptoz A, Lelli A, Dégardin J, Dupont T, Aghaie A, Oficjalska-Pham D, Picaud S, Najmabadi H, Smith RJ, Bowl MR, Brown SD, Avan P, Petit C, El-Amraoui A. CIB2, defective in isolated deafness, is key for auditory hair cell mechanotransduction and survival.EMBO Mol Med. 2017; 9: 1711-1731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 32. | Lv J, Fu X, Li Y, Hong G, Li P, Lin J, Xun Y, Fang L, Weng W, Yue R, Li GL, Guan B, Li H, Huang Y, Chai R. Deletion of Kcnj16 in Mice Does Not Alter Auditory Function. Front Cell Dev Biol. 2021;9:630361. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 33. | Qian F, Wang X, Yin Z, Xie G, Yuan H, Liu D, Chai R. The slc4a2b gene is required for hair cell development in zebrafish. Aging (Albany NY). 2020;12:18804-18821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 34. | He Y, Lu X, Qian F, Liu D, Chai R, Li H. Insm1a Is Required for Zebrafish Posterior Lateral Line Development. Front Mol Neurosci. 2017;10:241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | He Z, Fang Q, Li H, Shao B, Zhang Y, Han X, Guo R, Cheng C, Guo L, Shi L, Li A, Yu C, Kong W, Zhao C, Gao X, Chai R. The role of FOXG1 in the postnatal development and survival of mouse cochlear hair cells. Neuropharmacology. 2019;144:43-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 36. | Fang Q, Zhang Y, Da P, Shao B, Pan H, He Z, Cheng C, Li D, Guo J, Wu X, Guan M, Liao M, Sha S, Zhou Z, Wang J, Wang T, Su K, Chai R, Chen F. Deletion of Limk1 and Limk2 in mice does not alter cochlear development or auditory function. Sci Rep. 2019;9:3357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | Jarysta A, Tarchini B. Multiple PDZ domain protein maintains patterning of the apical cytoskeleton in sensory hair cells. Develop. 2021;148:dev199549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 38. | Ji Y, Qiao L, Liang X, Zhu L, Gao Y, Zhang J, Jia Z, Wei QP, Liu X, Jiang P, Guan MX. Leber's hereditary optic neuropathy is potentially associated with a novel m.5587T>C mutation in two pedigrees. Mol Med Rep. 2017;16:8997-9004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Zhang Y, Li W, He Z, Wang Y, Shao B, Cheng C, Zhang S, Tang M, Qian X, Kong W, Wang H, Chai R, Gao X. Pre-treatment With Fasudil Prevents Neomycin-Induced Hair Cell Damage by Reducing the Accumulation of Reactive Oxygen Species. Front Mol Neurosci. 2019;12:264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 40. | Zhong Z, Fu X, Li H, Chen J, Wang M, Gao S, Zhang L, Cheng C, Zhang Y, Li P, Zhang S, Qian X, Shu Y, Chai R, Gao X. Citicoline Protects Auditory Hair Cells Against Neomycin-Induced Damage. Front Cell Dev Biol. 2020;8:712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 41. | Zhou H, Qian X, Xu N, Zhang S, Zhu G, Zhang Y, Liu D, Cheng C, Zhu X, Liu Y, Lu L, Tang J, Chai R, Gao X. Disruption of Atg7-dependent autophagy causes electromotility disturbances, outer hair cell loss, and deafness in mice. Cell Death Dis. 2020;11:913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 42. | Gao S, Cheng C, Wang M, Jiang P, Zhang L, Wang Y, Wu H, Zeng X, Wang H, Gao X, Ma Y, Chai R. Blebbistatin Inhibits Neomycin-Induced Apoptosis in Hair Cell-Like HEI-OC-1 Cells and in Cochlear Hair Cells. Front Cell Neurosci. 2019;13:590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 43. | Li A, You D, Li W, Cui Y, He Y, Chen Y, Feng X, Sun S, Chai R, Li H. Novel compounds protect auditory hair cells against gentamycin-induced apoptosis by maintaining the expression level of H3K4me2. Drug Deliv. 2018;25:1033-1043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 44. | Sun S, Sun M, Zhang Y, Cheng C, Waqas M, Yu H, He Y, Xu B, Wang L, Wang J, Yin S, Chai R, Li H. In vivo overexpression of X-linked inhibitor of apoptosis protein protects against neomycin-induced hair cell loss in the apical turn of the cochlea during the ototoxic-sensitive period. Front Cell Neurosci. 2014;8:248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 45. | Yu X, Liu W, Fan Z, Qian F, Zhang D, Han Y, Xu L, Sun G, Qi J, Zhang S, Tang M, Li J, Chai R, Wang H. c-Myb knockdown increases the neomycin-induced damage to hair-cell-like HEI-OC1 cells in vitro. Sci Rep. 2017;7:41094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 46. | Guan M, Fang Q, He Z, Li Y, Qian F, Qian X, Lu L, Zhang X, Liu D, Qi J, Zhang S, Tang M, Gao X, Chai R. Inhibition of ARC decreases the survival of HEI-OC-1 cells after neomycin damage in vitro. Oncotarget. 2016;7:66647-66659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 47. | Liu L, Chen Y, Qi J, Zhang Y, He Y, Ni W, Li W, Zhang S, Sun S, Taketo MM, Wang L, Chai R, Li H. Wnt activation protects against neomycin-induced hair cell damage in the mouse cochlea. Cell Death Dis. 2016;7:e2136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 115] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 48. | Li R, Ishikawa K, Deng JH, Heman-Ackah S, Tamagawa Y, Yang L, Bai Y, Ichimura K, Guan MX. Maternally inherited nonsyndromic hearing loss is associated with the T7511C mutation in the mitochondrial tRNASerUCN gene in a Japanese family. Biochem Biophys Res Commun. 2005;328:32-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 49. | Tekin M, Duman T, Boğoçlu G, İncesulu A, Çomak E, Fitoz S, Yılmaz E, İlhan I, Akar N. Frequency of mtDNA A1555G and A7445G mutations among children with prelingual deafness in Turkey. Eur J Pediatr. 2003;162:154-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 50. | Hyslop SJ, James AM, Maw M, Fischel-Ghodsian N, Murphy MP. The effect on mitochondrial function of the tRNA Ser (UCN)/COI A7445G mtDNA point mutation associated with maternally-inherited sensorineural deafness. Biochem Mol Biol Int. 1997;42:567-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 51. | Estivill X, Govea N, Barceló E, Badenas C, Romero E, Moral L, Scozzri R, D'Urbano L, Zeviani M, Torroni A. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment of aminoglycosides. Am J Hum Genet. 1998;62:27-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 359] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 52. | Lu J, Qian Y, Li Z, Yang A, Zhu Y, Li R, Yang L, Tang X, Chen B, Ding Y, Li Y, You J, Zheng J, Tao Z, Zhao F, Wang J, Sun D, Zhao J, Meng Y, Guan MX. Mitochondrial haplotypes may modulate the phenotypic manifestation of the deafness-associated 12S rRNA 1555A>G mutation. Mitochondrion. 2010;10:69-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 6.0] [Reference Citation Analysis (0)] |